Abstract
Human African trypanosomiasis (HAT, commonly known as African sleeping sickness) is categorized as a neglected disease, as it afflicts > 50,000 people annually in sub-saharan Africa, and there are few formal programs in the world focused on drug discovery approaches for this disease. In this study, we examined the crude extracts of two fungal strains (Aspergillus fumigatus and Nectria inventa) isolated from deep water sediment which provided >99% growth inhibition at 1 μg/mL of Trypanosoma brucei, the causative parasite of HAT. A collection of fifteen natural products was supplemented with six semi-synthetic derivatives and one commercially available compound. Twelve of the compounds, each containing a diketopiperazine core, showed excellent activity against T. brucei (IC50 = 0.002 - 40 μM), with selectivity over mammalian cells as great as 20-fold. The trypanocidal diketopiperazines were also tested against two cysteine protease targets Rhodesain and TbCatB, where five compounds showed inhibition activity at concentrations less than 20 μM. A preliminary activity pattern is described and analyzed.
Keywords: Marine-derived fungi, Trypanosoma brucei, Rhodesain, TbCatB, marine natural products
1. Introduction
The quest for new generation therapeutics to treat human African trypanosomiasis (HAT) has been re-invigorated through the collaborative endeavors of several academic teams. HAT, designated as a neglected tropical disease (NTD) by the World Health Organization (WHO), is caused by the protozoan parasite Trypanosoma brucei, and as of 2009 affects more than 50,000 people in 36 countries of sub-Saharan Africa.1 Recent reports2,3 summarize that the four current clinical therapeutics consisting of suramin, pentamidine, melarsoprol, and eflornithine are inadequate. Furthermore, we believe that the pharmaceutical industry is minimally engaged in either fine-tuning these agents or conceptualizing new ones. An encouraging new development is the launch of an eflornithine - nifurtimox combination therapy by the Drugs for Neglected Disease Initiative (DNDi).4 While this mixed medication represents an improvement in both dosing schedule and cost, new therapeutic leads are still needed. Another approach, also fostered by DNDi, was the re-exploration of nitroimidazole compounds first examined in the 1980's,5 resulting in fexinidazole entering Phase 1 clinical trials (2009) as an oral treatment for HAT. 6
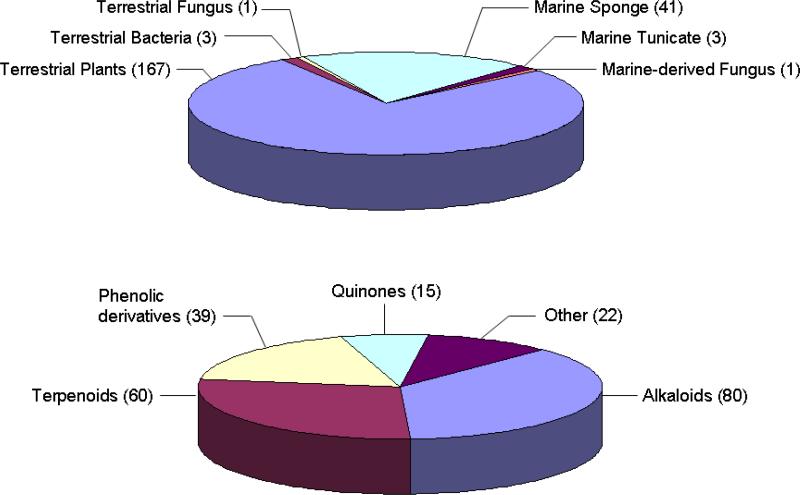
Natural products derived from varying sources have been explored for their potential to operate against T. brucei. A review published in 20047 surveyed the structures and properties of actives discovered from the mid-1980's to 2003, and this along with our supplemental literature searches provide the synopsis of Figure 1.8 Significantly, there are 215 natural products described that inhibit T. brucei, primarily composed of alkaloid, terpenoid and phenolic derivatives. Macro organisms, especially higher plants, constitute the major focal points of such research with 167 such compounds reported to date. Of interest to us are that marine organisms, including sponges (41 compounds) and tunicates (three compounds), have also been examined. Our nascent program to explore sponge-derived actives is being driven by medium throughput extract screening and it constituted an important proof-of-concept step. The first set of trypanocidal leads we discovered in this campaign exhibit divergent structures consisting of peroxiterpenes from Diacarnus bismarckensis,9 and unique sesquiterpenes from Cacospongia mycofijiensis.10
Figure 1.
Organism sources, structure types and tally of natural products with activity against T. brucei
Another part of the discovery record is compounds derived from synthesis shown to be active against T. brucei. An especially effective tactic involves combining active site mapping of cysteine proteases alongside creation and evaluation of synthetic libraries. Examples of lead scaffolds emerging from such research begun ten years ago consist of purine-nitrile11 and thiosemicarbazone12,13 inhibitors of T. brucei brucei, T. brucei rhodesiense, and their cathepsin B-like14 and L-like15 cysteine proteases (TbCatB and rhodesain, respectively).
The preceding discussions provide important background for the developments responsible for the research results described herein. The success in using the T. brucei screen for sponge crude extracts provided the transition to our investigation of marine-derived fungal extracts. A secondary screen step involving proteases (TbCatB and rhodesian) also seem appropriate. Inexplicably, microorganisms, an important source of numerous therapeutic leads, have received negligible consideration in screening programs focused on T. brucei, as only five active compounds have been reported and just two are from fungi. The latter includes an antimicrobial alkaloid, ascosalipyrrolidinone A (1a) (T. brucei rhodesiense: MIC=30 μg/mL) obtained from culture of an obligate marine-derived fungus Aschochyta salcorniae 16 and the heteroatom-rich ascofuranone (1b) (T. brucei brucei: IC100 = 0.03 μM) from the culture of a phytopathogenic fungus Aschochyta viciae.17 These two structures, while diverse, do not overlap or even represent the breadth of scaffold variety that we have observed in studies of marine-derived fungi. Furthermore, in more recent explorations, we have focused on strains cultivated from deep water sediments, and found that intriguing, bioactive compounds are often encountered.18-20 The account below outlines our comprehensive evaluation of diketopiperazine-containing compounds produced by deep water sediment-derived fungi whose isolation was facilitated by T. brucei screens.
2. Results and discussion
2.1. Initial screening
The foundation for this study was the screening of 151 crude marine fungal culture extracts for T. brucei growth inhibition (GI) responses. Five of these samples (3%) showed GI>99% at 1 μg/mL and two obtained through deep water sampling were selected for further study. One was Aspergillus fumigatus from a Vanuatu sediment collected below 33 meters using a hand-launched mud grabber. The other, identified as Nectria inventa, was from Monterey Bay dredged sediment obtained below 600 meters. Ensuing parallel studies on their three large-scale cultures, summarized in Figure S1 (Supplemental Information), involved A. fumigatus in 10L saltwater (coded 030402dZa), 14L of A. fumigatus in deionized water (030402dZi), and 20L of N. inventa in deionized water (049207cMi). Each of their crude extracts was examined by LCMS, 1H NMR, and UV max data. The preliminary dereplication conclusions were that indole and other nitrogen-containing diketopiperazines (DKPs) were abundant in these extracts and might account for the initial activity responses.
