Mechanisms that monitor the folding states of polypeptides are found throughout the cell. In the cytosol, how misfolded proteins are recognized and degraded is poorly understood. Here, misfolded proteins are shown to traffic to the nucleus where they are ubiquitinated by the San1p E3 ubiquitin ligase and degraded by the 26S proteasome.
Abstract
Intracellular quality control systems monitor protein conformational states. Irreversibly misfolded proteins are cleared through specialized degradation pathways. Their importance is underscored by numerous pathologies caused by aberrant proteins. In the cytosol, where most proteins are synthesized, quality control remains poorly understood. Stress-inducible chaperones and the 26S proteasome are known mediators but how their activities are linked is unclear. To better understand these mechanisms, a panel of model misfolded substrates was analyzed in detail. Surprisingly, their degradation occurs not in the cytosol but in the nucleus. Degradation is dependent on the E3 ubiquitin ligase San1p, known previously to direct the turnover of damaged nuclear proteins. A second E3 enzyme, Ubr1p, augments this activity but is insufficient by itself. San1p and Ubr1p are not required for nuclear import of substrates. Instead, the Hsp70 chaperone system is needed for efficient import and degradation. These data reveal a new function of the nucleus as a compartment central to the quality control of cytosolic proteins.
INTRODUCTION
The central dogma of molecular biology—DNA to RNA to protein—concisely describes the information flow of protein synthesis. Errors arising at any step can disrupt protein folding and lead to potentially toxic products. Although DNA replication is highly accurate, transcriptional and translational error rates can be as high as 10−4 and 10−3, respectively (Zaher and Green, 2009). Even with the correct protein sequence, the need for chaperones and modifying enzymes for folding makes an already complex process even more precarious. The consequences of accumulating aberrant proteins are so serious that numerous sophisticated protein quality control mechanisms (PQC) have evolved to protect cells.
Although found everywhere proteins are made, the best understood PQC mechanisms are in the endoplasmic reticulum (ER). As the site of secretory protein synthesis all the factors needed for folding reside there. Accordingly, ER quality control mechanisms have the added responsibility to control trafficking to prevent the premature exit of folding intermediates (Vembar and Brodsky, 2008). For proteins failing to fold, the integration of ER-associated degradation (ERAD) pathways removes and destroys aberrant products. ERAD includes the involvement of general folding factors like BiP and protein disulfide isomerase as well as specialized factors that recognize and target misfolded proteins to ERAD processing sites. These sites, made up of factors organized by E3 ubiquitin ligases, function to translocate and ubiquitinate substrates before they are degraded by the cytosolic 26S proteasome (Carvalho et al., 2006; Denic et al., 2006; Gauss et al., 2006).
At first glance, protein quality control in the cytosol is expected to be simpler, but it presents its own set of challenges. Because protein synthesis is not limited to a controlled compartment like the ER, folding intermediates and aberrant products could encounter a wider array of molecules and interfere with their functions. A recurring strategy to prevent inappropriate interactions is to partition aberrant proteins to discrete sites or compartments. Proteins that aggregate in the cytosol form large complexes called “aggresomes” and segregated to a perinuclear site (Johnston et al., 1998). In yeast, some aberrant proteins are localized to structures termed insoluble protein deposits (IPODs) and juxtanuclear quality controls (JUNCs) when degradation is blocked (Kaganovich et al., 2008). In the ER and cytosol, protein aggregates are segregated and delivered to lysosomes using an autophagic mechanism (Iwata et al., 2005; Kruse et al., 2006). Compartmentalization appears to be a practical tactic to deal with proteins that cannot be degraded immediately.
To be effective, quality control mechanisms must deploy a reliable means to differentiate misfolded proteins from normal proteins, folding intermediates, and presecretory proteins. Cytosolic protein quality control mechanisms, recently termed CytoQC (Metzger et al., 2008), seem to have this capability as observed for a variety of model substrates (McClellan et al., 2005; Park et al., 2007; Metzger et al., 2008). Although less understood than ER quality control, there are important similarities between the two systems. Both use major stress-regulated chaperones and the ubiquitin-proteasome system (UPS) to degrade substrates. In the ER, these include the Hsp70 homolog Kar2p/BiP and the ER DnaJ homologues Scj1p and Jem1p (Nishikawa et al., 2001; Kabani et al., 2003). For CytoQC, the Hsp70 isoforms of the Ssa family and the DnaJ homolog Ydj1 are required (Zhang et al., 2001; McClellan et al., 2005; Park et al., 2007; Metzger et al., 2008). Interestingly, about half of Ydj1p is anchored to nuclear/ER membranes with the rest in the cytosol (Caplan and Douglas, 1991). Heat shock protein 70 (Hsp70) proteins are localized in the cytosol and nucleus (Shulga et al., 1996; Chughtai et al., 2001). For some substrates, additional chaperones may be needed. Quality control of mutant von Hippel-Lindau (VHL) tumor suppressor protein in budding yeast also requires Hsp90 and the Hsp70 cochaperone Sti1p (McClellan et al., 2005).
Despite these recent advancements, the mechanisms used to sort, ubiquitinate, and degrade substrates in CytoQC remain unclear. In this study, detailed analyses of CytoQC revealed a dynamic system where nuclear compartmentalization of key functions plays a central role in substrate recognition and turnover.
MATERIALS AND METHODS
Plasmids Used in This Study
Plasmids were constructed using standard cloning protocols (Sambrook et al., 1989). All genes encoding expression constructs were sequenced in their entirety. All substrate proteins contain an engineered single hemagglutinin (HA) epitope tag at C-termini. The expression plasmids for all proteins were constructed by placing coding sequences under the strong constitutive TDH3 (encoding glyceraldehyde-3-phosphate dehydrogenase), inducible GAL1, or moderate constitutive GAS1 promoters as indicated, in yeast centromeric vectors. A plasmid list and oligonucleotide primers used in plasmid construction can be found in Tables S2 and S3, respectively.
pRP21.
pES76 encodes full-length CPY*-HA in pRS315 (Sikorski and Hieter, 1989). pRP21 encodes ΔssCPY*-HA and was constructed by deleting the N-terminal 20 residues of CPY*-HA by site-directed mutagenesis using pES76 and primer RP06.
pRP58 and pRP61.
The PEP4 proteinase A (PrA) gene was amplified from pKK247, which expresses wild-type PrA-HA, using primers RP57 and RP61 and Pfu polymerase (Stratagene, La Jolla, CA). The fragment was digested with BamHI and SmaI and inserted into pDN420 cut with BamHI and XbaI (treated with T4 DNA polymerase) generating pRP58. pRP58 contains the PrA-HA coding sequence followed by the ACT1 terminator in pRS313.pRP61 is similar to pRP58, except it contains the coding sequence for green fluorescent protein (GFP). GFP sequence was amplified from pDN291 with RP63and RP64 (HA tag was encoded in the reverse primer). The fragment was digested with BamHI and XbaI and inserted into pDN420 digested by the same enzymes.
pRP42 and pRP44.
pRP42 (ΔssPrA-HA) was constructed by deleting sequences encoding the first 22 residues of PrA-HA by site-directed mutagenesis using primer RP29 and pRP58 as the template. pRP44 (Δ2GFP-HA) was made by deleting sequences encoding amino acids 25 through 36 by site-directed mutagenesis on pRP61 using primer RP65.
pRP51 and pRP52.
pRP51 and pRP52 expresses ΔssPrA-HA and Δ2GFP-HA from GAS1 promoter, respectively. pRP51 and pRP52 were constructed by substituting THD3 promoter sequence with GAS1 promoter sequence in pRP42 and pRP44, respectively.
pSK112.
