Ubr1 and Ubr2 ubiquitin ligases are shown to promote degradation of misfolded cytosolic polypeptides in vivo and in a purified system in association with Hsp70.
Abstract
Quality control systems facilitate polypeptide folding and degradation to maintain protein homeostasis. Molecular chaperones promote folding, whereas the ubiquitin/proteasome system mediates degradation. We show here that Saccharomyces cerevisiae Ubr1 and Ubr2 ubiquitin ligases promote degradation of unfolded or misfolded cytosolic polypeptides. Ubr1 also catalyzes ubiquitinylation of denatured but not native luciferase in a purified system. This activity is based on the direct interaction of denatured luciferase with Ubr1, although Hsp70 stimulates polyubiquitinylation of the denatured substrate. We also report that loss of Ubr1 and Ubr2 function suppressed the growth arrest phenotype resulting from chaperone mutation. This correlates with increased protein kinase maturation and indicates partitioning of foldable conformers toward the proteasome. Our findings, based on the efficiency of this quality control system, suggest that the cell trades growth potential to avert the potential toxicity associated with accumulation of unfolded or misfolded proteins. Ubr1 and Ubr2 therefore represent E3 components of a novel quality control pathway for proteins synthesized on cytosolic ribosomes.
INTRODUCTION
The relative contribution of molecular chaperones and ubiquitin ligases to cellular quality control promotes proteome homeostasis (Wickner et al., 1999; Balch et al., 2008). Their combined action manifests in a quality control process that is best characterized for the endoplasmic reticulum (ER), where several chaperone and ubiquitin ligase assemblies ensure prompt degradation of misfolded proteins (Vembar and Brodsky, 2008). Less is known regarding quality control of polypeptides synthesized on cytosolic ribosomes. The ubiquitin ligase CHIP interacts with heat shock protein 70 (Hsp70) and Hsp90 molecular chaperones to promote degradation of misfolded or unfolded polypeptides (Connell et al., 2001; Meacham et al., 2001). Degradation still occurs in CHIP −/− cells, however, indicating that other ubiquitin ligases function in a redundant manner (Xu et al., 2002). In addition, there is no CHIP homologue in yeast despite rapid degradation of misfolded cytosolic proteins.
The role of molecular chaperones in promoting polypeptide folding is well established (Young et al., 2004). Results from recent studies also suggest that Hsp70, Hsp90, and several cochaperones function in polypeptide degradation (McClellan et al., 2005; Arndt et al., 2007). Recently, two molecular chaperones have been shown to protect newly synthesized protein kinases from degradation during or shortly after translation. Both Cdc37 and Ydj1 adopt this role and protect against a biphasic degradation process (Mandal et al., 2007, 2008). The first phase is very rapid on a timescale of 5 min or less (within the time taken for a pulse-labeling reaction) and appears to be the same “zero-point” effect as described by Varshavsky and colleagues (Baker and Varshavsky, 1991; Suzuki and Varshavsky, 1999; Xie and Varshavsky, 2002). Polypeptides degraded in the second phase have a half-life of ∼20 min. What remains unknown are the components that mediate the selection of misfolded proteins for the different types of degradation. Previous studies suggested that Ubc4 and Ubc5 ubiquitin-conjugating enzymes function in the clearance of misfolded polypeptides in the cytosol (Seufert and Jentsch, 1990), but the identities of the ubiquitin ligases, apart from CHIP, have not been reported.
Ubr1 is an ubiquitin ligase previously shown to function in N-end rule degradation (Bartel et al., 1990). This process, described by Varshavsky and colleagues, facilitates degradation of polypeptides by recognizing destabilizing N-terminal amino acids (Mogk et al., 2007). Bulky hydrophobic and basic amino acids such as phenylalanine or arginine, promote degradation via Ubr1, whereas others, such as methionine, are stabilizing and protect against degradation. In addition, Ubr1 also functions in N-end rule independent degradation (Du et al., 2002) and was shown recently by Eisele and Wolf (2008) to act on a misfolded protein in the cytosol. Ubr1 is a large protein with at least three distinct substrate-binding sites and functions in association with Ubc2/Rad6 and also Ubc4 (Byrd et al., 1998). Ubr1 has a paralog, Ubr2, which does not have the conserved sequence motifs required for N-end rule degradation but promotes ubiquitinylation of Rpn4, a transcription factor involved in regulating genes encoding proteasome subunits (Wang et al., 2004). In this report we describe Ubr1 and Ubr2 as having a general role in the clearance of misfolded cytosolic proteins and show that their action is stimulated by Hsp70.
MATERIALS AND METHODS
Chemicals and Antisera
Geldanamycin (GA) was purchased from InvivoGen (San Diego, CA) and stored at −20°C in dimethylsulphoxide. l-azetidine-2-carboxylic acid (AZC) was purchased from Sigma (A0760; St. Louis, MO) and stored in water at −20°C. MG132 was purchased from Calbiochem (La Jolla, CA). Nourseothricin was purchased from Werner BioAgents (Jena, Germany). G418 sulfate was obtained from Alexis Biochemicals (San Diego, CA). Complete protease inhibitors were purchased from Roche (Indianapolis, IN). Mouse monoclonal anti-myc (9E10) and anti-hemagglutinin (HA; 12CA5) were obtained from the Mount Sinai Hybridoma Facility (New York, NY). Anti-Flag was purchased from Sigma (F1804). Rabbit polyclonal anti-tap was prepared against the peptide: CSSGALDYDIPTTASENLYFQ by Covance Research (Madison, WI). Cdc28 was immunoprecipitated using anti-PSTAIRE (Santa Cruz Biotechnology, Santa Cruz, CA). Hsp70 mouse monoclonal antisera was purchased from Stressgen (spa-822; San Diego, CA). Anti-Hsp104 was from Stressgen (SPA-1040) and goat anti-luciferase from Chemicon (AB 3256; Temecula, CA). Anti–galactosidase (ß-Gal) was purchased from Promega (Z378A; Madison, WI).
Yeast Cell Growth Conditions
Saccharomyces cerevisiae was grown in either rich YPD media (2% [wt/vol] Bacto peptone (Difco), 1% Bacto yeast extract (Difco), 2% glucose) or in selective media (SD; 0.67% Yeast nitrogen base without amino acids (Difco), 2% glucose and appropriate amino acids). G418 sulfate and nourseothricin were added to a final concentration of 400 μg/ml and 100 μg/ml, respectively. S. cerevisiae strains used in this study are listed in Table 1.
Table 1.
Yeast strains used in this study
| Strain | MAT | Genotype | Source |
|---|---|---|---|
| BY4741 | a | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| S288C | a | his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| Tpk2-TAP | a | S288C, TPK2-TAP (HIS3) | Open Biosystems |
| Cmk2-TAP | a | S288C, CMK2-TAP (HIS3) | Open Biosystems |
| Cup9-TAP | a | S288C, CUP9-TAP (HIS3) | Open Biosystems |
| Rim11-TAP | a | S288C, RIM11-TAP (HIS3) | Open Biosystems |
| Kss1-TAP | a | S288C, KSS1-TAP (HIS3) | Open Biosystems |
| NNY1 | a | S288C, TPK2-TAP (HIS3) erg6::kanMX6 | This study |
| NNY2 | a | S288C, TPK2-TAP (HIS3) erg6::natMX4 | This study |
| NNY3 | a | S288C, CMK2-TAP (HIS3) erg6::kanMX6 | This study |
| NNY4 | a | S288C, CUP9-TAP (HIS3), erg6::natMX4 | This study |
| NNY5 | a | S288C, RIM11-TAP (HIS3) erg6::kanMX6 | This study |
| NNY6 | a | S288C, KSS1-TAP (HIS3) erg6::kanMX6 | This study |
| NNY7 | a | BY4741, erg6::natMX4, pTPK2 NT-HA 1.111 (LEU2) | This study |
| NNY8 | a | BY4741, ubr1::kanMX6, erg6::natMX4, pTPK2 NT-HA 1.111 (LEU2) | This study |
| NNY9 | a | BY4741, ubr2::kanMX6, erg6::natMX4, pTPK2 NT-HA 1.111 (LEU2) | This study |
| NNY10 | a | BY4741, ubr1::kanMX6, ubr2::HIS3, erg6::natMX4, pTPK2 NT-HA 1.111 (LEU2) | This study |
| NNY11 | a | S288C, TPK2-TAP (HIS3) cdc37S14A::URA3 erg6::kanMX6 | This study |
| NNY12 | a | BY4741, ubr1Δ::kanMX6, erg6::natMX4, pNTFlagUBR1 (LEU2) | This study |
| JN516 | α | his3-11, 3-15, leu2-3,2-112, ura3-52, trp1-Δ1, lys2, SSA1, ssa2::LEU2 ssa3::TRP1 ssa4::LYS2 | E. Craig |
| ssa1-45 | α | his3-11, 3-15, leu2-3,2-112, ura3-52, trp1-Δ1, lys2, ssa1-45, ssa2::LEU2 ssa3::TRP1 ssa4::LYS2 | E. Craig |
| NNY13 | a | S288C, TPK2-TAP (HIS3) erg6::natMX4 ubr1::kanMX6 | This study |
| NNY14 | a | S288C, RIM11-TAP (HIS3) erg6::natMX4 ubr1::kanMX6 | This study |
| NNY15 | a | S288C, KSS1-TAP (HIS3) erg6::natMX4 ubr1::kanMX6 | This study |
| NNY16 | a | S288C, TPK2-TAP (HIS3) erg6::natMX4 ubr2::kanMX6 | This study |
| NNY17 | a | S288C, RIM11-TAP (HIS3) erg6::natMX4 ubr2::kanMX6 | This study |
