This study highlights a connection between the eIF2B body and the regulation of translation initiation as a response to stress in Saccharomyces cerevisiae. Fusel alcohols are involved in signaling nitrogen scarcity to the cell and they inhibit protein synthesis by preventing the movement of the eIF2B body throughout the cell.
Abstract
Recycling of eIF2-GDP to the GTP-bound form constitutes a core essential, regulated step in eukaryotic translation. This reaction is mediated by eIF2B, a heteropentameric factor with important links to human disease. eIF2 in the GTP-bound form binds to methionyl initiator tRNA to form a ternary complex, and the levels of this ternary complex can be a critical determinant of the rate of protein synthesis. Here we show that eIF2B serves as the target for translation inhibition by various fusel alcohols in yeast. Fusel alcohols are endpoint metabolites from amino acid catabolism, which signal nitrogen scarcity. We show that the inhibition of eIF2B leads to reduced ternary complex levels and that different eIF2B subunit mutants alter fusel alcohol sensitivity. A DNA tiling array strategy was developed that overcame difficulties in the identification of these mutants where the phenotypic distinctions were too subtle for classical complementation cloning. Fusel alcohols also lead to eIF2α dephosphorylation in a Sit4p-dependent manner. In yeast, eIF2B occupies a large cytoplasmic body where guanine nucleotide exchange on eIF2 can occur and be regulated. Fusel alcohols impact on both the movement and dynamics of this 2B body. Overall, these results confirm that the guanine nucleotide exchange factor, eIF2B, is targeted by fusel alcohols. Moreover, they highlight a potential connection between the movement or integrity of the 2B body and eIF2B regulation.
INTRODUCTION
The regulation of protein synthesis allows cells to promptly modify protein levels. A key regulated step in the translation initiation pathway is mediated by the eukaryotic initiation factor (eIF) 2B (Proud, 2005). eIF2B supports a guanine nucleotide exchange reaction where GDP bound eIF2 is converted to the GTP-bound form. This form interacts with the methionyl initiator tRNA to form a “ternary complex” and hence sustains ongoing translation initiation. Inhibition of guanine nucleotide exchange on eIF2 therefore causes the global inhibition of protein production (Hinnebusch, 2005; Smirnova et al., 2005).
Probably the best characterized example of such a regulatory mechanism occurs after amino acid starvation in the yeast Saccharomyces cerevisiae (Hinnebusch, 2005). More specifically, amino acid starvation activates a protein kinase, Gcn2p, which phosphorylates the α subunit of eIF2, transforming it from a substrate of the eIF2B guanine nucleotide exchange factor to an inhibitor. Phosphorylated eIF2 interacts tightly and essentially sequesters eIF2B, causing it to become limiting (Rowlands et al., 1988). Severe starvation for amino acids therefore elicits a global inhibition of protein synthesis via the activation of an eIF2α kinase. This mechanism of translational control is highly conserved across different eukaryotic species. For example, in human cells there are four different eIF2α kinases that respond to a variety of stress conditions (Proud, 2005). Hence, eIF2B constitutes a key eukaryotic translation control point (Pavitt, 2005).
Even though global protein synthesis is inhibited after eIF2α phosphorylation and eIF2B inhibition, the GCN4 mRNA is translationally activated under such conditions via a complex mechanism involving short upstream open reading frames. The increase in Gcn4p serves to effectively reverse the starvation as this protein is a transcriptional inducer of many amino acid biosynthetic enzymes (Hinnebusch, 2005). In a mechanism analogous to that of yeast GCN4 regulation, translation of the ATF4 transcription factor is activated under stress conditions that repress global translation (Lu et al., 2004; Vattem and Wek, 2004). The induction of ATF4 serves to promote an integrated response to stress in terms of the reorganization of the gene expression program (Harding et al., 2003). Therefore this regulatory system closely parallels that identified in yeast.