The SSA1 gene was amplified by PCR using primers SK165 and SK166 and cloned into the pYes2.1 vector (Invitrogen, Carlsbad, CA). The SK165 primer encodes the FLAG tag fused to the N-terminus of Ssa1p. FLAG-Ssa1 coding sequences were amplified by using SK232 and SK233. The resulting fragment was digested with BamHI and XbaI and was placed under the control of the TDH3 promoter in pRS316 vector to generate pSK112.
pSK145 and pSK146.
The SAN1 gene was amplified using primers SK246 and SK247. The fragment was digested with BamHI and XbaI and was placed under the control of the GAL1 promoter in pTS210 vector generating pSK145. pSK146 is similar to pSK145, except San1p contains the V5H6 tag at its C-terminus.
Strains and Antibodies
S. cerevisiae strains used in this study are described in Table S1. Anti-HA mAb (HA.11) was purchased from Covance Research Products (Princeton, NJ). Anti-Kar2p and anti-Sec61p antibodies were provided by Peter Walter (University of California, San Francisco, CA). Anti-PrA was a gift from Tom Stevens (University of Oregon, Eugene, OR). Anti-Gas1p rabbit antiserum was previously described (Spear and Ng, 2003). Polyclonal anti-ubiquitin antibody was purchased from Abcam (Cambridge, United Kingdom), monoclonal anti-3-phosphoglycerate kinase (PGK) and anti-V5 antibody from Invitrogen, polyclonal anti-GFP antibody from Clontech (Palo Alto, CA), and monoclonal anti-FLAG antibody from Sigma-Aldrich (St. Louis, MO). Secondary antibodies labeled with Alexa Fluor 488 or Alexa Fluor 596 were purchased from Molecular Probes (Eugene, OR).
Metabolic Pulse-Chase Assay
Cells were grown synthetic complete media (SC) lacking methionine, cysteine, and components for plasmid selection where applicable; 3.0 OD600 units of cells were labeled with 82.5 μCi of [35S]methionine/cysteine (EasyTag EXPRESS 35S, Perkin Elmer-Cetus, Waltham, MA) and chased with excess cold amino acids for times indicated. For galactose induction, cells were incubated for 4 h in 2% galactose media before experiments. Protein immunoprecipitation and resolution by SDS-PAGE is carried as described (Vashist et al., 2001). Gels were exposed to phophor screens for 24–48 h and scanned and quantified using the TyphoonTM phosphorimager and ImageQuant TL software (GE Healthcare Life Sciences, Uppsala, Sweden). All data plotted reflect three independent experiments with the SD of the mean indicated.
Cycloheximide-Chase Assay and Western Blotting
Cells were grown to midlog phase in synthetic media. Cessation of protein synthesis was initiated by adding cycloheximide to 200 μg/ml to begin the chase. At each time point, the chase was terminated by transferring an aliquot of cells into 1 ml ice-cold 10% trichloroacetic acid (TCA). Detergent lysates were prepared by mechanical cell disruption and TCA precipitation as described previously (Vashist et al., 2001). SDS-PAGE–separated proteins were transferred to nitrocellulose membranes and incubated in LI-COR blocking buffer (Lincoln, NE). After incubation with appropriate primary antibodies, membranes were washed with phosphate-buffered saline (PBS) containing 0.1% Tween 20. Secondary antibodies labeled with IRDye 680 (infrared dye) or IRDye 800 were diluted 15,000-times in blocking buffer containing 0.1% Tween 20 and used for detecting bound primary antibodies. After washing, membranes were scanned and quantified by using the Odyssey infrared imaging system (LI-COR Biosciences).
Substrate Ubiquitination Assay
Cells expressing misfolded proteins were resuspended in 10% TCA chilled on ice. After bead beating, precipitated proteins were pelleted by centrifugation at 14000 rpm for 10 min at 4°C. The pellet was resuspended in TCA resuspension solution (3% SDS, 100 mM Tris-base, 3 mM DTT). Protein sample, 50 μl, was mixed with 550 μl of IPS II (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, and 1% Triton X-100), 6 μl of protease inhibitor cocktail (Roche, Nutley, NJ), and 6 μl of 100 mM PMSF. Misfolded proteins were immunoprecipitated and detected by anti-HA antibody. The ubiquitinated proteins were detected by using anti-ubiquitin antibody.
Trypsin Sensitivity Assay
Cells expressing wild-type or mutant proteins were harvested and resuspended in cytosol buffer (20 mM HEPES, pH 7.4, 14% glycerol, 100 mM KOAc, and 2 mM MgOAc) and disrupted by bead beating for five 1-min full-speed cycles on a vortex mixer. For assays of PrA and ΔssPrA, Triton X-100 was added to lysates (to 1% vol/vol). After 5-min incubation at 30°C, trypsin was added at 5.0 μg/ml and incubated at 30°C. A portion was removed at each time point, and the reaction was terminated by adding TCA to 10% and proteins precipitated on ice. The recovered proteins were analyzed by SDS-PAGE/Western blotting analysis using the relevant antibodies.
Indirect Immunofluorescence Microscopy
Indirect immunofluorescence was performed as described previously (Spear and Ng, 2003) with minor modifications. Briefly, cells were fixed with 3.7% formaldehyde at 30°C for 90 min and spheroplasted by zymolyase digestion (1 mg/ml zymolyase 20T (United States Biological, Marblehead, MA), 0.1 M potassium phosphate, pH 7.5, 1.4 M sorbitol). Spherolplasts were applied to each well of a poly-l-lysine–coated slide for 10 min and washed. Slides were immersed in methanol for 6 min and in acetone for 30 s at −20°C. Each well was blocked with PBST (PBS containing 0.1% Triton X-100) containing 5% bovine serum albumin. Primary antibodies and secondary antibodies were incubated in this buffer incubated for 90 min each. Slides were washed twice with PBS buffer after each application. Primary antibodies HA.11 mAb (Covance, Princeton, NJ), anti-V5 (Invitrogen) and polyclonal anti-Kar2p were diluted to 1:200, 1:200, and 1:500, respectively. Kar2p is an ER-resident protein and an established marker for the ER and nuclear envelope (Vogel et al., 1990). Secondary antibodies Alexa Fluor 488 goat anti-mouse and Alexa Fluor 596 goat anti-rabbit were diluted to 1:500. Nuclei were visualized by DAPI (4′,6′-diamidino-2-phenylindole) staining. Samples were examined by confocal microscopy using Axio Imager.M1 microscope with 100× 1.4 NA oil Plan- Aprochromat objective (Carl Zeiss MicroImaging, Oberkochen, Germany). Images were archived by LSM Image Browser (Zeiss) and Adobe Photoshop (San Jose, CA).