| NNY18 | a | S288C, KSS1-TAP (HIS3) erg6::natMX4 ubr2::kanMX6 | This study |
| NNY19 | a | S288C, TPK2-TAP (HIS3) cdc37S14A::URA3 | This study |
| NNY20 | a | S288C, TPK2-TAP (HIS3) cdc37S14A::URA3, ubr1::kanMX6, | This study |
| NNY21 | a | S288C, TPK2-TAP (HIS3) cdc37S14A::URA3, ubr2::kanMX6, | This study |
| NNY22 | a | S288C, TPK2-TAP (HIS3) cdc37S14A::URA3, ubr1::kanMX6, ubr2::natMX4 | This study |
| ubr1Δ | a | BY4741, ubr1::kanMX6 | Open Biosystems |
| ubr2Δ | a | BY4741, ubr2::kanMX6 | Open Biosystems |
| NNY23 | a | BY4741, ubr1::kanMX6, ubr2::HIS3 | This study |
| NNY24 | a | BY4741, cdc37S14A::natMX4 | This study |
| NNY25 | a | BY4741, cdc37S14A::natMX4, ubr1::kanMX6 | This study |
| NNY26 | a | BY4741, cdc37S14A::natMX4, ubr2::kanMX6 | This study |
| NNY27 | a | BY4741, cdc37S14A::natMX4, ubr1::kanMX6, ubr2::HIS3 | This study |
| NNY28 | a | BY4741, pHis-Ste11ΔN K444R (URA3) | This study |
| NNY29 | a | BY4741, cdc37S14A::natMX4, pHis-Ste11ΔN K444R (URA3) | This study |
| NNY30 | a | BY4741, cdc37S14A::natMX4, ubr1::kanMX6, pHis-Ste11ΔN K444R (URA3) | This study |
| NNY31 | a | BY4741, cdc37S14A::natMX4, ubr2::kanMX6, pHis-Ste11ΔN K444R (URA3) | This study |
| NNY32 | a | BY4741, cdc37S14A::natMX4, ubr1::kanMX6, ubr2:HIS3, pHis-Ste11ΔN K444R (URA3) | This study |
| NNY33 | a | BY4741, erg6::natMX4 pRS313 TPK2 NT-HA (HIS3) | This study |
| NNY34 | a | BY4741, ubr1::kanMX6, erg6::natMX4 pRS313 TPK2 NT-HA (HIS3) | This study |
| NNY35 | a | BY4741, rad6::kanMX6, erg6::natMX4 pRS313 TPK2 NT-HA (HIS3) | This study |
| NNY36 | a | BY4741, ubc4::kanMX6, erg6::natMX4 pRS313 TPK2 NT-HA (HIS3) | This study |
| NNY37 | a | BY4741, ubc5::kanMX6, erg6::natMX4 pRS313 TPK2 NT-HA (HIS3) | This study |
| NNY38 | a | BY4741, ate1::kanMX6, erg6::natMX4 pRS313 TPK2 NT-HA (HIS3) | This study |
| nta1Δ | a | BY4741, nta1::kanMX6 | Open Biosystems |
| NNY39 | a | BY4741 cdc37S14A::natMX4, nta1::kanMX6 | This study |
| ate1Δ | a | BY4741, ate1::kanMX6 | Open Biosystems |
| NNY40 | a | BY4741 cdc37S14A::natMX4, ate1::kanMX6 | This study |
| rad6Δ | a | BY4741, rad6::kanMX6 | Open Biosystems |
| NNY41 | a | BY4741, cdc37S14A::natMX4, rad6::kanMX6 | This study |
| ubc4Δ | a | BY4741, ubc4::kanMX6 | Open Biosystems |
| NNY42 | a | BY4741, cdc37S14A ::natMX4, ubc4::kanMX6 | This study |
| ubc5Δ | a | BY4741, ubc5::kanMX6 | Open Biosystems |
| NNY43 | a | BY4741, cdc37S14A::natMX4, ubc5::kanMX6 | This study |
| NNY44 | a | BY4741, pRS425 CUP1.mycUB (LEU2) | This study |
| NNY45 | a | BY4741, ubr1::kanMX6, pRS425 CUP1.mycUB (LEU2) | This study |
| NNY46 | a | BY4741, ubr2::kanMX6, pRS425 CUP1.mycUB (LEU2) | This study |
| NNY47 | a | BY4741, ubr1::kanMX6, ubr2::HIS3, pRS425 CUP1.mycUB (LEU2) | This study |
| NNY48 | α | JN516, pRS425 CUP1.mycUB (LEU2) | This study |
| NNY49 | α | ssa1-45, pRS425 CUP1.mycUB (LEU2) | This study |
| NNY50 | a | BY4741, pRS315 CPY* (LEU2) | This study |
| NNY51 | a | BY4741, ubr1::kanMX6, pRS315 CPY* (LEU2) | This study |
| NNY52 | a | BY4741, ubr2::kanMX6, pRS315 CPY* (LEU2) | This study |
| NNY53 | a | BY4741, erg6::natMX4, pHis-Ste11ΔN K444R (URA3), pRS425 CUP1.mycUB (LEU2) | This study |
| NNY54 | a | BY4741, ubr1::kanMX6, ubr2::HIS3 erg6::natMX4, pHis-Ste11ΔN K444R (URA3), pRS425 CUP1.mycUB (LEU2) | This study |
| NNY55 | a | BY4741, san1::kanMX6, erg6::natMX4, pHis-Ste11ΔN K444R (URA3) | This study |
| NNY56 | a | BY4741, erg6::natMX4, pGAL-Arg-β-Gal (URA3) | This study |
a Huntsville, AL.
Pulse-Chase Analysis and Immunoprecipitation
Cells were grown in selective media to midlog phase (A600 = 0.4–0.6), washed twice in water, and resuspended in SD-Met (0.67% yeast nitrogen base, 2% glucose, all amino acids minus methionine) at a concentration of 6 A600 units/ml. Cells were incubated at 30°C with shaking. GA (50 μM; 45 min), AZC (50 mM; 45 min), or MG132 (100 μM; 30 min) was also added at this time depending on the experiment. The cells were aliquoted for labeling, and [35S]methionine was added to 100 μCi/ml. Pulse-labeling was conducted for 10 min at 30°C with shaking. The pulse was quenched with cycloheximide (200 μg/ml) and cold methionine (1 mM) for chase reactions. Pulse and chase reactions (at various times as per individual experiments) were stopped by taking 400 μl of [35S]methionine-labeled culture and adding it to an equal volume of ice-cold 20% trichloroacetic acid until all chase samples had been processed.
Extracts were prepared by pelleting the cells and washing them twice in acetone (−20°C) before vacuum drying. The cells were resuspended in 200 μl ice-cold extract buffer (50 mM Tris-HCl, pH 7.5, 1% SDS, 1 mM EDTA, and 1× Complete protease inhibitors; Roche). Cells were added to an equal volume of glass beads (0.4 mm; Biospec Products, Bartlesville, OK) and broken in a bead beater at 4°C two times for 30 s. The extracts were quantified for 35S incorporation in a scintillation counter, and an equal number of counts was used for subsequent immunoprecipitations (IPs).
Extracts were prepared for IP by first diluting them at least 10-fold into IP dilution buffer (60 mM Tris-HCl, pH 7.5, 190 mM NaCl, 1.25% Triton-X-100, and 6 mM EDTA). Antisera was added, and the samples incubated overnight, rotating at 4°C. Immunoprecipitates were adsorbed onto protein A-Sepharose resin for 1 h and washed five times in IP dilution buffer. The samples were then dissolved in 1× SDS sample buffer and resolved by denaturing gel electrophoresis. The gels were fixed (10% acetic acid, 30% methanol) for 30 min, washed twice in water for 15 min, and incubated in 1 M sodium salicylate for 30 min before drying and exposing to x-ray film or phosphorimager screen.
Inhibition of N-end Rule Substrate-binding Sites of Ubr1 with Dipeptides
Dipeptide uptake and inhibition of Ubr1's N-end rule substrate-binding sites were conducted as described by Byrd et al. (1998) and Rao et al. (2001). Briefly, S. cerevisiae strains expressing either Tap-tagged Tpk2, Arg-ß-Gal (galactose-inducible promoter), or Tap-tagged Cup9 were grown in SHM medium lacking histidine (2% glucose, 0.1% allantoin [Acros, Pittsburgh, PA], and 0.17% yeast nitrogen base without amino acids and without ammonium sulfate) to an A600 of 0.6. Cells were harvested and washed twice with sterile water and once with SHM medium lacking methionine at a concentration of 6 A600 units/ml. Then the cells were incubated for 1.5 h at 30°C in SHM lacking methionine with 10 mM H-Leu-Ala-OH (Bachem, King of Prussia, PA) and 10 mM H-Arg-Ala-OH (Bachem). Pulse-chase analysis of Tap-tagged Tpk2 and Cup9 was carried out according to the protocol described above.
Tpk2 Enzyme Assay
PepTag nonradioactive detection system was used (Promega) according to the instructions from the manufacturer.
Denaturation and Reactivation of Firefly Luciferase
Yeast strains carrying a luciferase plasmid (pRS316) under control of a galactose promoter were grown overnight at 30°C to approximately A600 4–5. Samples were diluted back then induced with 2% galactose (wt/vol) for 2 h at midlog phase of growth. All samples were treated with 0.5 mg/ml cycloheximide, incubated at 42°C for 1 h, and then returned to 30°C. At the indicated time points, cells at 0.6 A600 were collected for lysis (in lysis buffer containing 0.1% Triton-X-100 and broken in bead beater) and protein analysis via Western blot. To analyze luciferase activity a 50-μl aliquot of intact cells was mixed with 50 μl of D-luciferin (Sigma) and buffered with 100 mM potassium phosphate, pH 8.0. Each sample was vortexed for 10 s, and luminescence was collected for 10 s with a TD20/20 luminometer (Turner Designs, Sunnyvale, CA).
Purification of Flag-tagged Ubr1
Flag-Ubr1 was affinity-purified from yeast as described (Du et al., 2002) with modifications. BY4741 ubr1Δ cells expressing pYEplac181NTFlagUBR1 was grown to A600 of ∼1.5, centrifuged, and washed once in water. Cell lysis was carried out in liquid nitrogen with glass beads. The ruptured cells were dissolved in lysis buffer (10% glycerol, 0.05% Nonidet P-40, 0.2 M KCl, 50 mM HEPES, pH 7.1, and 1× protease inhibitor cocktail from Roche). The affinity purification was carried out with anti-FLAG M2 beads (Sigma). The cell lysate was incubated on a nutator with the beads for 1 h at 4°C and washed five times with lysis buffer. Flag-Ubr1 was eluted with 200 μg/ml FLAG peptide in lysis buffer without Nonidet P-40. The eluate was concentrated with a 30,000 molecular-weight cutoff Amicon centricon (Millipore, Bedford, MA). The same procedure was carried out to purify Flag-Ubr1MR1 RING mutant.