At the outset we recognized that the results obtained from this research might be undermined as diketopiperazines are a well-studied class comprised of wide-ranging structures and diverse bioactivity properties.21 Alternatively, compounds in this class continue to be the seeds for important biomedical discoveries. At the top of our list of such examples is the phenylahistin (halimide, 2a) story. This simple DKP, isolated from Aspergillus sp.,22 shows cell cycle inhibition activity in the G2/M phase at 1 μM, and acts on the microtubule network of human lung carcinoma (A549) cells.23 An SAR optimized synthetic analogue, NPI-2358 (2b), is now being evaluated in a Phase I clinical trial as a vascular disrupting agent for the treatment of solid lung tumors or lymphoma.24
2.2. Isolation and semi-synthesis
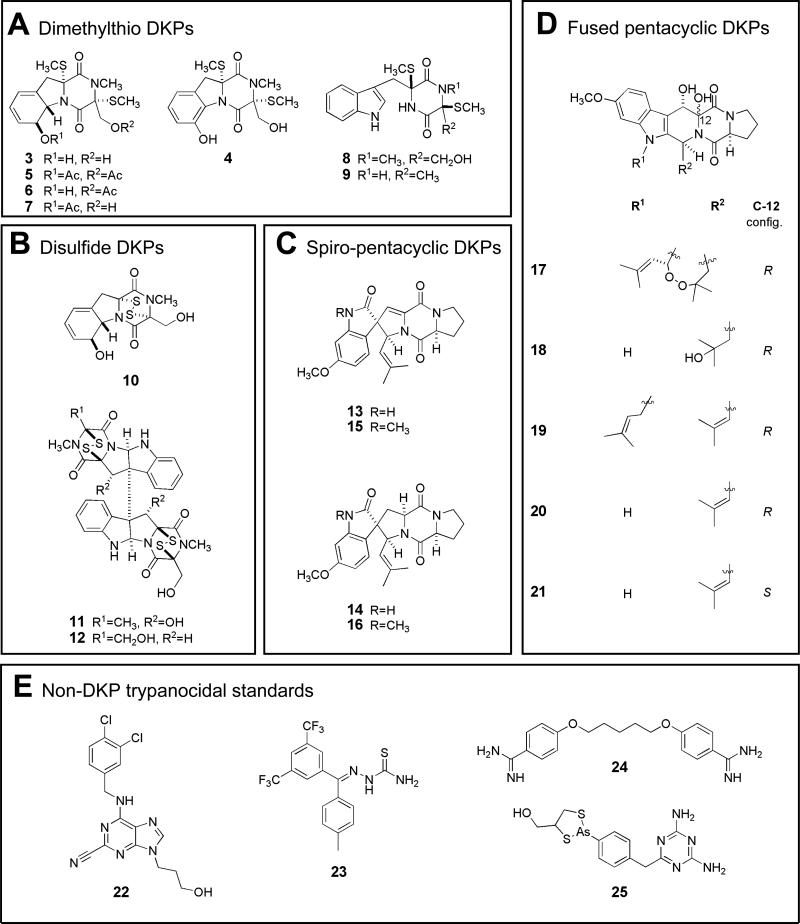
Combining traditional bioassay guided fractionation and LCMS evaluation in the parallel study of the extracts from all three cultures eventually led to the isolation and characterization of 12 DKP compounds. This result was anticipated from the initial dereplication efforts. Though all of these were previously described structures, surprisingly, there was no overlap in the constituents obtained from each source. An overview of the compounds isolated by source is provided in Figure S1 (Supporting Information), and the materials and procedures employed are as follows. The saltwater Aspergillus culture presented five compounds: bis(methylthio)gliotoxin25 (3), its dehydro derivative26 (4), 6-methoxyspirotryprostatin B27 (13), verruculogen TR-228 (18), and cyclotryprostatin A29 (21). Unexpectedly, the deionized water culture of this strain produced three different metabolites: verruculogen30 (17), fumitremorgin B31 (19), and 12, 13-dihydroxyfumitremorgin C32 (20). Lastly, the deionized water culture of N. inventa was a source of four sulfur-containing DKPs: chetoseminudin B33 (8), 3,6-bis(methylthio)-cyclo(alanyltryptophyl)34 (9), verticillin B35 (11), and chaetocin36 (12).
We were able to further expand the array of DKP structures for the follow-up bioassay testing through the preparation of three known and three novel semi-synthetic analogues of selected natural products isolated in multi milligram amounts. Two compounds, 3 and 13 (from 030402dZa), showing good activity in the first round of testing against T. brucei, were chosen as the scaffolds for further modification. Simple small scale reactions were chosen and carried out to manipulate existing functional group handles within these molecules and ultimately afforded six additional structures. The acetylation of compound 3 provided two known and one new compound - diacetylbis(methylthio)gliotoxin37 (5), 6-acetylbis(methylthio)gliotoxin38 (7), and 15-acetylbis(methylthio)gliotoxin (6). Compound 13 was a source of one known and two new substances through hydrogenation and methylation reactions- spirotryprostatin A39 (14), N-methyl-6-methoxyspirotryprostatin B (15), and N-methylspirotryprostatin A (16). It is important to summarize that 15-acetylbis(methylthio)gliotoxin (6) and the N-methyl derivatives (15, 16) are reported here for the first time and were characterized based on analogy to their congeners. A final compound added to the collection was the commercially available disulfide DKP, gliotoxin (10).
All of the known natural products isolated here, rich in N, O and in some cases S atoms, were characterized by comparing their properties to those in the literature. This process also involved verifying relative configurations at the various chiral sites previously assigned as shown in Figure 2. The one exception to this outline involved the absolute configuration of chetoseminudin B (8) because only its planar structure is reported in the literature.33 We employed circular dichroism, observing a negative Cotton effect at 212 nm (Δε -2.7) and a positive Cotton effect at 222 nm (Δε +6.7), to assign S, S absolute geometry at both DKP alpha carbons, based on the reported data of compound 9.34 In the case of compound 14, produced by hydrogenation of 13, the relative configuration was re-affirmed by an NOE correlation of H-9 to H-6. This proved 14 to be equivalent to the natural product spirotryprostatin A, originally isolated from A. fumigatus.39 Lastly, the configuration for semi-synthetic compounds 5, 6, 7, 15, and 16 is based on that of their respective starting materials (Figure S1).
Figure 2.
Compound Structure Groups A-E.
To simplify analysis of the subsequent structure vs. bioactivity data sets, the mini-library of 21 compounds was grouped into five framework categories. These are shown in Figure 2 as follows: (A) seven dithiomethyl DKPs (3-9), (B) three disulfide DKPs (10-12), (C) four spiro-pentacyclic DKPs (13 - 16), and (D) five fused pentacyclic DKPs (17-21). An additional structure group E, included in Figure 2, consists of four standard trypanocidal compounds. Two of these, purine-nitrile (22), and thiosemicarbazone (23), are members of combinatorial chemistry libraries shown to be inhibitors of TbCatB, while the other two are the clinical agents pentamidine (24) and melarsoprol (25).
2.3. Biological evaluation
The compound collection was rigorously evaluated for its trypanocidal potential to probe for a rough structure-activity pattern. The initial filter in the screening network employed the whole cell T. brucei assay, and compounds defined as active (IC50 < 25 μg/mL) were then tested for inhibition activity in a panel of the cysteine proteases. The most abundant enzyme isoform in the parasite is rhodesain (also known as brucipain, trypanopain), a cathepsin L-like protease.40 Also relevant and a target in our screen was the cathepsin B-like enzyme, TbCatB, a necessary factor for parasite survival, and a protease responsible for host protein degradation and iron acquisition.14 Although members of the rhodesain family are not essential for sustaining the parasite, they may play an important role in the migration of parasites across the blood-brain barrier in the second stage of host infection.15 Thus, compounds inhibiting these targets are worthy of further therapeutic evaluation. In the final arm of bioactivity assessment, trypanocidal active compounds were counter-screened against Jurkat cells (T-cell leukemia) to determine a selectivity index defined as IC50 Jurkat/IC50 T. brucei. A large amount of data was acquired during the screening phase; these results shown in Table 1 are discussed next according to the Structure Groups A-D. Our commentary will also include a comparison of the properties of these compounds vs. that of the standards from Structure Group E.
Table 1.