GFP Fluorescence Microscopy
Cells overexpressing NLS-GFP-NES or NLS-GFP-P12 were grown at room temperature and shifted to 23, 30, or 37°C for 1 h. Cells were incubated for 5 min with Hoechst 33342 (Invitrogen) before viewing. Cells were examined by confocal microscopy using Axio Imager.M1 microscope with 100 × 1.4 NA oil Plan- Aprochromat objective (Carl Zeiss MicroImaging). Images were archived by LSM Image Browser (Zeiss) and Adobe Photoshop.
Coimmunoprecipitation Assays
Cells expressing target substrates were harvested and suspended in lysis buffer (50 mM HEPES-KOH, pH 7.4, 250 mM sorbitol, 150 mM KOAc, and 5 mM MgOAc) containing 1 mM PMSF. Cells were mechanically disrupted with zirconium beads by vortexing at full speed (1 min vortex and 1 min on ice, 10 cycles). The lysate was clarified by centrifugation at 14,000 rpm for 5 min at 4°C twice. NaCl (final 0.5 M), Triton X-100 (final 0.5%), and protease inhibitor cocktail was added to the clarified lysate. IgG beads (GE Biosciences, Fairfield, CT) were added, and the mixture was gently rotated at 4°C for 2 h. Beads were washed three times with lysis buffer containing 0.5% Triton X-100. The bound proteins to beads were eluted, separated by SDS-PAGE, and detected on immunoblots.
RESULTS
The San1p-dependent Pathway Is a General Mechanism of Quality Control for Cytosolic Proteins
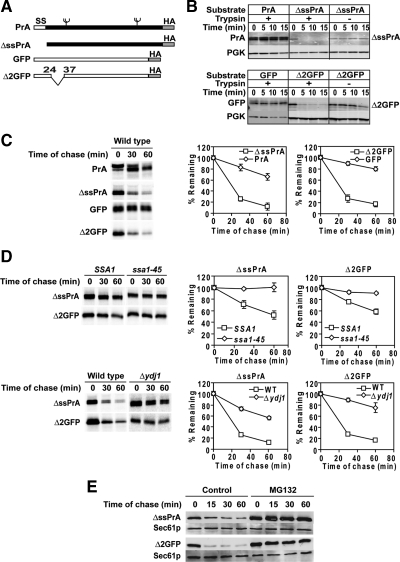
Wolf and colleagues created ΔssCPY*, a model CytoQC substrate, by deleting the CPY* signal sequence (Park et al., 2007). In our hands, ΔssCPY* degrades rapidly and is dependent on Hsp70 and Ydj1p as previously reported (Figure S1A and data not shown). We also found that a fraction of ΔssCPY* translocates into the ER, making CytoQC-specific analyses difficult (Figure S1A) This observation is consistent with the behavior of signal sequence-deleted wild-type CPY (Blachly-Dyson and Stevens, 1987). Using ΔssCPY* as a guide, two novel substrates were developed to overcome the limitation. The first was made by deleting the signal sequence of vacuolar proteinase A (PrA), to direct mislocalization to the cytosol and cause its misfolding (Figure 1A) (Klionsky et al., 1988; Park et al., 2007). Next, a short internal deletion (Figure 1A, residues 25 through 36) was engineered in the GFP to disrupt its well-ordered β-barrel structure to create Δ2GFP (Ormo et al., 1996).
Figure 1.
ΔssPrA and Δ2GFP are substrates of CytoQC. (A) Schematic representation of wild-type and mutant PrA and GFP proteins. All constructs are appended with a C-terminal HA epitope tag. ss, signal sequence. Branched symbols represent N-linked glycans. (B) Trypsin sensitivity assay. Postnuclear lysates prepared from wild-type cells expressing PrA, ΔssPrA, GFP, or Δ2GFP were incubated with 5.0 μg/ml trypsin for the times shown. Proteins were analyzed with immunoblots using monoclonal anti-HA antibody (PrA and ΔssPrA) and anti-GFP antibody (GFP and Δ2GFP). Endogenous 3-phophoglycerate kinase (PGK) was detected as an endogenous folded protein control. The GFP lysate was diluted 10-fold due to its higher steady-state level. (C) Stability of substrate proteins in vivo. Wild-type cells were pulse-labeled for 10 min and chased for the times indicated at 30°C. Immunoprecipitated proteins were resolved by SDS-PAGE and quantified using a phosphorimager. Representative phosphor screen scans are shown. Error bars, the SD of three independent experiments. (D) Turnover of ΔssPrA and Δ2GFP requires CytoQC chaperones. Pulse-chase analysis was performed in ssa1-45ts, Δydj1, and control cells expressing ΔssPrA and Δ2GFP as described in C, except stains SSA1 and ssa1-45ts were grown to log phase at 23°C and shifted to 37°C 30 min before labeling. (E) Cycloheximide decay experiment were performed in Δpdr5 cells in the absence and presence of MG132 (20 μM). Cell lysates were prepared, and proteins were resolved by SDS-PAGE. Substrates were detected by anti-HA antibody. The blot was stripped and probed with anti-Sec61p antiserum as a loading control.
An in vitro trypsinization assay was applied to assess the folding states of ΔssPrA and Δ2GFP. Unfolded proteins typically exhibit protease hypersensitivity compared with folded proteins (Taniuchi and Anfinsen, 1969). Consistent with this criterion for unfolded proteins, both molecules degraded by the first time point, whereas folded PrA and GFP were stable over the course of the experiment (Figure 1B). In vivo, a metabolic pulse-chase experiment showed that ΔssPrA and Δ2GFP are highly unstable compared with their stable, folded forms (Figure 1C). Thus far, all model CytoQC substrates require Hsp70, Hsp40, and the 26S proteasome for their degradation (Zhang et al., 2001; McClellan et al., 2005; Park et al., 2007; Metzger et al., 2008). To determine further whether ΔssPrA and Δ2GFP are representative model substrates of CytoQC, metabolic pulse-chase assays were performed in wild-type, ssa1-45ts, and Δydj1 cells. ssa1-45ts cells contain an ssa1 temperature sensitive allele with the SSA2, SSA3, and SSA4 genes deleted (Becker et al., 1996). This genetic background is necessary because SSA genes are essential only when simultaneously deleted. In addition, substrate stability was tested in the presence and absence of proteasome inhibitor MG132 in drug-sensitized Δpdr5 cells (Fleming et al., 2002; Lipford et al., 2005). As shown in Figure 1, D and E, the substrates are strongly stabilized in the mutant strains and in the presence of MG132. Together, these data show that ΔssPrA and Δ2GFP are bona fide substrates of CytoQC.