In contrast to the process of eIF2α phosphorylation, much less is known about eIF2α dephosphorylation and how it is regulated. In yeast, overexpression of a fragment of the type 1 protein phosphatase, Glc7p, suggests that this protein plays a key role in eIF2α dephosphorylation (Wek et al., 1992, 2006). In higher cells, GADD34 and CReP have been identified as proteins where overexpression leads to increased eIF2α dephosphorylation, and this most likely involves the recruitment of type 1 protein phosphatase catalytic subunits (Novoa et al., 2001; Jousse et al., 2003). GADD34, in particular, forms part of a negative feedback or recovery loop where activation of eIF2α phosphorylation and hence ATF4 expression transcriptionally induces the GADD34 gene, which then correlates with the subsequent reduction in phosphorylated eIF2α levels (Novoa et al., 2001). In yeast, the type 2A–related protein phosphatase Sit4p, which functions as part of the TOR pathway (Di Como and Arndt, 1996), has a number of links to the regulation of protein synthesis. For instance, the accumulation of phosphorylated eIF2α in response to the immunosuppressant and antifungal drug rapamycin requires inhibition of Sit4p (Cherkasova and Hinnebusch, 2003; Rohde et al., 2004). In addition, overexpression of Sit4p suppresses the effects of lithium stress on translation initiation (Montero-Lomeli et al., 2002).
eIF2B represents the target of phosphorylated eIF2 and is a heteropentameric complex (α to ε subunits). The α (Gcn3p), β (Gcn7p), and δ (Gcd2p) subunits are required for regulation via phosphorylated elF2α, whereas the γ (Gcd1p) and ε (Gcd6p) subunits form a catalytic subcomplex required for guanine nucleotide exchange (Pavitt et al., 1998). The GCN3 gene, which encodes the α subunit of eIF2B, represents the only nonessential eIF2B gene. However, this gene is essential for adaptation to cellular stresses that act via eIF2α phosphorylation (Hannig and Hinnebusch, 1988). The Gcn3p-Gcd7p-Gcd2p regulatory subcomplex has been shown to mediate the high-affinity interaction that is observed between eIF2B and phosphorylated eIF2 (Krishnamoorthy et al., 2001). Furthermore, in vitro experiments have shown that down-regulation of eIF2B activity by phosphorylated eIF2α is not observed in the absence of the Gcn3p subunit (Pavitt et al., 1998).
We have recently found that both eIF2B and eIF2 reside in a single eIF2B body (2B body) within yeast cells (Campbell et al., 2005; Campbell and Ashe, 2006). Fluorescent recovery after photobleaching (FRAP) studies suggest that while eIF2B is a resident feature, eIF2 rapidly transits through the 2B body. Indeed genetic and metabolic strategies to lower the in vivo rate of the eIF2B guanine nucleotide exchange reaction concomitantly reduce the rate of eIF2 transit (Campbell et al., 2005). These results are consistent with the 2B body participating in the guanine nucleotide exchange reaction.
A variety of factors outside of eIF2α phosphorylation have been found to impact on eIF2B guanine nucleotide exchange activity. For instance, in higher cells various protein kinases, including GSK3, CK1, CK2, and DYRK, have been described to phosphorylate specific eIF2Bε residues (Woods et al., 2001; Wang and Proud, 2008). In addition, mutations in all five eIF2B subunits have been identified as linked to the human inherited disorder leukoencephalopathy with vanishing white matter (VWM) or childhood ataxia with central hypomyelination (CACH; Scali et al., 2006). Furthermore, in both yeast and higher cells volatile anesthetics appear to inhibit protein synthesis via eIF2B (Palmer et al., 2005, 2006). Finally, in yeast fusel alcohols have been shown to inhibit translation initiation via a mechanism involving eIF2B (Ashe et al., 2001).
Fusel alcohols are byproducts of amino acid catabolism in yeast, and their production is associated with nitrogen scarcity. Fusel alcohols have been shown to have an impact on a range of cellular processes and structures including the cell cycle, mitochondria, the cell wall, and membrane transport (La Valle and Wittenberg, 2001; Martinez-Anaya et al., 2003; Kern et al., 2004). Previously, we have shown that fusel alcohols also inhibit translation initiation (Ashe et al., 2001). More specifically, we showed that in different isolates of the W303-1A yeast laboratory strain, 1-butanol led to variable effects on protein synthesis as measured by polysome analysis and pulse labeling. We defined this genetic variation as either butanol resistant (BUTR) or butanol sensitive (BUTS). Using genetic mapping, we showed that the explanation for the difference in the resistance/sensitivity to butanol lay in the GCD1 gene, which codes for the γ subunit of eIF2B. Ultimately, we found that a W303-1A isolate with serine at position 180 in eIF2Bγ is BUTS, whereas the isolate with proline at this position is BUTR. These results implicate eIF2B as an important component of the translational inhibition caused by fusel alcohols. This conclusion was further supported by the fact that overexpression of all five subunits of eIF2B partially suppressed the butanol-sensitive phenotype and that butanol led to a rapid induction of GCN4 mRNA translation. We have also compared butanol regulation to amino acid starvation in terms of those mRNAs that are translationally maintained under conditions where global protein synthesis is down-regulated. This analysis showed that consistent with the different physiological impact of the two stresses, a different set of mRNAs continue to be translated for each stress despite these stresses targeting the same factor eIF2B (Smirnova et al., 2005).