Heat Denaturation of Firefly Luciferase and In Vitro Ubiquitinylation
Recombinant firefly luciferase (Promega) was incubated at 4, 30, 37, and 42°C for 30 min. Each of the different temperature-treated luciferases was added at 1.6 μM to an ubiquitinylation reaction, which contained 230 nM affinity-purified Flag-Ubr1, 45 nM human recombinant UBE1 (Boston Biochem, Boston, MA), 900 nM human recombinant UbcH2 (Boston Biochem), 4.8 μM 32P-labeled PK-Ub, an ATP-regenerating system, 8.3 mM NaF, 1× phosphatase inhibitor cocktail 2 (Sigma), and 83 μM human recombinant ubiquitin aldehyde (BostonBiochem). The ubiquitin reaction was incubated at 30°C for 10 min before adding 32P-labeled PK-Ub. The ubiquitination was allowed to proceed for 1 h at 30°C on a neutator. The reaction was quenched with 1% SDS containing 10 mM NEM (Acros, Pittsburgh, PA) and 1× protease inhibitor cocktail (Roche). The quenched reaction was diluted to 0.1% SDS with IP dilution buffer (60 mM Tris-HCl, 190 mM NaCl, 6 mM EDTA, pH 8.0, 1.25% Triton X-100, 10 mM NEM), and 2.5 μl of 10 mg/ml goat anti-luciferase (Firefly) (Millipore) was added and incubated 3 h at 4°C on a nutator. This was followed by the addition of 20 μl of immobilized protein A/G beads (Thermo Scientific, Waltham, MA) for another hour at 4°C on a nutator. The beads were washed five times with IP dilution buffer and boiled in 2× sample buffer containing ß-mercaptoethanol. The immunoprecipitated luciferase was resolved on 4–20% SDS-PAGE (Bio-Rad, Richmond, CA) and fixed in 10% glacial acetic acid and 40% methanol for 15 min. The fixed gel was washed with water and treated with 1 M salicylic acid for 30 min before drying and exposing to film.
Chemical Denaturation of Firefly Luciferase and In Vitro Ubiquitinylation
Chemical denaturation was performed essentially as described (Silberg et al., 1998). Luciferase, 13 μg, was denatured by diluting it 10-fold in a luciferase denaturation solution (25 mM HEPES, pH 7.5, 50 mM KCl, 5 mm MgCl2, 5 mM DTT, 6 M guanidine hydrochloride). The mixture was incubated at 25°C for 1 h. As a control, 13 μg of luciferase was treated similarly with luciferase refolding solution (25 mM HEPES, pH 7.5, 50 mM KCl, 5 mm MgCl2, 1 mM DTT, 1 mM ATP). Then the denatured luciferase and the control was diluted 100 fold in luciferase refolding solution. The denatured and diluted luciferase, 85 μl, was used for the in vitro ubiquitination. The in vitro ubiquitination was conducted as described previously with the following modifications. The ubiquitination mixture contained 120 nM guanidine hydrochloride treated or nontreated (control) luciferase, 37 nM Flag-Ubr1, 8 nM human recombinant UBE1, 280 nM human recombinant UbcH2, 4.8 μM 32P-labeled PK-Ub, an ATP-generating system, 8.3 mM NaF, 1× phosphatase inhibitor cocktail 2, and 83 μM human recombinant ubiquitin aldehyde. The ubiquitinylation reaction was incubated for 3 h at 30°C. The reaction was quenched with 1% SDS. The IP with goat anti-luciferase was conducted as described above.
Luciferase Aggregation Assay
Luciferase (100 nM) was incubated in 100 μl assay buffer (25 mM HEPES, 25 mM KCL, 5 mM MgCl2, pH 7.4, 5 mM DTT, 2 mM ATP) with or without Ubr1 (2 μM) for 10 min at 25°C and shifted to 42°C for 10 min. The samples were centrifuged at 20,000 rpm for 10 min at 4°C. The supernatant and pellet fractions were separated by 10% SDS-PAGE and then transferred to nitrocellulose membrane. Luciferase were detected by Western blot.
Flag Ubr1 Pulldown and CoIP of Luciferase
Luciferase (100 nM) was incubated in 100 μl assay buffer (25 mM HEPES, 25 mM KCL, 5 mM MgCl2, pH 7.4, 5 mM DTT, 2 mM ATP) with Ubr1 (2 μM) for 10 min at 25°C and then shifted to 42°C for 10 min. The reaction was diluted to 300 μl with assay buffer, followed by addition anti-FLAG resin (Sigma) for a 20-min incubation with rocking at 4°C. The resin was washed with assay buffer Ubr1 eluted with the FLAG peptide at 0.2 mg/ml.
Yeast carrying a galactose inducible luciferase plasmid along with either an empty vector, or a plasmid containing Flag-Ubr1 under the ADH promoter were allowed to grow to log phase at 30°C. Luciferase expression was induced with 2% galactose. After 1 h of induction, 0.5 mg/ml cycloheximide was added to samples. Half of each sample was allowed to remain at 30°C, and the remaining yeast were heat-shocked at 42°C for 30 min. Yeast cultures were pelleted, and whole cell lysate was extracted via bead breaking in a lysis buffer containing 0.1% Triton. After normalization of whole cell lysates, Flag-Ubr1 was immunoprecipitated with anti-Flag antibody. Protein samples were run on 7% SDS-PAGE gels, and Western blotting was carried out, where membranes were probed for various proteins as indicated. Inputs are 15% of total.
In Vitro Refolding and Ubiquitinylation of Firefly Luciferase
Firefly luciferase, 200 nM, was mixed with 1 μM Ssa1 and 3 μM Ydj1 in the luciferase refolding solution. The mixture was incubated at 42°C for 15 min to denature the luciferase. The refolding reaction was carried out for 3 h at the respective temperatures. Luciferase activity was measured at the end of the refolding reaction using the luciferase assay system (Cat. no. E1501) from Promega. After the refolding reaction, 30 nM Flag-Ubr1, 8 nM human recombinant UBE1, 280 nM human recombinant UbcH2, 4.8 μM 32P-labeled PK-Ub, an ATP-generating system, 8.3 mM NaF, 1× phosphatase inhibitor cocktail 2, and 83 μM human recombinant ubiquitin aldehyde were added to the mixture and incubated for 2 h at 30°C. The reaction was quenched with 1% SDS. Luciferase was immunoprecipitated with goat anti-luciferase. For the experiments shown in Figure 6F, luciferase was denatured at 42°C for 10 min with or without Ubr1 or HDM2. Subsequent ubiquitinylation assays were performed in reaction buffer (20 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 2.5 mM ATP, 2 mM DTT) containing bovine ubiquitin (10 μM), E1 (Calbiochem; 0.1 μM), and 4 μM E2 (ubc2; 4 μM or Ubc5 0.4 μM) Ubr1 (0.4 μM), or HDM2 (Boston Biochem; 0.4 μM) was also added if not added during the denaturation reaction. Reactions were incubated at 37°C for 2 h and terminated by the addition of 25 μl of SDS sample buffer to 25-μl reactions. Proteins were resolved on 10% SDS-PAGE gels, and Western blot analysis performed with anti-luciferase.
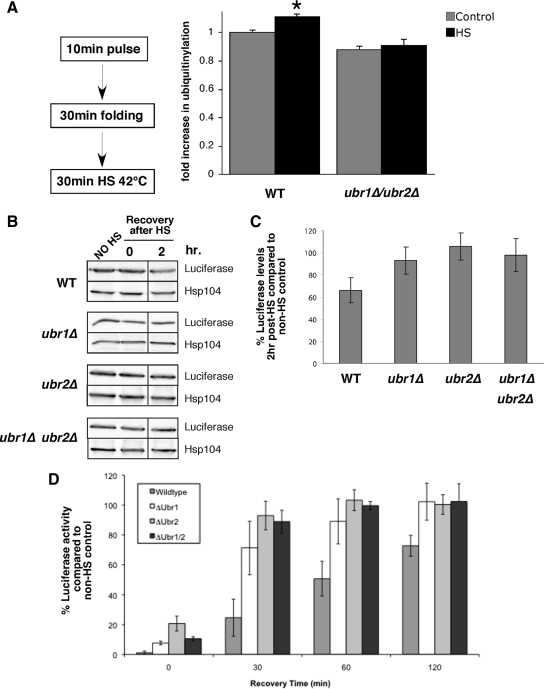
Figure 6.
Ubr1 and Ubr2 promote ubiquitinylation and degradation of mature proteins after heat stress. (A) Wild-type (WT) and ubr1Δ/ubr2Δ cells were pulse-labeled for 10 min and chased with cycloheximide and cold methionine for 30 min. Cells were then incubated at 30 (control) or 42°C (HS) for 30 min. 35S-labeled ubuiquitinylated proteins were immunoprecipitated with anti-Myc and resolved on denaturing gels. The relative amount of ubiquitinylation (fold increase after heat shock) is shown in the bar graph (n = 5; *p = 0.014). (B) Luciferase was expressed from a galactose-inducible promoter in WT, ubr1Δ, ubr2Δ, and ubr1Δ/ubr2Δ strains for 2 h. Samples were treated with 0.5 mg/ml cycloheximide, heat shocked (HS) for 60 min at 42°C, and then returned to 30°C for a recovery period. Total protein levels (B and C) of luciferase were monitored before HS, directly after heat shock, and at 2 h after heat shock. Hsp104 is shown as a loading control. (D) Luciferase activity was monitored at the same time points (as described in Materials and Methods). Activity is given as a percent of no heat shock control within each strain background. The data in C and D are plotted as means of three independent experiments. The Western blot shown in B is a representative of the averaged experiments.