IC50 Summary for Compounds in this Study.
| Structure Groupa | Compound | IC50 (μM) T. brucei | IC50 (μM) Cysteine Protease | IC50 (μM) Jurkat | Selectivity Index | |
|---|---|---|---|---|---|---|
| rho | Tbc | |||||
|
A Dimethylthio DKPs |
bis(methylthio)gliotoxin (3) | 40.2 ± 5.2 | NA | NA | 140 | 3.5 |
| dehydrobis(methylthio)gliotoxin (4) | 8.5 | 19.5 | NA | 141 | 16.6 | |
| diacetylbis(methylthio)gliotoxin (5o) | 35.5 | NA | NA | 56 | 1.6 | |
| 15-acetylbis(methylthio)gliotoxin (6o) | NAb | – c | – | – | – | |
| 6-acetylbis(methylthio)gliotoxin (7o) | 6.5 | 12.8 | NA | 125 | 19.2 | |
| chetoseminudin B (8) | 5.9 | NA | NA | 94.9 | 16.1 | |
| 3,6 bismethyl(thiocyclo)alanyltryptophyl (9) |
NA |
–
|
–
|
–
|
–
|
|
|
B Disulfide DKPs |
gliotoxin (10*) | 0.010 | NA | 0.5 | <1 | lowg |
| verticillin B (11) | 0.007 | NA | NA | <0.6 | low | |
| chaetocin (12) |
0.002 |
NA |
NA |
<0.6 |
low |
|
|
C Spiro-fused DKPs |
6-methoxyspirotryprostatin B (13) | 5.7 ± 2.7 | NA | NA | 21.1 | 3.7 |
| spirotryprostatin A (14o) | NA | – | – | – | – | |
| N-methyl-6-methoxyspirotryprostatin B (15o) | NA | – | – | – | – | |
|
N-methylspirotryprostatin A (16o) |
NA |
–
|
–
|
–
|
–
|
|
|
D Pentacyclic-fused DKPs |
verruculogen (17) | 12.9 ± 8.3 | 45.0 | NA | 97 | 7.5 |
| verruculogen TR-2 (18) | NA | – | – | – | – | |
| fumitremorgin B (19) | 19.5 ± 5.9 | 8 5 | NA | 104 | 5.3 | |
| 12,13-dihydroxyfumitremorgin C (20) | 6.4 ± 1.3 | 10.9 | NA | 97.2 | 17.6 | |
| cyclotryprostatin A (21) |
NA |
–
|
–
|
–
|
–
|
|
| E Non-DKP trypanocidal standards | purine-nitrile standard (22■) | 0.11d | – | 0.27 | 25d,e | 227 |
| thiosemicarbazone standard (23■) | 1.3d | 0.044 | 15.0 | 25d,e | 19 | |
| pentamidine (24■) | 0.007 | – | – | 3.80f | 542 | |
| melarsoprol (25■) | 0.004 | – | – | 7.78f | 1945 | |
IC50 values were calculated versus control wells and were performed in duplicate when sufficient material was available; values are reported ± the standard deviation of the mean.
Refer to Figure 3.
NA=Not active in assay (IC50 > 50 μM).
Not determined.
EC50.
HEK 293 cells.
L6 rat myoblast cells.
Toxic to Jurkat cells at <1 μM.
Semi-synthetic compound.
Commercially available compound.
2.3.1. Group A
The seven dimethylthio DKPs constitute a set of closely related structures. Their DKP cores, biogenetically derived from condensation of aromatic amino acids (phenylalanine or tryptophan) plus either serine or alanine, carry appendages of a methionine derived N-methyl group, and sulfur atoms from cysteine. The variation in DKP ring substituents influence the overall trypanocidal and enzymatic activity as five of the seven compounds showed positive responses. Compound 3, representing the first dimethylthio DKP isolated and tested, showed moderate trypanocidal activity (IC50= 40.2 ±5.2 μM) and inactivity against both cysteine protease targets, similar to that of pentamidine, while not being toxic to the Jurkat cells. The closely related benzene ring containing analogue 4 showed improved activity in both assays (IC50 = 8.5 μM T. brucei and IC50= 19.5 rhodesain), and the indole containing analogue 8 exhibited comparable trypanocidal activity (IC50=5.9 μM). It is relevant to note that 8 vs. 3, which exhibited similar activity against T. brucei, possess opposite configurations at both chiral centers of the DKP ring. This data set prompted our preparation of the acetylated analogues 5, 6, and 7 which provided additional information about the influence of alcohol functionalities at C-6 and C-15. The best profile for the closely related set 3 – 7 was exhibited by the C-6 acetyl derivative 7. Similarly the activity profile of 8 vs. 9 was sensitive to changes in the R appendages. All compounds with activity against T. brucei showed some selectivity versus mammalian cells, but the best selectivity index profile (16.1 to 19.2) was observed for 4, 7, and 8, on par with the thiosemicarbazone standard (23). Collectively this trio constitutes the standout scaffolds from Structure Group A.
2.3.2. Group B
The three disulfide DKPs of this class exhibited exquisite GI potency (nM) against T. brucei. The bioactivity properties of this group are illustrated best by those of the less complex analogue, gliotoxin (10), a ubiquitous fungal metabolite whose properties have been extensively scrutinized by others (see Table S1, Supporting Information). Further, 10 is often compared side-by-side with its bis S-methyl analogue, compound 3, and the reduced compound is usually less potent - as we also observed against T. brucei (10, IC50 = 0.01 μM, 3, IC50 = 40.2 ± 5.2 μM). The nM activity level for 10-12 vs. the μM responses for 3, 4, and 8 confirmed the role of the disulfide bridge in imparting remarkable potency against T. brucei. Another important dimension to the activity pattern of 10 is that it was the only compound out of the 19 examined that showed inhibition of TbCatB. This is similar to the profile of the purine-nitrile inhibitor (22) specifically designed to inhibit this cysteine protease. Perhaps relevant is that cystamine (also disulfide containing) inactivates the cysteine protease papain by blocking the cysteine residue in the active site.41 Although the entities for Structure Group B showed potent enzymatic and trypanocidal activity, their high toxicity to mammalian cells probably precludes their value as a lead scaffold.
2.3.3. Group C
The four spiro-pentacyclic DKPs provided structural constructs with added complexity. The isolation of 6-methoxyspirotryprostatin B (13) followed by the positive assay response (IC50 = 5.7 ± 2.7 μM T. brucei) and modest selectivity (3.7) stimulated the preparation of analogues 14 - 16. However none of these derivatives were active. In comparing the overall structures of Group B vs. C, it appears that the lack of sulfur residues attached to the DKP seems to be responsible for the poor activity profile of this Structure Group.
2.3.4. Group D
The fused spiro-pentacyclic DKPs provided a different context, vs. that of Structure Group C, to evaluate the impact of variations in pentacyclic array devoid of attached sulfur groups. Three of the members of this family, 17, 19, and 20 showed IC50's < 20 μM against T. brucei. The similar profile of verruculogen (17) vs. that of fumitremorgin B (19) demonstrates that the peroxide ring is not the bioactive pharmacophore. However, these two compounds cannot be considered as appropriate candidates for further development as they cause tremors in animals.42 It is also hypothesized that these metabolites may be responsible for the virulence of terrestrial Aspergillus strains that cause aspergillosis in humans.43 The slightly less unsaturated analogue, 12,13-dihydroxyfumitremorgin C (20, IC50 = 6.4 ± 1.3 μM) showed the best selectivity index (17.6), and comparing its data vs. that for 21 underscored the importance of C-12 R configuration. Finally its activity profile was on a par with that of the sulfur containing DKP 7 from Group A.