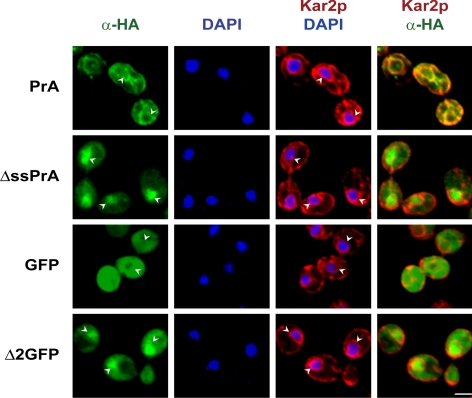
Next, the intracellular localization of the model substrates was analyzed by indirect immunofluorescence combined with confocal imaging. Surprisingly, ΔssPrA displayed strong nuclear staining in addition to the expected, but weaker, cytosolic staining (Figure 2, ΔssPrA). Δ2GFP also localized to the nucleus but in a less concentrated distribution (Figure 2, Δ2GFP). ΔssCPY* distribution is similar to ΔssPrA except that a fraction is also localized to the ER (data not shown). Nuclear localization observed for all three substrates suggested that it is a part of the CytoQC mechanism. Alternatively, it could be explained by the presence of cryptic NLSs. No classical NLS is predicted for Δ2GFP according to the PSORT II algorithm (Horton et al., 2007). ΔssPrA contains a sequence, PVRRK, that resembles the monopartite NLS motif P(B3X), where B is a basic amino acid and X is any residue. To test if the motif is functional, the conforming sequence was destroyed by mutation to PVAAA (ΔssPrA-3A). ΔssPrA-3A localizes to the nucleus, indicating that its trafficking is not dependent on the NLS-like sequence (Figure S1B). We next explored the functional significance of substrate nuclear localization.
Figure 2.
ΔssPrA and Δ2GFP are localized in the cytosol and nucleus. Cells were prepared for indirect immunofluorescence as described in Materials and Methods. Substrate proteins were detected using anti-HA antibody in the green channel. Anti-Kar2p was used to stain nuclear envelope/ER membranes and visualized in the red channel. Nuclear DNA staining is by DAPI. Images represent individual optical sections using confocal microscopy. Arrowheads indicate the position of nuclei. Scale bar, 2 μm.
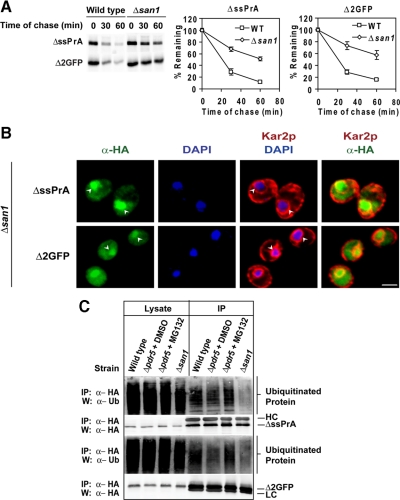
E3 ubiquitin ligases are a broad class of factors often responsible for substrate recognition (Kerscher et al., 2006). Gardner et al. (2005) demonstrated that the nuclear San1p E3 ligase is required for the degradation of damaged nuclear proteins. If misfolded cytosolic proteins specifically traffic to the nucleus, it seemed plausible that San1p also directs their turnover. To test this notion, wild-type and Δsan1 cells expressing the model substrates were analyzed by metabolic pulse-chase analysis. As shown in Figure 3A, their turnover is strongly curtailed in the Δsan1 mutant compared with wild type (similarly, ΔssPrA-3A degradation is also dependent on San1p; Figure S1C). In addition, stabilized ΔssPrA and Δ2GFP accumulated in Δsan1 nuclei, showing that substrate localization is independent of the E3 enzyme (Figure 3B). We next determined if San1p is required for substrate ubiquitination. ΔssPrA and Δ2GFP were immunoprecipitated from wild-type and Δsan1 cells, resolved by SDS-PAGE, blotted onto nitrocellulose, and probed with anti-ubiquitin antibodies. Both substrates displayed a significant reduction in polyubiquitination when expressed in the Δsan1 mutant compared with wild type (Figure 3C). These data show that San1p is required for efficient polyubiquitination of these substrates. However, residual degradation was observed in Δsan1 cells suggesting other pathways can partially compensate in its absence (see Figure 6).
Figure 3.
San1p is required for ΔssPrA and Δ2GFP ubiquitination and degradation. (A) Substrate turnover in wild type and Δsan1 was determined by pulse chase analysis as described in Figure 1C. (B) ΔssPrA and Δ2GFP stabilized in Δsan1 cells localize to the nucleus. Δsan1 cells expressing misfolded proteins were fixed, stained, and visualized as described in Figure 2. Arrowheads indicate positions of nuclei. Scale bars 2 μm. (C) Cells were treated with DMSO or MG132 (final 50 mM) for three hours before harvest. Misfolded proteins were immunoprecipitated from the cell lysates using anti-HA antibody and complexes resolved by SDS-PAGE were transferred to nitrocellulose. Ubiquitinated proteins were detected using α-ubiquitin antibody. For MG132-treated and Δsan1 cells, greater substrate stability required the load to be decreased twofold. Bands corresponding to IgG heavy chain (HC) and light chain (LC) are indicated.
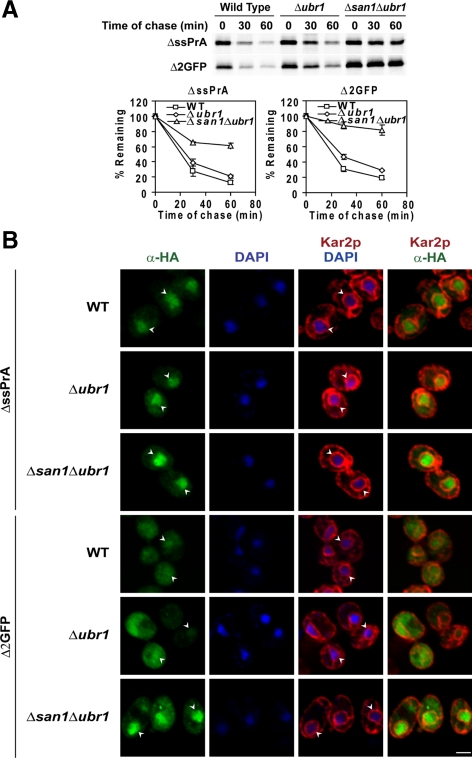
Figure 6.
Ubr1p augments the San1p system. (A) Turnover rates of ΔssPrA and Δ2GFP in wild type, Δubr1, and Δsan1Δubr1 were determined by pulse chase analysis as in Figure 1C. Error bars reflect the SD of three independent experiments. (B) Wild-type, Δubr1, and Δsan1Δubr1 cells expressing ΔssPrA and Δ2GFP were processed and stained as described in Figure 2. Identical parameters were used for image acquisition. Arrowheads indicate the positions of nuclei. Scale bar, 2 μm.
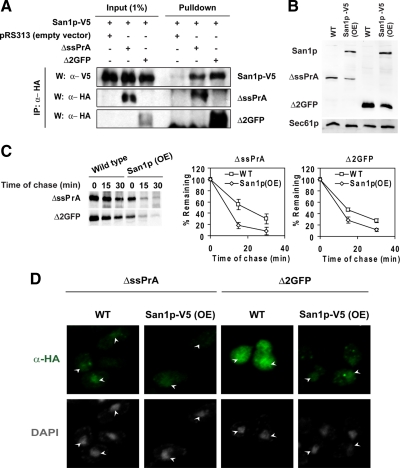
To determine if San1p interacts with substrates, coimmunoprecipitation experiments were performed. For this purpose, a functional V5-tagged San1p was introduced into MG132-sensitized cells expressing ΔssPrA, Δ2GFP, or no substrate. Cells were treated with MG132 for 4 h to stabilize substrates, and detergent extracts were prepared under nondenaturing conditions. Cell extracts were next subjected to immunoprecipitation with anti-HA antibody. Isolated proteins were resolved by SDS-PAGE and blotted onto nitrocellulose filters probed with anti-V5 and anti-HA antibodies. As shown in Figure 4A, San1p-V5 is specifically detected in complexes containing ΔssPrA and Δ2GFP (top panel).