Gap1 Permease Assays
Yeast strains were grown overnight in YPD media, washed with water, resuspended in minimal media with proline to an A600 of 0.5, and grown for 90 min at 30°C. The cells were harvested, washed, and resuspended in 10 mM citric acid buffer (pH 4.5) containing 2% glucose to an A600 of 2. Cell suspension, 0.5 ml, was incubated with 1 μCi [l-14C]citrulline (56.3 mCi/mmol) for 20 min at room temperature. After the reaction the cells were collected on glass microfiber filters (Whatman, Clifton, NJ) and washed three times with ice-cold water. The filters were dried, and the radioactivity on each filter was measured after addition of scintillation fluid.
RESULTS
Ubr1 and Ubr2 Function in Degradation of Newly Synthesized Unfolded Protein Kinases
A reverse genetic approach was used to identify ubiquitin ligases that inhibit degradation of newly synthesized protein kinases in yeast (S. cerevisiae) cells treated with GA, the Hsp90 inhibitor. GA is a competitive inhibitor of Hsp90's ATPase, and its action results in the rapid degradation of protein kinases and other clients by the ubiquitin/proteasome system (Whitesell and Lindquist, 2005; Caplan et al., 2007b). Yeast cells treated with GA were pulse-labeled with [35S]methionine followed by a chase period. Protein kinases were then immunoprecipitated from labeled cell extracts (the input was normalized so that the same number of counts per minute were in each sample) and resolved by denaturing gel electrophoresis, and their levels visualized by fluorography (Figure 1A). Tpk2 (a yeast homologue of cAMP-dependent protein kinase) levels were similar in untreated and GA-treated samples, although the predominant form in GA-treated cells was the immature (nonphosphorylated; slightly faster migration in a denaturing gel) compared with the mature form from solvent-treated cells. After a 1-h chase, Tpk2 levels were greatly diminished in the presence of GA, and this effect was reversed by treatment with the proteasome inhibitor, MG132 (Figure 1A). Tpk2 from solvent treated control cells was stable over the 1-h chase period. Cmk2, a protein kinase whose stability is chaperone-independent was not affected by GA treatment (Mandal et al., 2007; Supplementary Figure S1A).
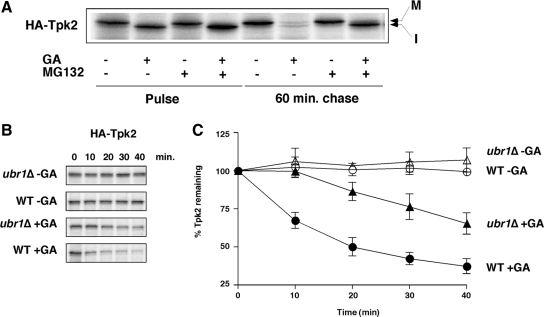
Figure 1.
Ubr1 functions in the degradation of newly synthesized protein kinases when Hsp90 is inhibited with geldanamycin. (A) Pulse-chase analysis of HA-Tpk2 in wild-type cells treated with geldanamycin (GA; 50 μM) and the proteasome inhibitor, MG132 (100 μM) or solvent alone. (B) Pulse-chase analysis of HA-tagged Tpk2 in wild-type and ubr1Δ cells treated with GA. Chase times shown in minutes. (C) Graph of HA-Tpk2 degradation (n = 3; error bars, ±SE).
Yeast cells deleted for genes encoding different ubiquitin ligases were then studied using the pulse-chase assay with Tpk2 protein kinase as a substrate. Initial studies focused on the N-end rule ubiquitin ligase, Ubr1. Although previous studies suggested that Ubr1 does not have a general function in the stress response (Bartel et al., 1990), those studies did not test directly whether this ubiquitin ligase promoted degradation of unfolded polypeptides. GA-induced degradation of Tpk2 was therefore studied in an ubr1Δ strain after pulse-chase analysis. The results of these experiments showed that UBR1 deletion led to a reduced rate of Tpk2 degradation in the presence of GA (Figure 1, B and C). Pulse-chase analysis demonstrated that the defect in degradation was restricted to the first 10 min of chase and that a wild-type rate of degradation resumed thereafter. This finding suggests that Ubr1 promotes Tpk2 degradation shortly after translation and was confirmed with studies of other protein kinases including Cdc28, Kss1, and Rim11 (Supplementary Figure S1B). Further studies with the UBR1 paralog, UBR2, revealed that it did not play the same role in GA-induced degradation of Tpk2. UBR2 deletion failed to protect Tpk2 kinase at the 10-min chase period, although the overall rate of degradation was reduced slightly in the ubr1Δ/ubr2Δ double mutant cells compared with the ubr1Δ mutant alone (Supplementary Figure S1C). It is likely that Ubr2 does a play a role in protein kinase degradation that is distinct from Ubr1, and this will be discussed in greater detail below.
We tested whether Ubr1 functioned in protein kinase degradation independently of GA treatment using a mutant of the kinase-specific chaperone, CDC37. This molecular chaperone protects the majority of newly made protein kinases from degradation. In a cdc37S14A mutant, newly made protein kinases are rapidly degraded via the ubiquitin proteasome system (UPS), primarily during pulse-labeling in a zero-point effect (Baker and Varshavsky, 1991; Suzuki and Varshavsky, 1999; Xie and Varshavsky, 2002; Mandal et al., 2007) Newly synthesized kinases such as Tpk2 appear after translation as a mixture of mature and immature forms (Mandal et al., 2007, 2008, see also Figure 1A). The immature form is quickly degraded by the proteasome (Figure 2, A and C), whereas the mature form is stable. UBR1 deletion in cdc37S14A resulted in increased Tpk2 levels after pulse-labeling, but in qualitative terms little was changed: the kinase was predominantly in the immature form after the pulse and in the mature form after a 1-h chase (cf. lanes 5 and 6 in Figure 2A). UBR2 deletion largely failed to prevent degradation of Tpk2 in the cdc37S14A strain at the zero time point, but did result in some accumulation of the immature form of the kinase after 1 h compared with the cdc37S14A strain alone (Figure 2A, cf. lanes 4 and 8). One possibility is that Ubr2 functions downstream of Ubr1 and promotes degradation of residual immature Tpk2 that was not targeted for degradation via Ubr1. Deletion of UBR2 in wild-type cells did not affect the rate of Tpk2 maturation (not shown).
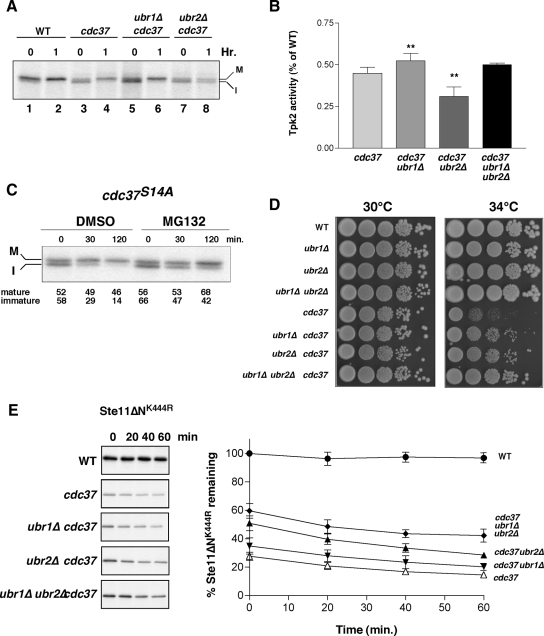
Figure 2.
Ubr1 and Ubr2 promote protein kinase degradation in a cdc37S14A mutant. (A) Pulse-chase analysis of Tap-tagged Tpk2 in wild-type (WT), cdc37S14A (cdc37) mutant cells in the presence and absence of UBR1 (ubr1Δ) and UBR2 (ubr2Δ). 0 and 1 represent times of chase after a 10-min pulse-labeling with [35S]methionine. M, mature and I, immature forms of Tap-tagged Tpk2. (B) Tpk2 activity was measured after IP of Tap-tagged Tpk2 from cell lysates. Assays performed using PepTag nonradioactive detection system. Lysates were from cdc37S14A (cdc37) with and without UBR1 and/or UBR2 as indicated. Results presented as percentage of activity in a wild-type strain. Statistical significance, **p = < 0.01; n = 8. (C) Effect of the proteasome inhibitor, MG132, on Tap-tagged Tpk2 maturation. Chase time shown in minutes. M, mature and I, immature form of the kinase. The relative amount of immature and mature forms are indicated beneath the panel. (D) Assay of yeast cell growth. Tenfold serial dilutions were plated onto YPD plates and incubated at 30 or 34°C as indicated for 3 d. (E) Pulse-chase analysis of Ste11ΔNK444R. Chase times indicated in minutes. Graph at right shows combined results from four independent experiments. Bars, ±SE.
The relative effect of deleting UBR1 or UBR2 genes on Tpk2 levels described above was correlated with changes in protein kinase activity using a peptide substrate. UBR1 deletion in cdc37S14A resulted in increased levels of Tpk2 activity by ∼ 15% over cdc37S14A alone, whereas enzyme activity was reduced in cdc37S14A/ubr2Δ mutants (Figure 2B). Deletion of both UBR1 and UBR2 led to an intermediate level of Tpk2 activity, but the apparent difference from the cdc37S14A mutant was not statistically significant. We also observed an increase in mature Tpk2 levels upon treatment with the proteasome inhibitor, MG132 (Figure 2C). In this case, proteasome inhibition led to an increase in mature Tpk2 by ∼1.5-fold compared with the untreated cdc37S14A cells. These results suggest that foldable conformers are degraded in the cdc37S14A mutant and that blocking the means of degradation can lead to some refolding.