In this current study, we have further analyzed the impact of fusel alcohols on protein synthesis and the eIF2B guanine nucleotide exchange factor. We investigate a variety of eIF2B mutants and identify butanol-resistant eIF2Bα mutants using DNA tiling array technology. We show that fusel alcohols lead to a Sit4p-dependent dephosphorylation of eIF2α, but that Sit4p does not play a role in the global inhibition of translation initiation. Finally, we describe the influence of fusel alcohols on the 2B body. Here the alcohols prevent the random diffusion of this body through the yeast cell cytoplasm, perhaps because the alcohols somehow stabilize a tethered form. This effect correlates well with the inhibition of protein synthesis. Therefore these studies highlight the possibility that the movement of the 2B body is somehow associated with eIF2B activity in recycling eIF2-GDP to eIF2-GTP.
MATERIALS AND METHODS
Strains Construction and Growth Conditions
Yeast strains (Table 1) were grown on standard yeast extract, peptone, glucose (YPD) media or synthetic complete media (SCD) at 30°C (Guthrie and Fink, 1991). Butanol and other alcohols were added at the specified concentrations in liquid culture for the indicated times at 30°C. Amino acid starvation was achieved via incubation in media lacking all amino acids for 10–30 min. In strains generated for localization or immunoprecipitation studies, genomic loci were tagged with enhanced green fluorescent protein (eGFP) or the Myc epitope using a PCR-based assay and plasmids (Janke et al., 2004). Tagged genomic loci were confirmed by both PCR and Western blot analysis.
Table 1.
Yeast strains used in this study
| Strain name | Genotype | Source/reference |
|---|---|---|
| yMK16 (BUT1-2) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 BUT1-2 (GCN3-R148K ) | This study |
| yMK23 (BUTR) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 | Ashe et al. (2001) |
| yMK36(BUTS) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 | Ashe et al. (2001) |
| yMK39(BUTS) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 p[HIS3 CEN] | Ashe et al. (2001) |
| yMK53 (BUT1-1) | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 BUT1-1 (GCN3-T41K) | This study |
| yMK54 (BUT1-1) | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 BUT1-1 (GCN3-T41K) | This study |
| yMK472 (BY4741) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| yMK644 (H1728) | MATa ura3-52 leu2-3,112 gcd6-1 | Bushman et al. (1993a) |
| yMK658 (H1730) | MATa ura3-52 leu2-3,112 | Bushman et al. (1993a) |
| yMK876 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 -GFP::G418 | This study |
| yMK878 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 GCD6-GFP::G418 | This study |
| yMK880 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180-GFP::G418 | This study |
| yMK882 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 GCD6-GFP::G418 | This study |
| yMK883 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 SUI2-GFP::G418 | This study |
| yMK914 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 SUI2-GFP::G418 | This study |
| yMK974 | MATa leu2-3,112 trp1-Δ63 ura3-52::HIS4-lacZ gcn2Δ gcd1::LEU2 p[GCD1 TRP1 CEN] | This study |
| yMK976 | MATa leu2-3,112 trp1-Δ63 ura3-52::HIS4-lacZ gcn2Δ gcd1::LEU2 p[GCD1-C483W TRP1 CEN] | This study |
| yMK1125 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180-MYC::TRP1 | This study |
| yMK1126 | MATa his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180-MYC::TRP1 | This study |
| yMK1211 | MATα his3–11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 GCD11-GFP::G418 | This study |
| yMK1212 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 GCD11-GFP::G418 | This study |
| yMK1235 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rts1::G418 | EUROSCARF |
| yMK1236 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tpd3::G418 | EUROSCARF |
| yMK1237 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pph3::G418 | EUROSCARF |
| yMK1238 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ptc3::G418 | EUROSCARF |
| yMK1239 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cnb1::G418 | EUROSCARF |
| yMK1240 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cmp2::G418 | EUROSCARF |