Figure 4.
San1p overexpression enhances substrate degradation. (A) To analyze San1p:substrate interactions, San1p-V5 was coexpressed with ΔssPrA or Δ2GFP in drug sensitized cells. Cells were treated with MG132 (20 μM) to stabilize substrates. Substrate proteins were immunoprecipitated under nondenaturing conditions and protein complexes were resolved by SDS-PAGE and transferred to nitrocellulose. Membranes were probed using anti-HA antibody to detect substrates and anti-V5 antibody to detect San1p-V5. Proteins were visualized by enhanced chemiluminescence. (B) Substrate steady state levels in wild-type and San1p-V5 (OE) cells were analyzed by immunoblotting. Substrates and San1p-V5 were detected by anti-HA and anti-V5 antibody respectively. Detection of Sec61p was used as a loading control. (C) Substrate turnover rates in wild-type and San1p-overexpressing (OE) cells were determined by pulse-chase analysis as in Figure 1C. San1p expression was induced by shifting cells grown in raffinose to galactose for 4 h before analyses. Error bars, the SD of three independent experiments. (D) Cells expressing the misfolded proteins were fixed, stained, and visualized as described in Figure 2 in wild-type and San1p-V5–overexpressing cells. Identical parameters were used for image scanning between samples. Arrowheads indicate the positions of nuclei.
In this study, ΔssPrA and Δ2GFP were expressed from the strong TDH3 promoter to facilitate localization and for consistency to prior studies using similarly active promoters (Kaganovich et al., 2008). We wondered if the San1p pathway is also used for moderate substrate loads. For this, substrate coding sequences were placed under the control of the moderate GAS1 promoter (Nuoffer et al., 1991). Under its control, ΔssPrA and Δ2GFP expression levels are reduced 4.7- and 5.2-fold, respectively (Figure S2A). Pulse-chase experiments showed their rapid degradation in wild-type cells and stabilization in ssa1-45, and Δydj1 cells (Figure S2, B and C). Importantly, degradation remains highly dependent on San1p (Figure S2D). From these data, we conclude that the San1p pathway is a constitutive mechanism of cytosolic quality control.
In the above experiment, substrate turnover rates were increased marginally in cells bearing reduced substrate loads, an indication of limiting factors in CytoQC (compare Figure S2D to Figure 1C). The steady state localization of high-expression substrates in the nucleus suggested that functions such as ubiquitination or proteasomal proteolysis within the nucleus could be a bottleneck (Figure 2). During ERAD, increasing the level of the rate-limiting Hrd1 E3 enzyme elevates substrate turnover rates (Gardner et al., 2000; Garza et al., 2009). To assess whether San1p-dependent ubiquitination might be rate limiting in CytoQC, SAN1 coding sequences were placed under the control of the inducible GAL1 promoter. Wild-type cells expressing ΔssPrA and Δ2GFP were transformed with the GAL1-SAN1 plasmid or empty vector. After a shift to galactose media, substrate degradation accelerated significantly in GAL1-SAN1 cells compared with vector control (Figure 4C). To determine if overexpression affects San1p localization, GAL1-SAN1 was appended with the V5 epitope tag. Under identical assay conditions, galactose-induced San1p-V5 accelerates substrate degradation as well as San1p and San1p-V5 localizes exclusively to nuclei (Figure S3). Intracellularly, substrate localization patterns did not change significantly, other than the expected reduced signal, suggesting that nuclear import is not rate-limiting (Figures 4B and 4D).
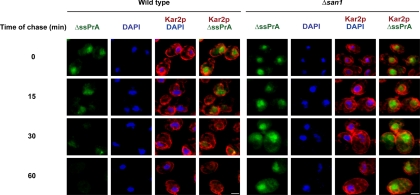
To visualize the site of decay, a cycloheximide chase assay was combined with indirect immunofluorescence. In wild-type cells, ΔssPrA localizes mostly to the nucleus before cycloheximide addition (Figure 5, wild type, 0 min). Nuclear localization is observed over the time course even as the signal diminishes, with staining virtually absent at 60 min. By contrast, the nuclear signal persists strongly throughout the time course when ubiquitination is disrupted by the Δsan1 mutation (Figure 5, Δsan1 panels). Substrate degradation was also analyzed in the xpo1–1 mutant that exhibits broad defects in nuclear export (Stade et al., 1997). XPO1/CRM1 encodes the major nuclear export receptor responsible for the trafficking of mRNA, preribosomal subunits, and proteins bearing nuclear export signals (Macara, 2001; Cullen, 2003; Zemp and Kutay, 2007). In cells deficient for XPO1/CRM1 function, CytoQC substrate degradation is as efficient as wild type, showing that this major export pathway is not required (Figure S4). However, the experiment does not rule out the existence of an XPO1/CRM1-independent pathway used for their transport. Taken together, these data show that some misfolded cytosolic proteins traffic to the nucleus where they are ubiquitinated by San1p and degraded by the 26S proteasome.
Figure 5.
Visualization of intracellular substrate decay. Logarithmically growing wild-type and Δsan1 cells expressing ΔssPrA were treated with cycloheximide (200 μg/ml final) to terminate translation. At times indicated, cell aliquots were taken, fixed, and stained for ΔssPrA and Kar2p as described in Figure 2. Image acquisition times and settings were identical for each series. Scale bars, 2 μm.
Ubr1p Augments, But Is Not Required for, ΔssPrA and Δ2GFP Degradation
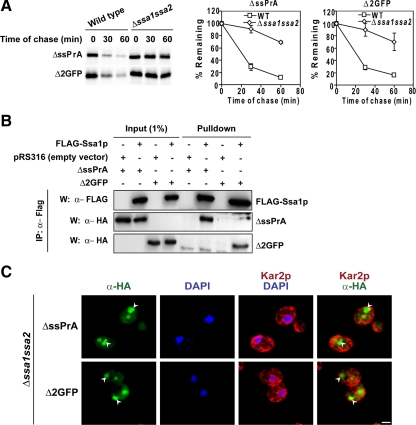
Ubr1p, the E3 ubiquitin ligase for substrates of the “N-end rule” degradation mechanism (Bartel et al., 1990), was recently reported to stabilize a CytoQC reporter protein called ΔssCL*myc (Eisele and Wolf, 2008). ΔssCL*myc is a hybrid containing the majority of ΔssCPY* fused to myc-tagged Leu2p. To assess the requirement of Ubr1p in ΔssPrA and Δ2GFP degradation, turnover assays were performed with wild-type, Δubr1, and Δubr1Δsan1 strains. By contrast to Δsan1, cells lacking UBR1 degraded these substrates nearly as well as wild type and did not alter substrate localization (Figure 6, A and B). However, deleting both genes stabilized the substrates to a greater extent than single gene knockouts, suggesting that Ubr1p can partially compensate if San1p is absent (cf. Figure 6A with Figure 3A). In addition, ΔssPrA and Δ2GFP accumulate strongly in nuclei of Δubr1Δsan1 cells (Figure 6B). Taken together, these data show that the San1p pathway is the major mechanism of quality control for ΔssPrA and Δ2GFP. Interestingly, neither E3 enzyme is required for substrate localization to the nucleus suggesting that the process is ubiquitin-independent.