Even though deletion of UBR1 or UBR2 resulted in different amounts of active Tpk2 kinase in a cdc37S14A background (Figure 2A), we noted that both mutants suppressed the growth arrest phenotype of cdc37S14A at a restrictive growth temperature of 34°C (Figure 2D). This effect was slightly more pronounced in cdc37S14A mutants that were deleted for both UBR1 and UBR2 suggesting an additive effect. The results described above suggested that Ubr2 had a subordinate role to Ubr1 with respect to Tpk2 degradation. However, we also tested whether the difference in their respective functions reflected substrate specificity. Studies with Rim11, Kss1, and Cdc28 suggested that Ubr2 did not play a general role in protein kinase degradation (not shown), but we did observe an effect with a recombinant catalytically inactive kinase, Ste11ΔNK444R (Flom et al., 2008). As with Tpk2, Ste11ΔNK444R levels are diminished in the cdc37S14A mutant during pulse-labeling in a zero-point effect (Figure 2E), with very little subsequent decrease in kinase levels. However, the zero-point degradation was partially suppressed by deletion of UBR1 or UBR2, albeit to different extents (and also by MG132; not shown). In this case, the effect of UBR2 deletion was more pronounced than with deletion of UBR1, thereby demonstrating substrate preference between these two E3 ligases. Western blot analysis confirmed these results and showed increased levels of Ste11ΔNK444R in cdc37S14A/ubr1Δ, cdc37S14A/ubr2Δ, and cdc37S14A/ubr1Δ/ubr2Δ mutants compared with the cdc37S14A mutant alone (not shown and Supplementary Figure S3F). These combined results show that Ubr1 and Ubr2 display substrate specificity and may also function in both zero point and slower degradation pathways.
We also analyzed whether Ubr1 and Ubr2 functioned in degradation pathways distinct from those originating from cytosolic ribosomes. To this end we investigated CPY*, a substrate for the ER-associated degradation pathway (ERAD; Hiller et al., 1996). However, neither Ubr1 nor Ubr2 function in ERAD because CPY* degradation was unaffected by deletion of either gene (Supplementary Figure. S1D). These findings also show that general attributes of the UPS are unaffected by deletion of UBR1 or UBR2.
Ubr1 functions via recognition of N-degrons that are revealed after removal of the N-terminal methionine, as well as internal degrons (Bartel et al., 1990; Mogk et al., 2007). We therefore analyzed how deletion of other N-end rule pathway components affected degradation of an unfolded Tpk2 synthesized in the presence of GA. Genes encoding the ubiquitin-conjugating enzyme (E2) RAD6, arginyl-tRNA protein transferase (ATE1) as well as two other E2 enzymes known to be important for unfolded protein degradation, UBC4 and UBC5 were studied. Tpk2 levels were monitored after pulse-labeling and after a 10-min and a 1-h chase in untreated cells and in cells treated with 50 μM GA. The pulse and chase reactions are aligned separately by strain in Figure 3, A and B, for direct comparison of the effects of each gene deletion on Tpk2 kinase levels. The data were quantified and presented in bar graph form in Figure 3, A and B. After a 10-min chase, there was a statistically significant rise in Tpk2 levels in strains deleted for UBR1 (by 19%) and RAD6 (by 10%) compared with the wild type (Figure 3A). After 1 h of chase there was a small but statistically significant rise in Tpk2 levels in the ubc4Δ strain (by 5%), in addition to ubr1Δ (by 17%) and rad6Δ (by 20%) strains compared with the wild type (Figure 3B). Previous studies have shown that the E2 Rad6 binds directly to Ubr1, whereas other studies suggest a functional association between Ubc4 and Ubr1 (Byrd et al., 1998; Xie and Varshavsky, 1999). These findings provide supporting evidence for a direct role of Ubr1 in the degradation of unfolded protein kinases, because its cognate E2, Rad6, is also involved. Ate1, by contrast, which acts in the N-end rule pathway does not appear to play a role in protein kinase degradation.
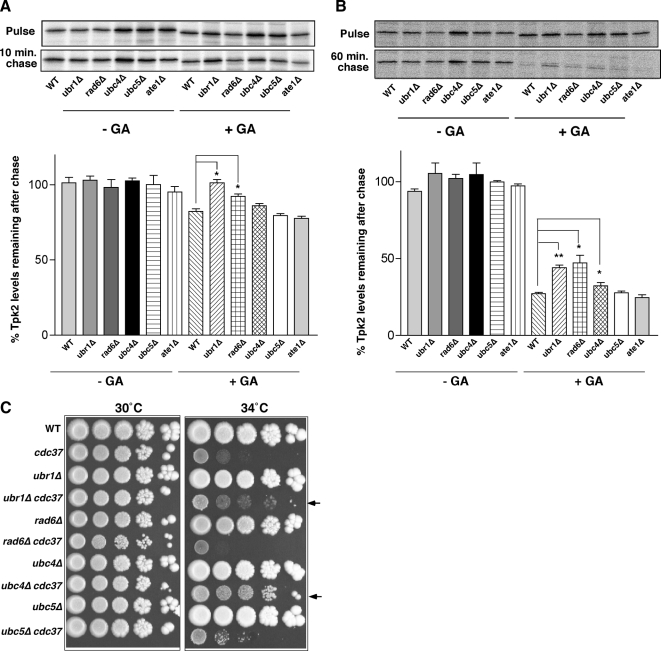
Figure 3.
Ubr1 functions in protein kinase degradation in association with Rad6. (A) Pulse and 10-min chase analysis of HA-Tpk2 in wild-type (WT) and mutant yeast strains as indicated in the absence and presence of 50 μM GA. Quantitation of band intensity shown in bar graph below panel. (B) As in A., except that chase period was 1 h. Data are from three independent experiments (*p < 0.05; **p < 0.01). (C) Growth of strains in 10-fold serial dilution at 30 and 34°C on YPD plates as indicated.
Deletion of UBR1 in the cdc37S14A mutant led to improved cell growth at 34°C, as shown in Figure 2. We anticipated that double mutants between cdc37S14A with rad6Δ and ubc4Δ would exhibit a similar phenotype because there were increased kinase levels in these strains due to defective degradation. This was tested by growth analysis on solid media (Figure 3C), and our prediction was borne out in the case of cdc37S14A/ubc4Δ cells (Figure 3C, arrowheads). In this case the growth suppression is more robust than for ubr1Δ/cdc37S14A cells. By contrast, double mutants of rad6Δ and cdc37S14A did not have better growth at 34°C compared with cdc37S14A alone. This may be due to the pleitotropic roles of Rad6 in genome integrity (Lee and Myung, 2008). Interestingly, the cdc37S14A mutant has synthetic growth defects with several other genes involved in genome integrity (Caplan et al., 2007a). Therefore, it is possible that any protein kinase refolding due to deletion of RAD6 in the cdc37S14A mutant is masked by other defects affecting cell growth. A synthetic growth defect was also observed in cdc37S14A/ate1Δ cells and cdc37S14A/nta1Δ cells (Supplementary Figure S2A).
Further studies investigated more directly whether the N-end rule pathway itself influenced Ubr1 function in protein kinase degradation. Previous studies demonstrated that dipeptides Arg-Ala and Leu-Ala inhibited N-end rule substrate degradation by binding to type I and type II sites, respectively. They also relieved autoinhibition of a third Ubr1 substrate-binding site for Cup9, a transcriptional repressor (Baker and Varshavsky, 1991; Du et al., 2002). Administration of both dipeptides stimulates Cup9 degradation in wild-type cells as shown in Figure 4A, as expected. However, these dipeptide inhibitors of the N-end rule sites had no effect on GA-dependent Tpk2 degradation, providing further support that Ubr1 functions independently of the N-end rule pathway. A similar conclusion can be drawn from a direct analysis of an N-end rule substrate, in this case, Arg-ß-Gal (Xie and Varshavsky, 1999; Arg-ß-Gal; Figure 4B). Arg-Ala and Leu-Ala dipeptides led to increased levels of Arg-ß-Gal at time zero, but had very little effect on Tpk2 degradation. Further evidence for N-end rule–independent degradation of unfolded Tpk2 came from studying this kinase in N- and C-terminal–tagged forms in the presence of GA. In each case deletion of UBR1 had a similar effect on suppressing the degradation (Figure 1, three used N-terminal HA-tagged Tpk2, and similar findings were made using C-terminally tagged Tpk2-TAP; Supplementary Figure S2B).
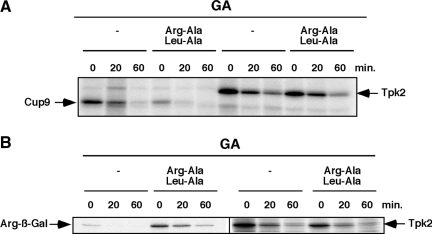
Figure 4.
Ubr1 functions in protein kinase degradation independently of the N-end rule. (A) Analysis of Cup9 and Tpk2-TAP degradation in the absence and presence of dipeptides Arg-Ala and Leu-Ala (10 mM each). Pulse-chase analysis shown at the indicated times in the presence of 50 μM GA. (B) As in A, except that the N-end rule substrate, Arg-ß-Gal was analyzed in comparison with HA-Tpk2 in the presence of GA (50 μM) and Arg-Ala/Leu-Ala dipeptides.
Ubr1 and Ubr2 Act Redundantly to Promote Ubiquitinylation of Unfolded Polypeptides
To test whether Ubr1 and Ubr2 function more broadly in degradation of newly synthesized proteins, we induced a stress response with AZC (Trotter et al., 2001, 2002). AZC is a proline analogue that incorporates competitively with l-proline; this reduces polypeptide thermal stability (Zagari et al., 1994) and results in increased levels of newly synthesized polypeptides binding to Hsp70 (Beckmann et al., 1990). Our approach was to pulse-label cells expressing myc-tagged ubiquitin with 35S-met in the absence and presence of 50 mM AZC. This was followed by IP of ubiquitinylated polypeptides with anti-myc. Newly synthesized polypeptides that became ubiquitinylated during or shortly after translation were visualized after denaturing gel electrophoresis and phosphorimaging (Figure 5). Wild-type cells treated with AZC under pulse-labeling conditions have increased amounts of polyubiquitinylated proteins by about twofold. The level of ubiquitinylation induced by AZC was decreased only slightly by deletion of UBR1 or UBR2 alone. By contrast, strains deleted for both genes were largely unable to respond to AZC, and the level of polyubiquitinylation in these cells was almost unchanged. Cells treated with AZC had similar viability (Supplementary Figure S3A). Uptake of [35S]methionine was similar in ubr1Δ/ubr2Δ compared with the wild type, although the profile of proteins synthesized in the presence of AZC was slightly different (Supplementary Figure S3B). In addition, similar amounts of unconjugated myc-Ubiquitin were observed in wild-type and ubr1Δ/ubr2Δ double mutant cells (Supplementary Figure S3B), and the levels of general ubiquitinylation appeared unaffected by AZC in either cell type (Supplementary Figure S3C). Further control experiments showed that AZC could enter the ubr1Δ/ubr2Δ cells in a manner similar to the wild type, based on its ability to compete for import of 14C-labeled citrulline (Lauwers et al., 2007), which like AZC is transported via the general amino acid transporter, Gap1 (Hoshikawa et al., 2003; Supplementary Figure S3D). Furthermore, the effect of AZC was different from the proteasome inhibitor MG132 alone, where no increase in ubiquitinylated proteins were observed in wild-type cells (Supplementary Figure S3E). Further experiments tested whether ubiquitinylation of a protein kinase was affected by deletion of UBR1 and UBR2. Ste11ΔNK444R was immunoprecipitated from wild-type and ubr1Δ/ubr2Δ strains expressing myc-tagged ubiquitin treated with and without GA and MG132 (Supplementary Figure S3F). The level of Ste11ΔNK444R was higher in the ubr1Δ/ubr2Δ strain compared with the wild type and was increased further by MG132 treatment. Nevertheless, there was ∼20% less ubiquitinylation of the protein kinase in the ubr1Δ/ubr2Δ strain compared with the wild type after normalization of kinase levels.