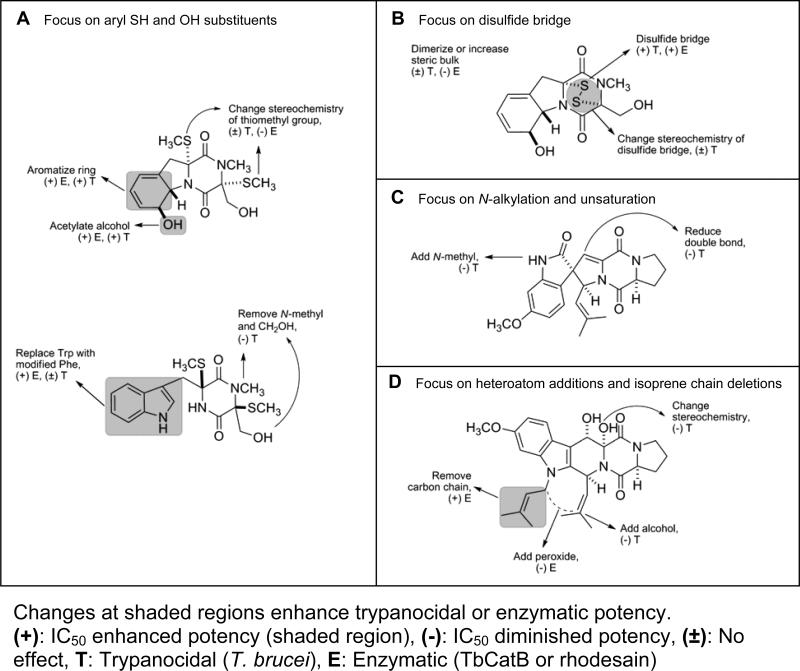
2.4. Activity patterns
A first draft of the structure activity patterns emerging for this study is shown in Figure 3. The shaded regions for Structure Groups A – D indicate structural space where changes can result in enhanced biological activity against T. brucei or the proteases (TbCatB, rhodesain). The data sets and structures for Group E, the trypanocidal non-DKP standards, provide important input for assessing the outcomes of this work. In particular, inhibitors 22 and 23 represent current benchmarks for one type of profile being sought in the quest to develop new therapeutic leads for HAT. The structural motifs shown in Figure 3 represent a contrast to that present in these standards. In this regard, the DKP framework has potential for future fine-tuning and DKPs with phenol (see 4), indole (see 20), and SR (see 4, 7, 8, 20), substituents appear to be keys for achieving a useful bioactivity profile. Each of these has selectivity indexes (though at the bottom range) comparable to that of the two standards (see 22 and 23). On the other hand, none of our library compounds achieved a selectivity index (SI) on par with that of pentamidine (24) or melarsoprol (25) (SI = 542, and 1945, respectively44), but there are still problems associated with the clinical use of these drugs. Pentamidine is only effective against the first stage of sleeping sickness, where symptoms often are undetected or misinterpreted. Melarsoprol is successful in treating patients during the second stage of sleeping sickness, however, this arsenic-containing drug has serious toxicity issues, the most deleterious side effect being encephalopathic syndrome, and a mortality rate of approximately 5%.1 In addition, T. brucei is evolving cross-resistance to both drugs by mutation of transporter proteins.45
Figure 3.
Activity patterns for compounds of groups A-D
3. Conclusions
We have identified DKP containing scaffolds from investigation of marine-derived fungi metabolites with favorable activity – selectivity profiles. There are three compounds, 4, 7, and 20 that are at the top of this list because each is trypanocidal in the low micromolar range, has a selectivity index >15 and binds to rhodesain. The other trypanocidal compound 8, possessing a good selectivity index (16) and inactivity against either cysteine protease is also of current interest because a number of other validated T. brucei protein targets could be affected by it. Examples of emerging targets include glycogen synthase kinase 3 (GSK-3),46 glucose-6-phosphate dehydrogenase47 and dolicholphosphate mannose synthase (DPMS).48 An especially striking observation was that the disulfide-containing diketopiperazines 10, 11, and 12 showed nanomolar activity against the whole cell parasite. This is counterbalanced by their substantial toxicity to mammalian cells and at this point precludes their further therapeutic assessment. However, we suggest these disulfide-containing scaffolds should be considered as substrates for target finding experiments. Overall, we have shown that structurally diverse diketopiperazines obtained from marine-derived fungi can function as inhibitors to trypanosomal cysteine proteases and cause death of the protozoan parasite T. brucei.
4. Experimental section
4.1. General experimental procedures
The 1D NMR spectra were obtained using a Varian Unity 500+ at 500 MHz for 1H and 125 MHz for 13C or a Varian Inova 600 at 600 MHz for 1H and 150 MHz for 13C, outfitted with a cryoprobe. High-resolution and low-resolution mass spectra were acquired with a bench top Mariner ESI-TOF-MS. The optical rotations were determined on a Jasco DIP 370 polarimeter. RP-HPLC was performed using MeCN or MeOH and H2O with 0.1% formic acid and a 10 μm (preparative) or 5 μm (semipreparative and analytical) ODS column. Gliotoxin (10) was purchased from BIOMOL. Compound purity was determined to be ≥ 95% by analytical HPLC.
4.2. Biological materials
The Aspergillus fumigatus (coll. no. 030402d) was isolated from sediment collected using a hand held deep water sampling device at >33 m depth in Vanuatu in 2003. The Nectria inventa strain (coll. no. 049207c) was isolated from sediment collected by dredge at >600 m in the Monterey Bay trench. These strains were taxonomically identified by molecular (ITS and D1/D2 regions of rDNA) and morphological methods at the University of Texas Fungus Testing Laboratory. They are maintained as cryopreserved glycerol stocks at UCSC.
4.3. Culture conditions
030402d was grown in two different culture conditions coded as 030402dZi and 030402dZa. 14L were grown in 3.5% Czapek-Dox liquid media made with deionized water, pH adjusted to 7.3, with ca. 50 g of prewashed XAD-16 resin added to each liter prior to autoclaving (030402dZi). 10L were also grown in 3.5% Czapek-Dox liquid media made with artificial seawater (030402dZa). 20L of 049207c were grown in liquid medium containing 1.5% malt extract broth made with deionized water (049207cMi). Cultures were inoculated and shaken at 150 rpm for 21 days at room temperature.
4.4. Extraction and Isolation
4.4.1. Isolation of Compounds 3, 4, 13, 18, and 21 from 030402dZa
The mycelia was separated from the culture broth by vacuum filtration with a Buchner funnel. The culture broth was then extracted 3X with ethyl acetate and concentrated. The resulting crude extract was partitioned between hexanes (EFH), CH2Cl2 (EFD), and MeOH (EFM) per our standard procedure. The EFD extract was then subjected to preparative HPLC affording eight fractions on a 10-100% MeCN/H2O gradient. All bioactivity was contained in fraction P3 (IC50 = 0.8 μg/mL) which was further purified. Reverse phase semipreparative HPLC produced 18 fractions in which three pure compounds were obtained: verruculogen TR-2 (18, H5, 3.8 mg), bisdethiobis(methylthio)gliotoxin (3, H14, 27.8 mg), and 6-methoxyspirotryprostatin B (13, H15, 9.3 mg). H18 (22.5 mg, IC50 = 4 μg/mL) was separated into 15 fractions by analytical HPLC giving pure compounds cyclotryprostatin A (21, H18H8, 1.2 mg) and dehydrobisdethiobis(methylthio)gliotoxin (4, H18H10 1.5 mg).
4.4.2. Isolation of Compounds 17, 19, and 20 from 030402dZi
The 14 L culture material was transferred to a glass column with a cotton plug, and the broth was drained. The residual mycelia and resin were washed with H2O, and the extract was eluted with MeOH followed by CH2Cl2. The resulting crude extract was partitioned between hexanes (CXH), CH2Cl2 (CXD), and MeOH (CXM) per the same procedure mentioned above. The CXD fraction was applied to a to normal-phase automated flash chromatography using a linear gradient on silica gel (CH2Cl2 to CH3OH, 40 min), yielding nine fractions (CXDC1−CXDC9). Fractions CXDC3 and CXDC4 were combined (CXDC3-4, 37.0 mg) as well as CXDC-5, -6, and -7 (CXDC5-7, 24.4 mg). CXDC3-4 was fractionated using semi-preparative HPLC affording twelve fractions. Verruculogen (17, H9, 1.6 mg) was obtained as a pure compound. Further purification of H10 provided fumitremorgin B (19, H10H3, 0.3 mg). CXDC5-7 was separated in a similar fashion providing first 10 fractions. H5 showed outstanding bioactivity and was further purified, providing 12,13-dihydroxyfumitremorgin C (20, H5H4, 0.7 mg).