The Hsp70 Chaperone System Is Required for Degradation and Nuclear Localization of ΔssPrA and Δ2GFP
The loss of cytoplasmic Hsp70 function shuts down the turnover of misfolded cytosolic proteins (Zhang et al., 2001; McClellan et al., 2005; Park et al., 2007; Metzger et al., 2008). Substrates aggregate under these conditions suggesting that Hsp70 proteins are required to maintain the solubility of substrates for degradation. To analyze the role of Hsp70 in CytoQC and various other functions, the ssa1-45ts mutant has proven to be particularly useful (Becker et al., 1996). However, due to the specific nature of the mutant (simultaneous deletion SSA2, SSA3, and SSA4), it remains unclear which Hsp70 isoforms are involved in PQC. To better understand the role Hsp70 isoforms, we analyzed the contribution of SSA genes in PQC systematically.
First, single deletion mutants were generated and tested for their ability to degrade ΔssPrA and Δ2GFP. Pulse-chase analysis shows that no single SSA gene is essential for CytoQC (Figure S5A). Next, double mutants were generated in every combination and turnover of the substrates again measured. Strikingly, only one combination displayed a clear defect. In strains deleted of SSA1 and SSA2, both substrates were dramatically stabilized at the normal growth temperature of 30°C (Figure 7A and Figure S5B). However, the extent of stabilization was less than the ssa1-45 strain at the restrictive temperature, which causes a complete loss of Hsp70 function (Figure 1D). This suggests that Ssa3p and Ssa4p (SSA3 and SSA4 genes strongly induce in a Δssa1Δssa2 strain (Werner-Washburne et al., 1987) can partially compensate, albeit poorly, in the absence of Ssa1p and Ssa2p. These data show that Ssa1p and Ssa2p are the primary Hsp70 isoforms in constitutive CytoQC and are functionally redundant.
Figure 7.
Ssa1p and Ssa2p are required for substrate degradation and nuclear localization. (A) Stability of ΔssPrA and Δ2GFP was examined in the wild type and Δssa1Δssa2 by pulse-chase analysis as described in Figure 1C. Error bars, the SD of three independent experiments. (B) Ssa1p coimmunoprecipitates with substrate proteins. ΔssPrA and Δ2GFP were immunoprecipitated from Δssa1Δssa2 cells expressing FLAG-tagged Ssa1p under nondenaturing conditions. Protein complexes were resolved by SDS-PAGE and analyzed on immunoblots. Anti-HA antibody was used to detect substrates and anti-FLAG antibody to detect Ssa1p. The bands migrating below Δ2GFP in lanes 5–7 are nonspecific. (C) Substrates accumulate as cytosolic inclusions in the absence of SSA1 and SSA2. Δssa1Δssa2 cells expressing ΔssPrA and Δ2GFP were processed and stained as described in Figure 2. Arrowheads indicate positions of cytosolically localized substrates. Scale bar, 2 μm.
To determine whether Ssa proteins bind CytoQC substrates before degradation, FLAG-tagged Ssa1p was introduced into the Δssa1Δssa2 strain. In this strain, substrate degradation was fully restored confirming that the tagged protein is functional (data not shown). In coimmunoprecipitation experiments, Ssa1-FLAG was recovered in a specific complex with ΔssPrA and Δ2GFP (Figure 7B, bottom panels).
Next, we sought to understand the roles Ssa1p and Ssa2p play in CytoQC. For this, we performed indirect immunofluorescence to visualize the fate of substrates stabilized in the Δssa1Δssa2 strain. Through this analysis, two major observations were made. The first shows substrates forming large intracellular inclusions instead of the normally diffuse patterns (Figure 7C). This is consistent with the established role of Hsp70 in keeping unfolded proteins soluble (Skowyra et al., 1990; Cyr, 1995). More notable is the distribution of substrates. More of the proteins were found in large cytosolic inclusions instead of the nucleus. Although the induction of SSA3 and SSA4 genes in this mutant is sufficient for viability (Werner-Washburne et al., 1987), it is not enough to facilitate efficient transport of the substrates into the nucleus. The defect is not due to substrate excess because they accumulate primarily in the nucleus in the Δsan1Δubr1 strain (Figures 6B). These data show that Ssa1p and Ssa2p are required for efficient transport of ΔssPrA and Δ2GFP into the nucleus for degradation.
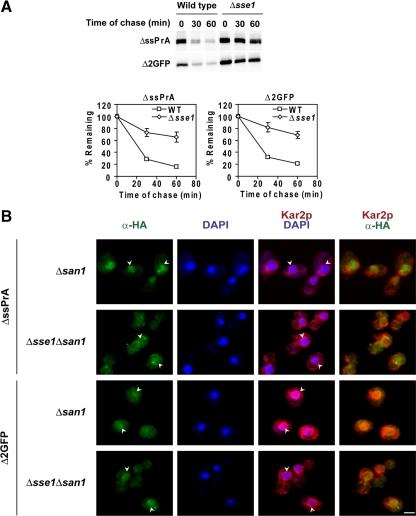
The Hsp70 nucleotide exchange factor Sse1p (Hsp110) is required for the degradation of misfolded VHL tumor suppressor in yeast cells (McClellan et al., 2005). Sse1p is a nucleotide exchange factor for Hsp70 that mediates the switch from high- to low-affinity binding. Sse1p also has the capacity to bind peptide substrates (Goeckeler et al., 2008; Polier et al., 2008). For these reasons, we wanted to learn if it is required for nuclear import and degradation of ΔssPrA and Δ2GFP. In a Δsse1 mutant, both substrates are stabilized as strongly as in Δsan1 (Figure 8A). To assess its role in substrate nuclear localization, indirect immunofluorescence was performed using Δsan1 and Δsse1Δsan1 mutants. The Δsan1 background was used to stabilize proteins able to bypass an import block, if one exists. In Δsan1 cells, substrate accumulation in nuclei was observed as usual (Figure 8B). In Δsse1Δsan1 cells, however, we observed substrate-specific effects. For ΔssPrA, the distribution becomes slightly more cytosolic suggesting that Sse1p may play a role for its efficient nuclear import. For Δ2GFP, there is no discernable difference between the strains (Figure 8B). It should be noted that nuclear import is not strongly blocked for either substrate in the absence of Sse1p and that the resulting intracellular distribution becomes similar for both substrates in the Δsse1Δsan1 strain background. Although Sse1p is required for substrate degradation, we conclude that, at best, it enhances the import of some substrates.
Figure 8.
Sse1p is required for substrate degradation. (A) Turnover rates of ΔssPrA and Δ2GFP in wild-type and Δsse1 cells were determined by pulse-chase analysis as in Figure 1C. (B) Δsan1 and Δsse1Δsan1 cells expressing substrate proteins were processed and stained as described in Figure 2. Arrowheads indicate the position of nuclei. Scale bar, 2 μm.