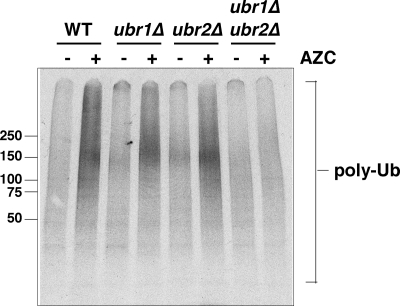
Figure 5.
Ubiquitinylation of newly synthesized polypeptides in the presence of AZC requires Ubr1 and Ubr2. Analysis of ubiquitinylation of newly synthesized polypeptides after a 10-min pulse-labeling in the absence (−) and presence (+) of 50 mM AZC. Polyubiquitinylated polypeptides (poly-Ub) were immunoprecipitated with anti-myc and resolved in a 4–20% denaturing gel before phosphorimaging. Strains used were wild-type (WT), ubr1Δ, ubr2Δ, and ubr1Δ/ubr2Δ double mutants.
Ubr1 and Ubr2 Promote Degradation of Mature Proteins That Become Denatured
Ubr1 and Ubr2 were shown above to function in quality control of newly synthesized polypeptides. We next tested whether this property was exclusive or whether Ubr1/Ubr2 function extended to previously folded proteins that become denatured. Our first approach was to adapt the assay involving IP of myc-Ub from cells pulse-labeled with 35S-Met (Figure 5). In this case, however, the pulse-labeled cells were chased with cold methionine and cycloheximide for 30 min to allow sufficient time for folding of the 35S-labeled proteins. These cells were subsequently heat stressed at 42°C for 30 min. In a recent study, heat stress was found to induce a modest increase in protein degradation, by ∼10–13% (Medicherla and Goldberg, 2008). In this case, newly made proteins remained sensitive to degradation for 30–60 min. Our results are consistent with this (Figure 6A), and the amount of 35S-labeled protein immunoprecipitated with anti-Myc was increased by 10% after heat shock compared with a non-heat-shocked control (n = 5, p = 0.014). By contrast there was no statistically significant change in ubiquitinylation of heat-shocked proteins in ubr1Δ/ubr2Δ cells. This finding suggests that Ubr1 and Ubr2 function is not restricted to newly synthesized proteins.
In another test, we monitored luciferase levels and activity after heat shock in vivo at 42°C, which is a denaturing condition. Cycloheximide was added to the cells before heat shock, and luciferase recovery was measured after 2 h. This was performed in cells deleted for UBR1 and UBR2 and the double mutant strains, and the data compared with a non-heat-shocked control for each strain. Our findings showed that luciferase levels and activity were higher in the ubr1Δ, ubr2Δ, and double mutant cells compared with the wild type after the 2-h recovery after heat shock (Figure 6, B–D). Notably, increased levels of luciferase occurred in each of the single mutants, although no additive effect was observed. These findings may suggest collaborative action between Ubr1 and Ubr2 in some circumstances.
Ubr1 Displays Specificity for a Denatured Substrate
The function of Ubr1 and Ubr2 could be direct or indirect based on our genetic studies. We therefore investigated whether Ubr1 had preference for unfolded versus native protein substrates in vitro. Luciferase was used as a test substrate because it denatures readily. FLAG-Ubr1 was affinity-purified from yeast (Supplementary Figure S4A) and added to ubiquitinylation reactions containing human Ube1 (E1), UbcH2 (human Rad6 homolog), and 32P-labeled ubiquitin (Tan et al., 1999). Reactions containing equal amounts of luciferase were incubated at 30°C followed by IP with anti-luciferase, followed by denaturing gel electrophoresis and fluorography. Studies with native luciferase revealed very little ubiquitinylation (Figure 7A). However, when luciferase was first denatured with guanidinium hydrochloride, there was a substantial increase in the amount of mono-, di-, and polyubiquitinylated species. The latter are observed as a smear above the mono- and diubiquitinylated forms of luciferase. The specificity of this reaction was determined by adding each reaction component separately. As shown in Figure 7B, a small amount of monoubiquitinylated luciferase was observed in the presence of the E1 and E2 alone. However, this was increased by addition of Ubr1 to the reaction. Furthermore, a previously described RING domain mutant of Ubr1, MR1 (C1220S; Xie and Varshavsky, 1999), had a reduced ability to polyubiquitinylate denatured luciferase in the purified system (Figure 7B). The specificity of the E2 enzyme was addressed by performing the reactions with different Ubc enzymes. We observed that only UbcH2 was functional in this assay, although a small amount of activity occurred with UbcH5 enzymes, which are the human orthologues of Ubc4 and Ubc5 (Jensen et al., 1995; Supplementary Figure S4B).
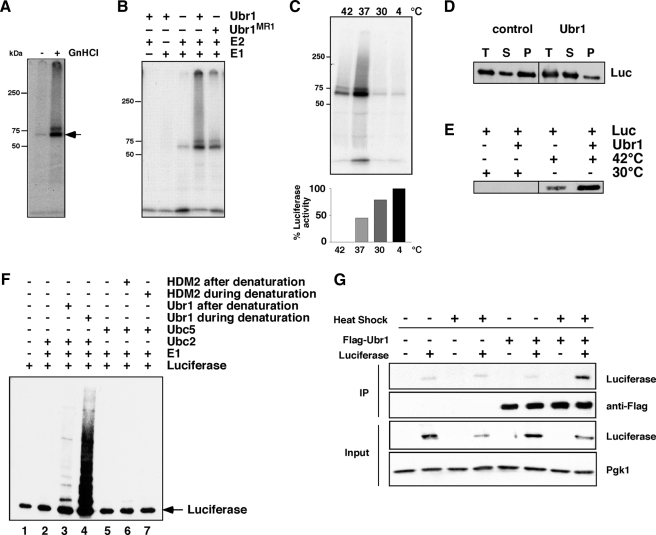
Figure 7.
Binding and ubiquitinylation of denatured luciferase by Ubr1. (A) Purified luciferase was kept native or denatured with guanidinium hydrochloride (GnHCl) before an ubiquitinylation reaction with Ubr1, Ube1, and UbcH2 and 32P-labeled ubiquitin. Reactions were immunoprecipitated with anti-luciferase and were resolved on a denaturing gel before being visualized by autoradiography. Arrow denotes monoubiquitinylated luciferase. (B) Denatured luciferase was incubated with different components of the ubiquitinylation reaction as indicated. A RING mutant of Ubr1, MR1, was also used in this experiment. (C) Purified luciferase was preincubated at the indicated temperatures for 30 min before being split into two aliquots. One was added to an ubiquitinylation reaction containing Ubr1, Ube1 (E1), UbcH2 (E2), and 32P-labeled ubiquitin (top panel). The second aliquot was used to measure remaining luciferase activity (graph in bottom panel). The bars represent luciferase activity as a percentage of the aliquot maintained at 4°C. (D) Purified luciferase was incubated with or without Ubr1 before denaturation at 42°C. The total reaction (T) was separated into supernatant (S) and pellet (P) fractions after centrifugation. The amount of luciferase in each fraction was determined by Western blot. (E) Luciferase was kept native or denatured at 42°C in the absence or presence of Ubr1. Ubr1 was immunoprecipitated, and the amount of luciferase that coimmunoprecipitated was determined by Western blot. (F) Ubiquitinylation of luciferase when Ubr1 is present during the denaturation step. Luciferase was denatured by incubation at 42°C by itself (lane 1) or in the presence of Ubr1 (lane 4) or HDM2 (lane 7). The luciferase was then added to reactions containing E1 and Ubc2 (lane 2); E1, Ubc2, and Ubr1 (lane 3); E1 and Ubc5 (lane 5); and E1, Ubc5, and HDM2 (lane 6). All reactions contained ATP and ubiquitin. Luciferase was visualized by Western blot. Arrow denotes nonubiquitinylated luciferase. (G) IP of Flag-Ubr1 and Western blot for coimmunoprecipitating luciferase before and after heat shock at 42°C for 30 min.
Further studies addressed luciferase ubiquitinylation using heat as a denaturing factor (Schroder et al., 1993). Native luciferase was preincubated at temperatures ranging from 4 to 42°C for 30 min and then split into two aliquots: one for measurement of luciferase activity and one for an ubiquitinylation reaction performed at 30°C. As shown in Figure 7C, there was a substantial incorporation of 32P-ubiquitin into luciferase preincubated at 37 and 42°C, but not at 4 or 30°C. Activity measurements of luciferase after the preincubations revealed that all luciferase was denatured after the 42°C preincubation and 55% was denatured after the 37°C incubation. Notably, the 42°C preincubation reaction was turbid, suggesting that the luciferase was largely aggregated. Turbidity was noted also in the 37°C preincubation but to a lesser extent.