4.4.3. Isolation of Compounds 8, 9, 11, and 12 from 049207cMi
The mycelia were separated from the culture broth by vacuum filtration with a Buchner funnel. XAD-16 was added to the culture broth and was shaken for 2 hrs. The absorbent resin was then extracted 3X with acetone and this crude extract was then partitioned between hexanes (EFH), CH2Cl2 (EFD), and MeOH (EFM) per the same procedure mentioned above. The EFD fraction was then subjected to normal-phase automated flash chromatography using a linear gradient on silica gel (CH2Cl2 to EtOAc, 40 min), yielding nine fractions (EFDC1−EFDC9). EFDC-3, -4, -5, and -6 were combined (EFDC3-6, 135.2 mg) and then further fractionated via normal phased HPLC into 18 subfractions, one of which contained pure verticillin B (11, H2, 3.2 mg) 1H NMR (CDCl3, 500 MHz) δ 7.84 (1H, d, 3J = 7.5 Hz, H-10), 7.83 (1H, d, 3J = 7.5 Hz, H-10’), 7.17 (1H, dd, 3J = 7.5 Hz, 3J = 7.5 Hz, H-8), 7.16 (1H, dd, 3J = 7.5 Hz, 3J = 7.5 Hz, H-8’), 6.86 (1H, dd, 3J = 7.5 Hz, 3J = 7.5 Hz, H-9), 6.85 (1H, dd, 3J = 7.5 Hz, 3J = 7.5 Hz, H-9’), 6.68 (1H, d, 3J = 7.5 Hz, H-7), 6.67 (1H, d, 3J = 7.5 Hz, H-7’), 5.76 (1H, s, H-5a), 5.74 (1H, s, H-5a’), 5.70 (1H, s, H-6), 5.68 (1H, s, H-6’), 5.17 (1H, s, H-11), 5.14 (1H, s, H-11’), 5.10 (1H, br s, OH-11), 5.03 (1H, br s, OH-11’), 4.28 (1H, dd, 2J = 12.5 Hz, 3J = 5.5 Hz, H-13a), 4.18 (1H, dd, 2J = 12.5 Hz, 3J = 11.0 Hz, H-13b), 3.30 (1H, dd, 3J = 11.0 Hz, 3J = 5.5 Hz, OH-13), 3.11 (3H, s, H3-12), 3.01 (3H, s, H3-12’), 1.9 (3H, s, H3-13’); 13C NMR (CDCl3, 125 MHz) δ 166.6 (C, C-1), 166.4 (C, C-1’), 162.9 (C, C-4), 162.6 (C, C-4’), 148.8, (C, C-6a), 148.6 (C, C-6a’), 130.3, (2CH, C-8, C-8’), 129.5 (C, C-10a), 129.4 (C, C-10a’), 128.4 (2CH, C-10, C-10’), 120.9 (CH, C-9), 120.7 (CH, C-9’), 111.1 (CH, C-7), 111.0 (CH, C-7’), 83.1 (2CH, C-11, C11’), 82.2 (CH, C-5a), 82.0 (CH, C-5a’), 76.7 (C, C-11a), 76.3 (C, C-11a’), 75.6 (C, C-3), 73.1 (C, C-3’), 66.2 (C, C-10b), 66.0 (C, C-10b’), 60.5 (CH2, C-13), 27.4 (CH3, C-12), 27.2 (CH3, C-12’), 17.7 (CH3, C-13’); HRESIMS [M + Na]+ m/z 735.08529 (calculated for C30H28N6O7NaS4, 735.07946). Subfraction H9 was subjected to orthogonal chromatography and 3,6 bismethylthiocyclo-(alanyltryptophyl) was obtained (9, H9H1, 0.7 mg). H13 was also purified further by RP-HPLC to provide chaetocin, (12, H13H1, 1.2 mg). H14 was purified by normal phase HPLC to give chetoseminudin B (8, H14H6, 5.7 mg).
4.5. Semi-synthesis
4.5.1. Acetylation of bis(methylthio)gliotoxin
2.5 mg of bis(methylthio)gliotoxin was dissolved in pyridine and was submerged in an ice bath. 0.5 mL of acetyl chloride was added and the mixture was stirred for 1 hour. The reaction was quenched with water and extracted with pentanes. The organic layer was purified by RP-HPLC providing diacetylbis(methylthio)gliotoxin (5, 1.1 mg) 1H NMR (CDCl3, 600 MHz) δ 6.26 (1H, dm, 3J=14.0 Hz, H-6), 5.99 (2H, m, H-8, H-9), 5.62 (1H, m, H-7), 5.23 (1H, d, 3J=14.0 Hz, H-5a), 4.69 (1H, d, 2J=11.4 Hz, H-15a), 4.45 (1H, d, 2J=11.4 Hz, H-15b), 3.11 (1H, d, 2J=14.0, Hz, H-10a), 3.07 (3H, s, H3-13), 2.83 (1H, d, 2J=14.0, Hz, H-10a), 2.31 (3H, s, H3-12), 2.28 (3H, s, H3-14), 2.14 (3H, s, H3-6Ac), 1.98 (3H, s, H3-15Ac); 13C NMR detected indirectly by HMQC and HMBC (CDCl3, 150 MHz) δ 171.1 (C, C-6Ac), 169.8 (C, C-15Ac), 166.3 (C, C-1), 162.8 (C, C-4), 133.8 (C, C-9a), 128.1 (CH, C-7), 125.1 (CH, C-8), 120.0 (C, C-9), 75.1 (CH, C-6), 73.1 (C, C-11), 69.9, (C, C-3), 65.3 (CH, C-5a), 64.5 (CH2, C-15), 40.3 (CH2, C-10), 28.9 (CH3, C-13), 21.2 (CH3, C-6Ac), 20.5 (CH3, C-15Ac), 14.8 (CH3, C-12), 13.0 (CH3, C-14); HRESIMS [M + Na]+ m/z 463.09323 (calculated for C19H24N2O6NaS2, 463.09680);15-acetylbis(methylthio)gliotoxin (6, 0.7 mg), [α]28D - 43.8 (c 0.04, CH3OH); 1H NMR (CDCl3, 600 MHz) δ 5.98 (1H, d, 3J=10.0 Hz, H-9), 5.91 (1H, dd, 3J=10.0, 5.0 Hz, H-8), 5.79 (1H, m, H-7), 4.95 (2H, m, H-5a, H-6), 4.75 (1H, d, 2J, 11.6 Hz, H-15a), 4.52 (1H, d, 2J, 11.6 Hz, H-15b), 3.12 (1H, d, 2J, 16.0 Hz, H-10a), 3.08 (3H, s, CH3-13), 2.91 (1H, d, 2J, 16.0 Hz, H-10b), 2.33 (3H, s, CH3-14), 2.26 (3H, s, CH3-12), 2.02 (3H, s, CH3-15Ac); 13C NMR detected indirectly by HMQC and HMBC (CDCl3, 150 MHz) δ 169.8 (C, C-15Ac), 167.8 (C, C-4), 165.3 (C, C-1), 131.4 (C, C-9a), 130.6 (CH, C-7), 122.8 (CH, C-8), 120.3 (CH, C-9), 74.3 (CH, C-6), 71.4 (C, C-11), 69.7 (CH, C-5a), 69.6 (C, C-3), 64.3 (CH2, C-15), 38.9 (CH2, C-10), 28.7 (CH3, C-13), 20.6 (CH3, C-15Ac), 14.8 (CH3, C-12), 13.7 (CH3, C-14); HRESIMS [M + H]+ m/z 399.1090 (calculated for C17H23N2O5S2, 399.1043); and 6-acetylbis(methylthio)gliotoxin (7, 0.5 mg), 1H NMR (CDCl3, 600 MHz) δ 6.22 (1H, d, 3J=14.0 Hz, H-6), 5.97 (2H, m, H-8, H-9), 5.60 (1H, d, 3J=8.0 Hz, H-7), 5.21 (1H, d, 3J=14.0 Hz, H-5a), 4.33 (1H, dd, 2J=12.0 Hz, 3J=5.0 Hz, H-15a), 3.83 (1H, dd, 2J=12.0 Hz, 3J=9.0 Hz, H-15b), 3.13 (3H, s, CH3-13), 3.11 (1H, d, 2J=16.0 Hz, H-10a), 2.88 (1H, d, 2J=16.0 Hz, H-10b), 2.30 (3H, s, CH3-12), 2.21 (3H, s, CH3-12), 2.09 (3H,s, CH3-6Ac), 2.09 (1H, dd, 3J=9.0, 5.0 Hz, H-15OH); 13C NMR detected indirectly by HMQC and HMBC (CDCl3, 150 MHz) δ 170.8 (C, C-6Ac), 166.4 (C, C-1), 164.1 (C, C-4), 133.8 (C, C-9a), 127.9 (CH, C-7), 125.2 (CH, C-8), 119.9 (CH, C-9), 75.2 (CH, C-6), 72.7 (C, C-11), 71.8 (C, C-3), 65.2 (CH, C-5a), 63.5 (CH2, C-15), 40.1 (CH2, C-10), 28.7 (CH3, C-13), 21.2 (CH3, C-6Ac), 14.8 (CH3, C-12), 12.7 (CH3, C-14); HRESIMS [M + Na]+ m/z 421.09003 (calculated for C17H23N2O5NaS2, 421.08624).