DISCUSSION
The accumulation of aberrant proteins in the cytosol and nucleus is associated with numerous human diseases including Parkinson's and Huntington's (Rubinsztein, 2006). Surprisingly, our understanding of cytosolic protein quality control remains rudimentary despite significant recent advances (McClellan et al., 2005; Park et al., 2007; Kaganovich et al., 2008; Metzger et al., 2008). In this study, we demonstrate a new role of the nucleus in the management of misfolded cytosolic proteins. Importantly, the nucleus is not just a site of proteolysis but also substrate recognition by the E3 ligase San1p. San1p was previously established as a key mediator in the recognition and recycling of damaged nuclear proteins (Gardner et al., 2005). Taken together, the data show that the inspection of cytosolic proteins is part of a general, broader system of intracellular protein quality control.
The nucleus accounts for over 80% of proteasomes at steady state throughout the cell cycle (Russell et al., 1999; Laporte et al., 2008). This distribution provides the simplest explanation for why misfolded proteins traffic there. However, the ability of proteasomes to redistribute according to need suggests an additional purpose for segregating ubiquitination and degradation functions in CytoQC (Laporte et al., 2008). One of the fundamental principles established for ERAD might provide a hint. In the glycan-dependent pathway, sequential trimming of specific N-linked carbohydrates provides a timing mechanism for the folding of the attached polypeptide. Should the protein fail to fold by the time the last enzyme encounters the substrate, hydrolysis of a specific residue generates a terminal α1,6-linked mannose that is the ligand of the Yos9p ERAD receptor (Quan et al., 2008; Clerc et al., 2009). By analogy, the spatial segregation of synthesis and degradation could support a kinetic partitioning mechanism to regulate CytoQC. The localization of Hsp70 in the cytosol and nucleus suggests shuttling between the compartments (Shulga et al., 1996; Chughtai et al., 2001). Therefore, it is possible that misfolded proteins enter the nucleus by “piggyback” through their chaperone association. This mode contrasts with the classical NLS-dependent pathways that transits fully folded polypeptides (Talcott and Moore, 1999). For CytoQC substrates, partial cytosolic localization at steady state suggests that their rate of nuclear import is even slower than the NLS-dependent pathways, which would allow ample time for folding. In this model, CytoQC makes use of Hsp70 proteins' ability to bind unfolded proteins so that only folding failures are delivered to the nucleus for degradation. Folded proteins, on the other hand, do not enter via this route because they no longer bind Hsp70.
In the ER, multiple quality control pathways are deployed to handle the diversity of proteins that traffic through. Each is composed of specialized factors structured around a specific E3 ubiquitin ligase to handle a defined set of client substrates (Nakatsukasa and Brodsky, 2008). Although the boundaries are less clear, a similar theme may underlie CytoQC. As mentioned earlier, Ubr1p is essential to degrade the hybrid reporter ΔssCL*myc (Bartel et al., 1990). During the preparation of this manuscript, Hampton and coworkers reported that a version of ΔssCPY* appended with folded GFP (CPY†-GFP) is only partially dependent on San1p for its degradation (Heck et al., 2010). In a genetic screen for alternative factors, a UBR1 mutation combined with the Δsan1 allele strongly stabilized CPY†-GFP. Through a dosage suppression screen, novel substrates of cytosolic quality control were discovered. Truncated versions of Fas1p, Gnd1p, and an uncharacterized cytosolic protein require both E3s for efficient turnover. Interestingly, the original truncated Gnd1p fused to GFP is mostly dependent on San1p, whereas a different truncation variant called stGnd1 is completely dependent on Ubr1p. From an independent study, Caplan and coworkers showed that the Ubr1p/Ubr2p system is required to degrade conformationally compromised protein kinases (Nillegoda et al., 2010). In vitro, Ubr1p specifically ubiquitinates unfolded luciferase, suggesting an intrinsic ability to recognized unfolded proteins.
In addition to San1p and Ubr1p/Ubr2p, evidence exists for a third CytoQC pathway. Doa10p, the ERAD E3 ubiquitin ligase that recognizes misfolded cytosolic domains of membrane proteins, is also required for the cytosolic degradation of mutant Ura3p (Ura3-2p and Ura3-3p) and a Ura3p-CL1 fusion protein (Vashist and Ng, 2004; Metzger et al., 2008; Lewis and Pelham, 2009). Although Ura3p-CL1 is not misfolded per se, the CL1 peptide degron appears to mimic a determinant recognized by CytoQC (Metzger et al., 2008). Interestingly, appending folded GFP to Ura3-2p and Ura3-3p reduces their dependence on Doa10p while making them dependent on San1p (Lewis and Pelham, 2009). Although the nature of this change is unclear, it suggests significant cross-talk between ERAD and CytoQC depending on the substrate. It should be noted that Doa10p is localized to the inner nuclear envelope, suggesting a common site of ubiquitination between the San1p and Doa10p pathways (Deng and Hochstrasser, 2006). Despite its localization being well situated to ubiquitinate ΔssPrA and Δ2GFP, these substrates are degraded independently of Doa10p (Figure S7). These data provide yet another line of evidence that San1p, Ubr1p, and Doa10p can each recognize distinct client substrates.
A recent study proposed that misfolded proteins partition between subcellular compartments termed JUNC and IPOD, depending on their physical states before they are degraded (Kaganovich et al., 2008). Triton X-100–soluble substrates localized to JUNCs, whereas detergent insoluble aggregates were found in IPODs. The redistrubution of proteasomes to JUNCs suggested that these might be sites of degradation. In our experiments, structures resembling JUNCs or IPODs were not observed under normal growth conditions (e.g., Figures 4 and 5). Initially, we assumed that the difference is substrate-specific. However, our experiments were generally performed at 30°C, whereas IPODs and JUNCs were observed primarily at 37°C, a temperature classically defined as heat shock in budding yeast (Ingolia et al., 1982; Kaganovich et al., 2008). When ΔssPrA was expressed at 37°C in wild-types cells, we observed a dramatic redistribution to cytosolic inclusions resembling JUNCs and IPODs (Figure S6). Similar structures were also observed for Δ2GFP when expressed at 37°C (data not shown). Taken together, the data support the view that the San1p and Ubr1p pathways represent constitutive mechanisms of CytoQC that manages misfolded proteins under normal conditions. Under conditions of stress, when intracellular levels of unfolded proteins can elevate, an auxiliary mechanism is activated that partitions some aberrant proteins into JUNCs and IPODs until they can be degraded.
It is becoming clear that protein quality control in the cytosol follows the general paradigm established for ERAD pathways. That is, multiple pathways and mechanisms exist to handle the diversity of substrates. However, unlike ERAD, where topological constraints makes the rationale for divergent mechanisms more transparent, the need for multiple mechanisms in CytoQC is unclear. Perhaps the answer could be found in how the E3 enzymes recognize their substrates. The three E3s are not functionally redundant. All substrates display degradation defects in at least one of the singly deleted strains. For example, stGnd1p is entirely Ubr1p-dependent, whereas ΔssPrA and Δ2GFP are almost entirely San1p-dependent (Heck et al., 2010 and Figure 6A). Are specific signals embedded in cytosolic proteins exposed upon unfolding? The CL1 degron has been proposed to be such a determinant that might emerge from a normal protein due to a frameshift event (Metzger et al., 2008). With the development of well-characterized model substrates paired to their respective pathways, the answer to this question and others will be shortly forthcoming.