The findings described above show that Ubr1 displays specificity toward denatured luciferase over the native enzyme. Further studies addressed whether it does so by direct interaction with the denatured form of luciferase. To this end, luciferase was denatured in the absence or presence of Ubr1. The reactions were centrifuged to pellet aggregates, and these were resuspended to determine the amount of luciferase in both pellet and supernatant fractions. These studies revealed that Ubr1 largely prevented luciferase aggregation as demonstrated by the reduced amount in the pellet fraction (Figure 7D). Direct binding of denatured luciferase to Ubr1 was also observed in a pulldown reaction (Figure 7E). On the basis of these observations, we propose that Ubr1 acts to promote ubiquitinylation of denatured proteins via direct chaperone-like interactions. We next determined whether Ubr1 interaction with the substrate during denaturation influenced its activity. As shown in Figure 7F, there was a dramatic increase in luciferase ubiquitinylation when Ubr1 was present during the denaturation reaction compared with when it was added afterward (cf. Figure 7F, lanes 3 and 4). Heat treatment of Ubr1 did not increase its activity (data not shown), so Ubr1 appeared to maintain denatured luciferase in a ubiquitinylation competent state during heating. Importantly, another E3 ligase, HDM2 was unable to ubiquitinylate luciferase when added during or after the denaturation (Figure 7F, lanes 6 and 7), although it had a similar capacity as Ubr1 to catalyze ubiquitin chain assembly nonspecifically (not shown). This demonstrates the specificity of Ubr1 toward unfolded proteins.
To further address whether Ubr1 has a direct function in luciferase degradation in vivo, we tested whether denatured luciferase could be specifically immunoprecipitated with Ubr1 from cell extracts. Our approach was to use heat shock as a source of denatured luciferase, and our findings were that only this form of the enzyme coimmunoprecipitated with Ubr1, whereas the native enzyme from non-heat-shocked cells could not (Figure 7G).
Hsp70 Stimulates Polyubiquitinylation by Ubr1
Previous studies demonstrated that some molecular chaperones function in both folding and degradation pathways, suggesting that decisions regarding this fate will be determined in part by them. A likely candidate in this regard is Hsp70, which interacts with many newly synthesized polypeptides. We therefore addressed whether Hsp70 was required for ubiquitinylation in reactions known to involve Ubr1 and Ubr2. To this end, we used the ssa1-45 mutant (Becker et al., 1996) that was shown previously to be defective for degradation of proteins in the cytosol and those exported from the ER (Huyer et al., 2004; McClellan et al., 2005; Han et al., 2007; Nakatsukasa et al., 2008). As shown in Figure 8A, ssa1-45 cells are also defective for ubiquitinylation of newly synthesized polypeptides induced by adding AZC to cells before pulse-labeling. This was not due to decreased protein synthesis, and general levels of ubiquitinylation were unaffected (Supplementary Figure S4C).
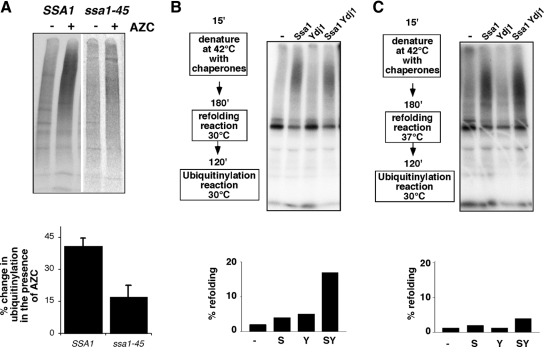
Figure 8.
Hsp70 promotes ubiquitinylation by Ubr1. (A) Ubiquitinylation of newly synthesized polypeptides in the absence (−) and presence (+) of AZC in strains expressing myc-tagged ubiquitin. IP of 35S-labeled polypeptides with anti-myc is shown. Strains used were wild type (JN516; SSA1) and ssa1-45. Bar graph below panel indicates levels of induction of ubiquitinylated newly synthesized polypeptides. Bars, SE; n = 3. (B) Ubiquitinylation reaction of heat-denatured luciferase using 32P-labeled ubiquitin. Purified Ssa1 (5:1 with luciferase) or Ydj1 (15:1 with luciferase) were added to the heat denaturation reaction, followed by a further incubation (“chaperone incubation”) and the subsequent ubiquitinylation reaction using 32P-labeled ubiquitin as indicated. In this reaction, the denaturation was followed by chaperone and ubiquitinylation reactions both at 30°C. The reaction was split into two before the ubiquitinylation reaction. One-half was used for the ubiquitinylation reaction, and the second half used for assays of luciferase activity, shown in the graph below the panel. (C) Same as B, except that the chaperone reaction was performed at 37°C.
We also analyzed whether purified Hsp70 promoted ubiquitinylation of luciferase by Ubr1 in the purified system. As shown in Figure 8B, incubation of luciferase with Hsp70 (yeast Ssa1) during denaturation but before the ubiquitinylation reaction promoted subsequent polyubiquitinylation by Ubr1. A similar finding was made with chemically denatured luciferase, although in this case the chaperone was not present during the denaturation itself, but in the subsequent incubation (not shown). This effect was not observed with Ydj1, an Hsp40 chaperone partner of Hsp70 (Cyr, 1995). Furthermore, when the incubation was performed with both chaperones even under refolding conditions (Figure 8B, left panel), we observed partitioning between both refolding and ubiquitinylation fates during the subsequent ubiquitinylation reaction. The actions of Hsp70 in promoting ubiquitinylation of luciferase were enhanced when the chaperone incubation was performed at 37°C, where refolding is not favored (Figure 8C). These findings suggest that Hsp70 is important for ubiquitinylation by Ubr1.
DISCUSSION
Quality control processes contribute to the etiology of several late-onset diseases from neurodegenerative conditions to diabetes and cancer. The best-described quality control process is ERAD, for which several components of the UPS have been shown to act in a specific manner (Vembar and Brodsky, 2008). By contrast, much less is known about quality control of polypeptides emerging from cytosolic ribosomes. Eisele and Wolf (2008) recently showed that Ubr1 functioned in the degradation of a mislocalized ER protein. While this article was in preparation, Hampton and colleagues revealed that Ubr1 and San1 ubiquitin ligases also promote degradation of unfolded cytosolic proteins (Heck et al., 2010). In this report, we describe the actions and mechanisms underlying Ubr1 and Ubr2 in the cytosol. Perhaps most importantly, we demonstrate here that Ubr1 recruits unfolded polypeptides directly (Figure 7). Similarly, the chaperone action of Hsp70 also contributes to substrate ubiquitinylation via Ubr1.
Our findings support a novel role for Ubr1 in the quality control process that is independent of the N-end rule. Ubr1 selectively binds to nonnative proteins and suppressed their aggregation. It is also active in the degradation of misfolded proteins under conditions where action of the N-end rule pathway is blocked. For example, we can inhibit N-end rule type I and type II binding sites with dipeptides, but this does not affect GA-dependent protein kinase degradation. Tpk2 kinase degradation is partially inhibited in the ubr1Δ strain independently of having epitope tags on the N- or C-terminus. Nor is there any role for Ate1 in the cytosolic quality control system (Figure 3).
The biochemical mechanisms underlying Ubr1 action are not limited to its own chaperone-like function by interacting directly with unfolded substrates. Hsp70 also plays a role in this process, and our experiments with luciferase underscore the point because the chaperone promotes Ubr1-dependent polyubiquitinylation. The most likely explanation for the role of Hsp70 is that it prevents luciferase aggregation and presents the substrate to Ubr1. However, not all chaperones have the same ability. Ydj1, for example, was unable to promote luciferase ubiquitinylation (Figure 8). This is consistent with our prior observations that Ydj1 protects newly synthesized protein kinases from degradation in vivo (Mandal et al., 2008). It is clear, however, that Ydj1's protective actions are not universal, because several earlier reports showed that it is required for protein disposal (Lee et al., 1996; Huyer et al., 2004; McClellan et al., 2005; Han et al., 2007; Nakatsukasa et al., 2008). This has been demonstrated for normally cytosolic proteins and ERAD substrates. Similar findings have been made for Hsp90 (Gusarova et al., 2001). In our case, we observed that Hsp70 could participate in both folding and degradation reactions at the same time. It seems likely, therefore, that different chaperone assemblies display substrate specificity for folding or degradation targeting functions.
In addition to the biochemical characterization of Ubr1/Ubr2 function in cytosolic quality control, our results also reveal how this process contributes to growth. This was evident in the way that deletion of UBR1 and UBR2 resulted in phenotypes that may be viewed as opposite to those observed in strains deleted for molecular chaperones. Deletion of UBR1 or UBR2 partially suppressed the growth arrest phenotype of a cdc37S14A mutant strain. One interpretation of these findings is that foldable conformers of unfolded proteins were saved from rapid degradation because their ubiquitinylation was prevented. Alternatively, it is also possible that changes in transcription due to stabilization of Cup9 and Rpn4, in ubr1Δ and ubr2Δ strains, may also play a role. Evidence for the first explanation comes from direct assessment of Tpk2 kinase activity in cdc37S14A cells that were also deleted for UBR1. In this case we observed a small but significant rise in enzyme levels and activity when degradation was inhibited (Figure 2). In addition, greater levels of luciferase activity were recovered subsequent to a denaturing heat shock in strains deleted for UBR1 or UBR2 (Figure 6). A similar finding was made for a mutant of the cystic fibrosis transmembrane regulator (CFTR) in animal cells, because blocking its ubiquitinylation led to greater levels of active CFTR (Younger et al., 2004). Furthermore, a recently characterized juxtanuclear quality control compartment, named JUNQ, accumulates misfolded proteins in a soluble form that is largely, although not completely dependent on their ubiquitinylation (Kaganovich et al., 2008).
Suppression of the cdc37S14A mutant growth arrest phenotype by UBR1 or UBR2 deletion supports the hypothesis that protein kinase degradation in this strain leads to poor growth. However, given that foldable conformers are destroyed, the pathway must be very efficient; indeed, there have been reports of rapid biphasic degradation of unstable recombinant proteins (Xie and Varshavsky, 2002) and protein kinases in yeast (Mandal et al., 2007, 2008). This suggests that there is a trade-off between growth potential and accumulation of unfolded or misfolded proteins built into cytosolic quality control. In the case of the cdc37S14A mutant, the quality control system so efficiently removes unfolded newly synthesized protein kinases that the cell has reduced growth, even though their potential for folding still exists (Figure 2). We speculate that the efficiency of the quality control system matches the potential for toxicity of unfolded or misfolded proteins. Our model, therefore, is that Hsp70 plays a central role in cytosolic quality control. Without protection from other chaperones such as Ydj1 or Cdc37 or under stress, the degradation pathway is favored by direct interaction of the polypeptide with Ubr1/2 or via Hsp70 itself. The physiological consequence, as deduced from studies with Cdc37, is reduced growth as degradation increases.