Other physical properties for compounds 5 and 7 matched reported data.37,38
4.5.2. Hydrogenation of 6-methoxyspirotryprostatin B
1 mg of 6-methoxy spirotryprostatin B was dissolved in 1 mL of dry EtOH. 3 mg of palladium on carbon was then added, and the mixture was stirred for 30 minutes under H2 at 1 atm. The reaction was quenched with H2O and the catalyst was filtered through Celite. Purification of the resultant product was performed with analytical HPLC at 32% MeOH/H2O, providing spirotryprostatin A (14, 1 mg). Physical properties match reported literature.39 Surprisingly, no reduction was observed for the isoprenyl alkene.
4.5.3. Methylation of 6-methoxyspirotryprostatin B
1 mg of 6-methoxyspirotryprostatin B was dissolved in 1 mL of dry THF. 2 mg of NaH (60% dispersed in mineral oil) was then added, and the mixture was stirred for 10 minutes. 0.8 mL of iodomethane was then added and the mixture was stirred for 30 minutes. The reaction was quenched with H2O, and then 5 mL of MeOH were added and then aqueous mixture was extracted with pentanes. The aqueous layer was evaporated to dryness and then partitioned between H2O and EtOAc. The organic layer was subjected to HPLC to afford N-methyl 6-methoxyspirotryprostatin B as a white solid (15, 0.7 mg); [α]25D -115 (c 0.2, CHCl3); 1H NMR (CDCl3, 600 MHz) δ 6.98 (1H, d, 3J = 8.4 Hz, H-4), 6.53 (1H, dd, 3J = 8.4 Hz, 4J = 2.4 Hz, H-5), 6.41 (1H, d, 4J = 2.4 Hz, H-7), 5.73 (1H, s, H-8), 5.35 (1H, d, 3J = 9.0 Hz, H-18), 5.20 (1H, ds, 3J = 9.0 Hz, 4J = 1.2, H-19), 4.34 (1H, dd, 3J = 10.8 Hz, 4J = 6.0, H-12), 3.84 (3H, s, OCH3), 3.83 (1H, m, H-15a), 3.57 (1H, m, H-15b), 3.21 (3H, s, NCH3), 2.49 (1H, m, H-13a), 2.12 (1H, m, H-14a), 1.99 (2H, m, H-13b, H-14b), 1.58 (3H, d, 4J = 1.2, H3-21), 1.27 (3H, d, 4J = 1.2, H3-22); 13C NMR detected indirectly by HMQC and HMBC (CDCl3, 150 MHz) δ 177.1 (C, C-2), 162.6 (C, C-11), 161.1, (C, C-6), 155.4 (C, C-17), 145.1 (C, C-7a), 137.9 (C, C-20), 137.8 (C, C-9), 128.0 (CH, C-4), 120.4 (CH, C-19), 118.6 (C, C-3a), 117.0 (CH, C-8), 106.4 (CH, C-5), 96.0 (CH, C-7), 64.3 (CH, C-18), 61.6 (CH, C-12), 61.0 (C, C-3), 55.5 (CH3, OCH3), 44.7 (CH2, C-15), 29.0 (CH2, C-13), 26.8 (CH3, NCH3), 25.3 (CH3, C-21), 21.9 (CH2, C-14), 18.1 (CH3, C-22); HRESIMS [M + Na]+ m/z 430.1763 (calculated for C23H25N3O4Na, 430.1737).
4.5.4. Methylation of spirotryprostatin A
1 mg of spirotryprostatin A was dissolved in 1 mL of dry THF. 2 mg of NaH (60% dispersed in mineral oil) was then added, and the mixture was stirred for 10 minutes. 0.8 mL of iodomethane was then added and the mixture was stirred for 30 minutes. The reaction was quenched with H2O, and then 5 mL of MeOH were added and the aqueous mixture was extracted with pentanes. The aqueous layer was evaporated to dryness and then partitioned between H2O and EtOAc. The organic layer was subjected to HPLC to afford N-methyl spirotryprostatin A as a white solid (16, 0.9 mg); [α]26D -138 (c 0.2, CHCl3); 1H NMR (CDCl3, 600 MHz) δ 6.96 (1H, d, 3J = 8.4 Hz, H-4), 6.50 (1H, dd, 3J = 8.4 Hz, 4J = 2.4 Hz, H-5), 6.39 (1H, d, 4J = 2.4 Hz, H-7), 5.04 (2H, m, H-9, H-19), 4.72 (1H, d, 3J = 9.0 Hz, H-18), 4.34 (1H, dd, 3J = 8.4 Hz, 4J = 7.8, H-12), 3.83 (3H, s, OCH3), 3.62 (1H, m, H-15a), 3.57 (1H, m, H-15b), 3.21 (3H, s, NCH3), 2.61 (dd, 2J = 13.2 Hz, 3J = 10.8, H-8a), 2.33 (dd, 2J = 13.2 Hz, 3J = 7.2, H-8b), 2.33 (1H, m, H-13a), 2.26 (1H, m, H-13b), 2.05 (1H, m, H-14a), 1.97 (1H, m, H-14b), 1.66 (3H, d, 4J = 1.2, H3-21), 1.16 (3H, d, 4J = 1.2, H3-22); 13C NMR detected indirectly by HMQC and HMBC (CDCl3, 150 MHz) δ 179.1 (C, C-2), 167.1 (C, C-17), 166.8 (C, C-11), 160.6 (C, C-6), 145.1 (C, C-7a), 138.1 (C, C-20), 121.6 (CH, C-19), 126.7 (CH, C-4), 118.2 (C, C-3a), 105.9 (CH, C-5), 95.6 (CH, C-7), 61.1 (CH, C-12), 60.4 (CH, C-18), 58.4 (CH, C-9), 55.4 (CH3, OCH3), 55.1 (C, C-3), 45.3 (CH2, C-15), 34.2 (CH2, C-8), 27.0 (CH2, C-13), 26.1 (CH3, NCH3), 25.5 (CH3, C-21), 23.8 (CH2, C-14) 18.0 (CH3, C-22); HRESIMS [M + Na]+ m/z 432.1852 (calculated for C23H27N3O4Na, 432.1894).