Supplementary Material
ACKNOWLEDGMENTS
We thank Randy Hampton, Richard Gardner, and Avrom Caplan for communicating their unpublished data. We thank the members of the Ng Lab for discussion and comments. We are grateful to Karsten Weis, Jeff Brodsky, Reid Gilmore, Tom Stevens, and Peter Walter for strains and antibodies. This work is supported by funds from the Temasek Trust and a National University of Singapore Graduate Scholarship to R.P.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-02-0111) on May 12, 2010.
REFERENCES
- Bartel B., Wunning I., Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J., Walter W., Yan W., Craig E. A. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachly-Dyson E., Stevens T. H. Yeast carboxypeptidase Y can be translocated and glycosylated without its amino-terminal signal sequence. J. Cell Biol. 1987;104:1183–1191. doi: 10.1083/jcb.104.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan A. J., Douglas M. G. Characterization of YDJ1, a yeast homologue of the bacterial dnaJ protein. J. Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- Chughtai Z. S., Rassadi R., Matusiewicz N., Stochaj U. Starvation promotes nuclear accumulation of the hsp70 Ssa4p in yeast cells. J. Biol. Chem. 2001;276:20261–20266. doi: 10.1074/jbc.M100364200. [DOI] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., Sommer T., Aebi M. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 2009;184:159–172. doi: 10.1083/jcb.200809198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- Cyr D. M. Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]
- Deng M., Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006;443:827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Denic V., Quan E. M., Weissman J. S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Eisele F., Wolf D. H. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008;582:4143–4146. doi: 10.1016/j.febslet.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Fleming J. A., Lightcap E. S., Sadis S., Thoroddsen V., Bulawa C. E., Blackman R. K. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl. Acad. Sci. USA. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. G., Nelson Z. W., Gottschling D. E. Degradation-mediated protein quality control in the nucleus. Cell. 2005;120:803–815. doi: 10.1016/j.cell.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Gardner R. G., Swarbrick G. M., Bays N. W., Cronin S. R., Wilhovsky S., Seelig L., Kim C., Hampton R. Y. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza R. M., Sato B. K., Hampton R. Y. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. J. Biol. Chem. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R., Sommer T., Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler J. L., Petruso A. P., Aguirre J., Clement C. C., Chiosis G., Brodsky J. L. The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 2008;582:2393–2396. doi: 10.1016/j.febslet.2008.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heck J. W., Cheung S. K., Hampton R. Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA. 2010;107:1106–1111. doi: 10.1073/pnas.0910591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia T. D., Slater M. R., Craig E. A. Saccharomyces cerevisiae contains a complex multigene family related to the major heat shock-inducible gene of Drosophila. Mol. Cell. Biol. 1982;2:1388–1398. doi: 10.1128/mcb.2.11.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A., Christianson J. C., Bucci M., Ellerby L. M., Nukina N., Forno L. S., Kopito R. R. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl. Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J. A., Ward C. L., Kopito R. R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Kelley S. S., Morrow M. W., Montgomery D. L., Sivendran R., Rose M. D., Gierasch L. M., Brodsky J. L. Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol. Biol. Cell. 2003;14:3437–3448. doi: 10.1091/mbc.E02-12-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O., Felberbaum R., Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., Banta L. M., Emr S. D. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol. Cell. Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse K. B., Brodsky J. L., McCracken A. A. Autophagy: an ER protein quality control process. Autophagy. 2006;2:135–137. doi: 10.4161/auto.2.2.2388. [DOI] [PubMed] [Google Scholar]
- Laporte D., Salin B., Daignan-Fornier B., Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. J., Pelham H. R. Inefficient quality control of thermosensitive proteins on the plasma membrane. PLoS One. 2009;4:e5038. doi: 10.1371/journal.pone.0005038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford J. R., Smith G. T., Chi Y., Deshaies R. J. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Macara I. G. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan A. J., Scott M. D., Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell. 2005;121:739–748. doi: 10.1016/j.cell.2005.03.024. [DOI] [PubMed] [Google Scholar]
- Metzger M. B., Maurer M. J., Dancy B. M., Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J. Biol. Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J. L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda N. B., Theodoraki M. A., Mandal A. K., Mayo K. J., Ren H. Y., Sultana R., Wu K., Johnson J., Cyr D. M., Caplan A. J. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell. 2010 doi: 10.1091/mbc.E10-02-0098. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S. I., Fewell S. W., Kato Y., Brodsky J. L., Endo T. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 2001;153:1061–1070. doi: 10.1083/jcb.153.5.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C., Jeno P., Conzelmann A., Riezman H. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 1991;11:27–37. doi: 10.1128/mcb.11.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormo M., Cubitt A. B., Kallio K., Gross L. A., Tsien R. Y., Remington S. J. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- Park S. H., Bolender N., Eisele F., Kostova Z., Takeuchi J., Coffino P., Wolf D. H. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell. 2007;18:153–165. doi: 10.1091/mbc.E06-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier S., Dragovic Z., Hartl F. U., Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 2008;133:1068–1079. doi: 10.1016/j.cell.2008.05.022. [DOI] [PubMed] [Google Scholar]
- Quan E. M., Kamiya Y., Kamiya D., Denic V., Weibezahn J., Kato K., Weissman J. S. Defining the glycan destruction signal for endoplasmic reticulum-associated degradation. Mol. Cell. 2008;32:870–877. doi: 10.1016/j.molcel.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D. C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Russell S. J., Steger K. A., Johnston S. A. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 1999;274:21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. M., Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Shulga N., Roberts P., Gu Z., Spitz L., Tabb M. M., Nomura M., Goldfarb D. S. In vivo nuclear transport kinetics in Saccharomyces cerevisiae: a role for heat shock protein 70 during targeting and translocation. J. Cell Biol. 1996;135:329–339. doi: 10.1083/jcb.135.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra D., Georgopoulos C., Zylicz M. The E. coli dnaK gene product, the hsp70 homolog, can reactivate heat-inactivated RNA polymerase in an ATP hydrolysis-dependent manner. Cell. 1990;62:939–944. doi: 10.1016/0092-8674(90)90268-j. [DOI] [PubMed] [Google Scholar]
- Spear E. D., Ng D. T. Stress tolerance of misfolded carboxypeptidase Y requires maintenance of protein trafficking and degradative pathways. Mol. Biol. Cell. 2003;14:2756–2767. doi: 10.1091/mbc.E02-11-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K., Ford C. S., Guthrie C., Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- Talcott B., Moore M. S. Getting across the nuclear pore complex. Trends Cell Biol. 1999;9:312–318. doi: 10.1016/s0962-8924(99)01608-6. [DOI] [PubMed] [Google Scholar]
- Taniuchi H., Anfinsen C. B. An experimental approach to the study of the folding of staphylococcal nuclease. J. Biol. Chem. 1969;244:3864–3875. [PubMed] [Google Scholar]
- Vashist S., Kim W., Belden W. J., Spear E. D., Barlowe C., Ng D. T. Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 2001;155:355–368. doi: 10.1083/jcb.200106123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S., Ng D. T. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vembar S. S., Brodsky J. L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. P., Misra L. M., Rose M. D. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J. Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M., Stone D. E., Craig E. A. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher H. S., Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I., Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. doi: 10.1016/j.febslet.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Nijbroek G., Sullivan M. L., McCracken A. A., Watkins S. C., Michaelis S., Brodsky J. L. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.