Comparison of our results with the recent studies of Heck et al. (2010) shows many similarities and some differences. The main difference is that we observed a role for Ubr2 in cytosolic quality control processes whereas they did not. Furthermore, they described how Ydj1 promotes degradation while we showed previously that Ydj1 protects newly synthesized protein kinases from that fate (Mandal et al., 2008). One clear difference between our studies and those of Heck et al. (2010) is that we studied predominantly wild-type proteins (e.g., Tpk2) whose folding pathways are blocked (with the exception of the AZC experiment shown in Figure 5), while they studied mutant proteins or proteins mislocalized from other compartments. Heck et al. (2010) also demonstrated a role for the nuclear ubiquitin ligase, San1, in quality control of cytosolic proteins. Indeed, we also observed reduced degradation of a protein kinase in san1Δ cells (Supplementary Figure S5).
In mammalian cells, rapid degradation of newly synthesized proteins has also been observed, but in a constitutive manner (Schubert et al., 2000; Qian et al., 2006). Although different interpretations on the significance of this finding have been made (Vabulas and Hartl, 2005; Yewdell and Nicchitta, 2006), it seems likely that the cytosolic quality control system is conserved. Several Ubr-related proteins are expressed in mammalian cells (Tasaki et al., 2005). Testing whether age-related decreases in mammalian quality control processes contribute to late-onset disease states is a clear direction for future studies.
The identification of Ubr1 and Ubr2 in cytosolic quality control process represents a novel function for these ubiquitin ligases. It is clear that they do not act alone, however, and that other ubiquitin ligases must function to clear misfolded polypeptides. Such is the case for ERAD where several distinct E3 ubiquitin ligases promote degradation, sometimes acting on distinct domains of substrate polypeptides (Younger et al., 2006). The identities of other ubiquitin ligases that function alongside Ubr1 and Ubr2 remain to be determined.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. Jeff Brodsky (University of Pittsburgh), Elizabeth Craig (University of Wisconsin-Madison), Jeanne Hirsch (Mount Sinai School of Medicine) and Youming Xie (Wayne State University) for strains and plasmids. We thank Dr. Zhen-Qiang Pan (Mount Sinai School of Medicine) for generous gift of purified ubiquitin and to Jaskirat Singh for valuable discussion. This work was supported by National Institutes of Health (NIH) Grants R01GM70596 and U54CA132378 (A.J.C.), NIH Grant R01GM067785 (D.M.C.), and the NIH National Center for Research Resources Grant 5G12-RR03060 (CCNY).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0098) on May 12, 2010.
REFERENCES
- Arndt V., Rogon C., Hohfeld J. To be, or not to be—molecular chaperones in protein degradation. Cell Mol. Life Sci. 2007;64:2525–2541. doi: 10.1007/s00018-007-7188-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. T., Varshavsky A. Inhibition of the N-end rule pathway in living cells. Proc. Natl. Acad. Sci. USA. 1991;88:1090–1094. doi: 10.1073/pnas.88.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch W. E., Morimoto R. I., Dillin A., Kelly J. W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Bartel B., Wunning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E. A. Functional interaction of cytosolic hsp70 and a Dnaj-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R. P., Mizzen L. E., Welch W. J. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- Byrd C., Turner G. C., Varshavsky A. The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J. 1998;17:269–277. doi: 10.1093/emboj/17.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. J., Ma'ayan A., Willis I. M. Multiple kinases and system robustness: a link between Cdc37 and genome integrity. Cell Cycle. 2007a;6:3145–3147. doi: 10.4161/cc.6.24.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. J., Mandal A. K., Theodoraki M. A. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007b;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Connell P., Ballinger C. A., Jiang J., Wu Y., Thompson L. J., Hohfeld J., Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- Cyr D. M. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Du F., Navarro-Garcia F., Xia Z., Tasaki T., Varshavsky A. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA. 2002;99:14110–14115. doi: 10.1073/pnas.172527399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele F., Wolf D. H. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Flom G. A., Lemieszek M., Fortunato E. A., Johnson J. L. Farnesylation of Ydj1 is required for in vivo interaction with Hsp90 client proteins. Mol. Biol. Cell. 2008;12:5249–5258. doi: 10.1091/mbc.E08-04-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarova V., Caplan A. J., Brodsky J. L., Fisher E. A. Apoprotein B degradation is promoted by the molecular chaperones hsp90 and hsp70. J. Biol. Chem. 2001;276:24891–24900. doi: 10.1074/jbc.M100633200. [DOI] [PubMed] [Google Scholar]
- Han S., Liu Y., Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J. Biol. Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- Heck J. W., Cheung S. K., Hampton R. Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M. M., Finger A., Schweiger M., Wolf D. H. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- Hoshikawa C., Shichiri M., Nakamori S., Takagi H. A nonconserved Ala401 in the yeast Rsp5 ubiquitin ligase is involved in degradation of Gap1 permease and stress-induced abnormal proteins. Proc. Natl. Acad. Sci. USA. 2003;100:11505–11510. doi: 10.1073/pnas.1933153100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyer G., Piluek W. F., Fansler Z., Kreft S. G., Hochstrasser M., Brodsky J. L., Michaelis S. Distinct machinery is required in Saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J. Biol. Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- Jensen J. P., Bates P. W., Yang M., Vierstra R. D., Weissman A. M. Identification of a family of closely related human ubiquitin conjugating enzymes. J. Biol. Chem. 1995;270:30408–30414. doi: 10.1074/jbc.270.51.30408. [DOI] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauwers E., Grossmann G., Andre B. Evidence for coupled biogenesis of yeast Gap1 permease and sphingolipids: essential role in transport activity and normal control by ubiquitination. Mol. Biol. Cell. 2007;18:3068–3080. doi: 10.1091/mbc.E07-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. H., Sherman M. Y., Goldberg A. L. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. Y., Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol. Cell. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- Mandal A. K., et al. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J. Cell Biol. 2007;176:319–328. doi: 10.1083/jcb.200604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A. K., Nillegoda N., Chen J. A., Caplan A. J. Ydj1 protects nascent protein kinases from degradation and controls the rate of their maturation. Mol. Cell. Biol. 2008;28:4434–4444. doi: 10.1128/MCB.00543-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A. J., Scott M. D., Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Medicherla B., Goldberg A. L. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J. Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A., Schmidt R., Bukau B. The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 2007;17:165–172. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S. B., Princiotta M. F., Bennink J. R., Yewdell J. W. Characterization of rapidly degraded polypeptides in mammalian cells reveals a novel layer of nascent protein quality control. J. Biol. Chem. 2006;281:392–400. doi: 10.1074/jbc.M509126200. [DOI] [PubMed] [Google Scholar]
- Rao H., Uhlmann F., Nasmyth K., Varshavsky A. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 2001;410:955–959. doi: 10.1038/35073627. [DOI] [PubMed] [Google Scholar]
- Schröder H., Langer T., Hartl F. U., Bukau B. Dnak, Dnaj and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U., Anton L. C., Gibbs J., Norbury C. C., Yewdell J. W., Bennink J. R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Seufert W., Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9:543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg J. J., Hoff K. G., Vickery L. E. The Hsc66-Hsc20 chaperone system in Escherichia coli: chaperone activity and interactions with the DnaK-DnaJ-grpE system. J. Bacteriol. 1998;180:6617–6624. doi: 10.1128/jb.180.24.6617-6624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Varshavsky A. Degradation signals in the lysine-asparagine sequence space. EMBO J. 1999;18:6017–6026. doi: 10.1093/emboj/18.21.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P., Fuchs S. Y., Chen A., Wu K., Gomez C., Ronai Z., Pan Z. Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol. Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Tasaki T., Mulder L. C., Iwamatsu A., Lee M. J., Davydov I. V., Varshavsky A., Muesing M., Kwon Y. T. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter E. W., Berenfeld L., Krause S. A., Petsko G. A., Gray J. V. Protein misfolding and temperature up-shift cause G1 arrest via a common mechanism dependent on heat shock factor in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2001;98:7313–7318. doi: 10.1073/pnas.121172998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter E. W., Kao C. M., Berenfeld L., Botstein D., Petsko G. A., Gray J. V. Misfolded proteins are competent to mediate a subset of the responses to heat shock in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44817–44825. doi: 10.1074/jbc.M204686200. [DOI] [PubMed] [Google Scholar]
- Vabulas R. M., Hartl F. U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science. 2005;310:1960–1963. doi: 10.1126/science.1121925. [DOI] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Mao X., Ju D., Xie Y. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J. Biol. Chem. 2004;279:55218–55223. doi: 10.1074/jbc.M410085200. [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S. L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Wickner S., Maurizi M. R., Gottesman S. Posttranslational quality control: folding, refolding, and degrading proteins. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
- Xie Y., Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Varshavsky A. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat. Cell Biol. 2002;4:1003–1007. doi: 10.1038/ncb889. [DOI] [PubMed] [Google Scholar]
- Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell J. W., Nicchitta C. V. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol. 2006;27:368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Young J. C., Agashe V. R., Siegers K., Hartl F. U. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. 2004;5:781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Younger J. M., Ren H. Y., Chen L., Fan C. Y., Fields A., Patterson C., Cyr D. M. A foldable CFTRΔF508 biogenic intermediate accumulates upon inhibition of the Hsc70-CHIP E3 ubiquitin ligase. J. Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagari A., Palmer K. A., Gibson K. D., Nemethy G., Scheraga H. A. The effect of the l-azetidine-2-carboxylic acid residue on protein conformation. IV. Local substitutions in the collagen triple helix. Biopolymers. 1994;34:51–60. doi: 10.1002/bip.360340107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.