4.6. Biological Assays
4.6.1. Cysteine protease assays
Recombinant cruzain, rhodesain and TbCatB were expressed as described previously.11,40,49 Compounds were first dissolved in DMSO to 5 mg/mL and serially diluted in DMSO in the range of 25 μg/mL – 0.001 μg/mL for IC50 determinations.
Protease inhibition assays for cruzain and rhodesain were carried out in 96 well plate format as described previously.50 Cruzain (4 nM), rhodesain (4 nM) or TbCatB (258 nM) was incubated with test compound in 100 mM sodium acetate, pH 5.5, containing 5 mM DTT and 0.001% Triton X-100 (buffer A), for 5 min at room temperature. Then buffer A containing Z-Phe-Arg-AMC (Bachem) was added to enzyme-compound mixture to give 10 μM substrate in a final assay volume of 200 μL. The rate of free AMC release was measured at excitation and emission wavelengths of 355 and 460 nm, respectively, with a microtiter plate spectrofluorimeter (SpectraMax M5, Molecular Devices) for 3 min. Percentage inhibition of test compound was calculated relative to the DMSO control (0% inhibition control). IC50 values were determined with Prism 4 software (GraphPad, San Diego, CA) using sigmoidal dose-response variable slope model.
4.6.2. Trypanosoma brucei brucei assay
The growth inhibition assay for T. brucei brucei was conducted as described previously.51 Bloodstream forms of the monomorphic T. b. brucei clone 427-221a were grown in complete HMI-9 medium containing 10% FBS, 10% Serum Plus medium (Sigma Inc. St. Louis Mo. USA), 50 U/mL penicillin and 50 μg/mL streptomycin (Invitrogen) at 37°C under a humidified atmosphere and 5% CO2. Compounds were screened at 12.5 μg/mL and 1.25 μg/mL for % inhibition values or serially diluted in the range of 25 μg/mL – 0.001 μg/mL for IC50 determinations. 5 μL of each dilution was added to 95 μL of diluted parasites (1x104 cells per well) in sterile Greiner 96-well flat white opaque culture plates such that the final DMSO concentration was 0.5%. 0% inhibition control wells contained 0.5% DMSO while 100% inhibition control wells contained 50 μM thimerosal (Sigma). After compound addition, plates were incubated for 40 hours at 37°C. At the end of the incubation period, 50 µL of CellTiter-Glo™ reagent (Promega Inc., Madison, WI, USA) was added to each well and plates were placed on an orbital shaker at room temperature for 2 minutes to induce lysis. After a 10 minute incubation to stabilize the signal, the ATP-bioluminescence of each well was determined using an Analyst HT plate reader (Molecular Devices, Sunnyvale, CA, USA). Raw values were converted to log10 and percentage inhibition calculated relative to the controls. IC50 curve fittings were performed with Prism 4 software as above.
4.6.3. Jurkat cell cytotoxicity assay
Jurkat cells (clone E6-1) were grown in complete RPMI-1640 medium containing 10% FBS, 50 μg/mL penicillin and 50 μg/mL streptomycin (Invitrogen) at 37°C under a humidified atmosphere and 5% CO2. For cytotoxicity testing, cells were diluted to 1x105 per mL in complete RPMI-1640 medium. Test compound stocks were prepared in 100% DMSO and screened at concentrations ranging from 25 μg/mL – 0.001 μg/mL against cells (1×104 cells per well) in sterile Greiner 96-well flat white opaque culture plates (0.5%. final DMSO concentration). 0% inhibition control wells contained 0.5% DMSO while 100% inhibition control wells contained 40 μM staurosporine. After compound addition, plates were incubated for 40 hr at 37°C. At the end of the incubation period, 50 μL of CellTiter-Glo™ reagent (Promega Inc., Madison, WI, USA) was added to each well and plates were placed on an orbital shaker at room temperature for 2 minutes to induce lysis. After a 10 minute incubation to stabilize the signal, the ATP-bioluminescence of each well was determined using an Analyst HT plate reader (Molecular Devices, Sunnyvale, CA, USA). Calculations of percentage inhibition and IC50 were performed similar to those in the T. brucei brucei assay.
Supplementary Material
Acknowledgement
This work was supported by grants from the Sandler Family Foundation, the California Institute for Quantitative Biosciences (QB3), NMR equipment grants NSF CHE-0342912 and NIH S10-RR19918, and MS equipment grant NIH S10-RR20939. We would like to thank Dr. J. Musser for suggesting semi-synthesis to expand our library. Gratitude is also extended to Prof. Greg Cailliet at the Moss Landing Marine Lab for the Monterey Bay expedition invitation. Thank you to Captain (S. Crusot) and Crew of the M/V Horizon for field assistance in Vanuatu and to the Monterey Bay Aquarium Research Institute (MBARI). The staff of the UT San Antonio Health Sciences Center Fungus Testing Laboratory, D. A. Sutton and B. Wickes, provided fungal ID's.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary material associated with this article can be found, in the online version, at [INSERT DOI].
References and notes
- 1.The World Health Organization [Jan 6, 2010];African Trypanosomiasis (Sleeping Sickness) http://www.who.int/trypanosomiasis_african/en/
- 2.Barrett MP, Boykin DW, Brun R, Tidwell RR. Br. J. Pharmacol. 2007;152:1155–1171. doi: 10.1038/sj.bjp.0707354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peter GEK. Annal. Neurol. 2008;64:116–126. [Google Scholar]
- 4.Priotto G, Kasparian S, Mutombo W, Ngouama D, Ghorashian S, Arnold U, Ghabri S, Baudin E, Buard V, Kazadi-Kyanza S, Ilunga M, Mutangala W, Pohlig G, Schmid C, Karunakara U, Torreele E, Kande V. The Lancet. 2009;374:56–64. doi: 10.1016/S0140-6736(09)61117-X. [DOI] [PubMed] [Google Scholar]
- 5.Jennings FW, Urquhart GM. Parasitol. Res. 1983;69:577–581. doi: 10.1007/BF00926669. [DOI] [PubMed] [Google Scholar]
- 6.United States National Institutes of Health [Jan 6, 2010];Human African Trypanosomiasis: First in Man Clinical Trial of a New Medicinal Product, the Fexinidazole. http://clinicaltrials.gov/ct2/show/NCT00982904.
- 7.Hoet S, Opperdoesb F, Brunc R, Quetin-Leclercqa J. Nat. Prod. Rep. 2004;21:353–364. doi: 10.1039/b311021b. [DOI] [PubMed] [Google Scholar]
- 8.A SciFinder search on August 10th, 2009 with the search terms Trypanosoma brucei, natural products, and 2003- were used to supplement Hoet's review.
- 9.Rubio BK, Tenney K, Ang K-H, Abdulla M, Arkin M, McKerrow JH, Crews P. J. Nat. Prod. 2009;72:218–222. doi: 10.1021/np800711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson TA, Amagata T, Sashidhara KV, Oliver AG, Tenney K, Matainaho T, Ang KK-H, McKerrow JH, Crews P. Org. Lett. 2009;11:1975–1978. doi: 10.1021/ol900446d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mallari JP, Shelat AA, Obrien T, Caffrey CR, Kosinski A, Connelly M, Harbut M, Greenbaum D, McKerrow JH, Guy RK. J. Med. Chem. 2008;51:545–552. doi: 10.1021/jm070760l. [DOI] [PubMed] [Google Scholar]
- 12.Mallari JP, Shelat A, Kosinski A, Caffrey CR, Connelly M, Zhu F, McKerrow JH, Guy RK. Bioorg. Med. Chem. Lett. 2008;18:2883–2885. doi: 10.1016/j.bmcl.2008.03.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallari JP, Shelat AA, Kosinski A, Caffrey CR, Connelly M, Zhu F, McKerrow JH, Guy RK. J. Med. Chem. 2009;52:6489–6493. doi: 10.1021/jm900908p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien TC, Mackey ZB, Fetter RD, Choe Y, O'Donoghue AJ, Zhou M, Craik CS, Caffrey CR, McKerrow JH. J. Biol. Chem. 2008;283:28934–28943. doi: 10.1074/jbc.M805824200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdulla M-H, O'Brien T, Mackey ZB, Sajid M, Grab DJ, McKerrow JH. PLoS Negl Trop Dis. 2008;2:e298. doi: 10.1371/journal.pntd.0000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osterhage C, Kaminsky R, Konig GM, Wright AD. J. Org. Chem. 2000;65:6412–6417. doi: 10.1021/jo000307g. [DOI] [PubMed] [Google Scholar]
- 17.Minagawa N, Yabu Y, Kita K, Nagai K, Ohta N, Meguro K, Sakajo S, Yoshimoto A. Mol. Biochem. Parasitol. 1997;84:271–280. doi: 10.1016/s0166-6851(96)02797-1. [DOI] [PubMed] [Google Scholar]
- 18.Boot CM, Gassner NC, Compton JE, Tenney K, Tamble CM, Lokey RS, Holman TR, Crews P. J. Nat. Prod. 2007;70:1672–1675. doi: 10.1021/np070307c. [DOI] [PubMed] [Google Scholar]
- 19.Gautschi JT, Amagata T, Amagata A, Valeriote FA, Mooberry SL, Crews P. J. Nat. Prod. 2004;67:362–367. doi: 10.1021/np030388m. [DOI] [PubMed] [Google Scholar]
- 20.Gautschi JT, Tenney K, Compton J, Crews P. Nat. Prod. Commun. 2007;2:541–546. [Google Scholar]
- 21.Li S-M. Nat. Prod. Rep. 2010;27:57–78. doi: 10.1039/b909987p. [DOI] [PubMed] [Google Scholar]
- 22.Kanoh K, Kohno S, Asari T, Harada T, Katada J, Muramatsu M, Kawashima H, Sekiya H, Uno I. Bioorg. Med. Chem. Lett. 1997;7:2847–2852. [Google Scholar]
- 23.Kanoh K, Kohno S, Katada J, Takahashi J, Uno I. J. Antibiot. 1999;52:131–141. doi: 10.7164/antibiotics.52.134. [DOI] [PubMed] [Google Scholar]
- 24.Mita AC, Yee LK, Papadopoulos KP, Heath EI, Romero O, Lloyd GK, Cropp G, Spear MA, Mita MM, LoRusso PM. J. Clin. Oncol. (Meeting Abstracts) 2008;26:3525. [Google Scholar]
- 25.Kirby GW, Robins DJ, Sefton MA, Talekar RR. J. Chem. Soc., Perk. Trans. 1. 1980:119–121. [Google Scholar]
- 26.Hanson JR, Oleary MA. J. Chem. Soc. Perkin 1. 1981:218–220. [Google Scholar]
- 27.Zhang M, Wang WL, Fang YC, Zhu TJ, Gu QQ, Zhu WM. J. Nat. Prod. 2008;71:985–989. doi: 10.1021/np700737g. [DOI] [PubMed] [Google Scholar]
- 28.Steyn PS, Vleggaar R, Rabies CJ. Phytochemistry. 1981;20:538–539. [Google Scholar]
- 29.Cui C-B, Kakeya H, Osada H. Tetrahedron. 1997;53:59–72. [Google Scholar]
- 30.Fayos J, Lokensgard D, Clardy J, Cole RJ, Kirksey JW. J. Am. Chem. Soc. 1974;96:6785–6787. doi: 10.1021/ja00828a054. [DOI] [PubMed] [Google Scholar]
- 31.Yamazaki M, Sasago K, Miyaki K. J. Chem. Soc., Chem. Commun. 1974:408–409. [Google Scholar]
- 32.Abraham WR, Arfmann HA. Phytochemistry. 1990;29:1025–1026. [Google Scholar]
- 33.Fujimoto H, Sumino M, Okuyama E, Ishibashi M. J. Nat. Prod. 2004;67:98–102. doi: 10.1021/np0302201. [DOI] [PubMed] [Google Scholar]
- 34.Son BW, Jensen PR, Kauffman CA, Fenical W. Nat. Prod. Lett. 1999;13:213–222. [Google Scholar]
- 35.Minato H, Matsumot M, Katayama T. J. Chem. Soc., Perkin Trans. 1. 1973:1819–1825. doi: 10.1039/p19730001819. [DOI] [PubMed] [Google Scholar]
- 36.Hauser D, Weber HP, Sigg HP. Helv. Chim. Act. 1970;53:1061–1073. doi: 10.1002/hlca.19700530521. [DOI] [PubMed] [Google Scholar]
- 37.Okamoto M, Yoshida K, Uchida I, Nishikawa M, Kohsaka M, Aoki H. Chem. Pharm. Bull. 1986;34:340–344. doi: 10.1248/cpb.34.340. [DOI] [PubMed] [Google Scholar]
- 38.Van der Pyl D, Inokoshi J, Shiomi K, Yang H, Takeshima H, Omura S. J. Antibiot. 1992;45:1802–1805. doi: 10.7164/antibiotics.45.1802. [DOI] [PubMed] [Google Scholar]
- 39.Cui C-B, Kakeya H, Osada H. Tetrahedron. 1996;52:12651–12666. [Google Scholar]
- 40.Caffrey CR, Hansell E, Lucas KD, Brinen LS, Alvarez Hernandez A, Cheng J, Gwaltney SL, Roush WR, Stierhof Y-D, Bogyo M, Steverding D, McKerrow JH. Mol. Biochem. Parasitol. 2001;118:61–73. doi: 10.1016/s0166-6851(01)00368-1. [DOI] [PubMed] [Google Scholar]
- 41.Sanner T, Pihl A. J. Biol. Chem. 1963;238:165–171. [PubMed] [Google Scholar]
- 42.Schroeder HW, Cole RJ, Hein H, Kirksey JW. Appl. Microbiol. 1975;29:857–858. doi: 10.1128/am.29.6.857-858.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Latge JP. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bakunova SM, Bakunov SA, Patrick DA, Kumar EVKS, Ohemeng KA, Bridges AS, Wenzler T, Barszcz T, Kilgore Jones S, Werbovetz KA, Brun R, Tidwell RR. J. Med. Chem. 2009;52:2016–2035. doi: 10.1021/jm801547t. [DOI] [PubMed] [Google Scholar]
- 45.de Koning HP. Trends Parasitol. 2008;24:345–349. doi: 10.1016/j.pt.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Ojo KK, Gillespie JR, Riechers AJ, Napuli AJ, Verlinde CLMJ, Buckner FS, Gelb MH, Domostoj MM, Wells SJ, Scheer A, Wells TNC, Van Voorhis WC. Antimicrob. Agents Chemother. 2008;52:3710–3717. doi: 10.1128/AAC.00364-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cordeiro AT, Thiemann OH, Michels PAM. Bioorg. Med. Chem. 2009;17:2483–2489. doi: 10.1016/j.bmc.2009.01.068. [DOI] [PubMed] [Google Scholar]
- 48.Smith TK, Young BL, Denton H, Hughes DL, Wagner GK. Bioorg. Med. Chem. Lett. 2009;19:1749–1752. doi: 10.1016/j.bmcl.2009.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eakin AE, Mcgrath ME, McKerrow JH, Fletterick RJ, Craik CS. J. Biol. Chem. 1993;268:6115–6118. [PubMed] [Google Scholar]
- 50.Greenbaum DC, Mackey Z, Hansell E, Doyle P, Gut J, Caffrey CR, Lehrman J, Rosenthal PJ, McKerrow JH, Chibale K. J. Med. Chem. 2004;47:3212–3219. doi: 10.1021/jm030549j. [DOI] [PubMed] [Google Scholar]
- 51.Mackey ZB, Baca AM, Mallari JP, Apsel B, Shelat A, Hansell EJ, Chiang PK, Wolff B, Guy KR, Williams J, McKerrow JH. Chem. Biol. Drug Des. 2006;67:355–363. doi: 10.1111/j.1747-0285.2006.00389.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.