| yMK1241 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ppz1::G418 | EUROSCARF |
| yMK1242 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ppz2::G418 | EUROSCARF |
| yMK1243 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pph22::G418 | EUROSCARF |
| yMK1244 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ppq1::G418 | EUROSCARF |
| yMK1245 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ptc2::G418 | EUROSCARF |
| yMK1246 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 sit4::G418 | EUROSCARF |
| yMK1247 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ptc4::G418 | EUROSCARF |
| yMK1248 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 cna1::G418 | EUROSCARF |
| yMK1249 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pph21::G418 | EUROSCARF |
| yMK1262 | MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 gcn3::URA3 | This study |
| yMK1347 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 GCD7-GFP::G418 | This study |
| yMK1355 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 GCN3-GFP::G418 | This study |
| yMK1356 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 GCN3-GFP::G418 | This study |
| yMK1363 | MATα his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 GCN7-GFP::G418 | This study |
| yMK1439 | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 gcn3::TRP1 | This study |
| yMK1441 | MATα ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 gcn3::TRP1 p[GCN3-R148K URA3 CEN] | This study |
| yMK1442 | MATα ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 gcn3::TRP1 p[GCN3 URA3 CEN] | This study |
| yMK1451 | MATα ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 gcn3::TRP1 p[GCN3-T41K URA3 CEN] | This study |
| yMK1465 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 BUT1-2 (GCN3-R148K) CHRXI 493505::URA3 | This study |
| yMK1475 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 CHRXI 493505::URA3 | This study |
| yMK1567 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 BUT1-1 (GCN3-T41K ) GCD6-GFP::G418 | This study |
| yMK1595 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 BUT1-2 (GCN3-R148K) GCD6-GFP::G418 | This study |
| yMK1596 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 GCD6-GFP::G418 | This study |
| yMK1597 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 GCD6-GFP::G418 | This study |
| yMK1613 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-S180 p[SUI3-Flag TRP1 CEN] | This study |
| yMK1615 | MATa ade2α1 his3α11,15 leu2-3,112 trp1-1 ura3-1 GCD1-P180 p[SUI3-Flag TRP1 CEN] | This study |
Plasmid Construction
The GCD1-C483W, GCN3-T41K, and GCN3-R148K plasmids were constructed using the QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA) on the pMK27 (pGCD1 TRP1 2μ; Ashe et al., 2001) and p1999 (pGCN3 LEU2 2μ; Yang and Hinnebusch, 1996) parent plasmids. Plasmid YAB502 [pSUI3-Flag (eIF2β-Flag) TRP1 CEN] was a gift from K. Asano (Kansas State University; Singh et al., 2007).
Analysis of Ribosome Distribution on Sucrose Gradients
Yeast cultures were grown to an OD600 of 0.7 and treated by the addition of butanol, as described above. Extracts were prepared in 100 μg/ml cycloheximide, and the extracts were subsequently layered onto 15–50% sucrose gradients. The gradients were sedimented via centrifugation at 40,000 rpm using a SW41 Beckman rotor for 2.5 h (Fullerton, CA), and the A254 was measured continuously to give the traces shown (Ashe et al., 2000).
Tiling Arrays
The protocol was carried out as described previously (Gresham et al., 2006) with some minor modifications. Tiling arrays in the forward direction were used, and cobalt chloride was omitted from the DNase digestion step. To obtain reliable results using SNPscanner (http://genomics-pubs.princeton.edu/SNPscanner/), new hybridizations of the control strain (FY3) and the polymorphic training strain (RM11-1a) were carried out. In total six FY3 arrays (five technical replicates from the same DNase I digestion and one biological replicate from a separate DNase I digestion) and three arrays of RM11a (from the same DNase I digestion) were used to train the SNPscanner software. After retraining, virtually identical results were obtained to previous studies from a control hybridization of FY3 at a prediction signal of >5 (Gresham et al., 2006). For hybridizations of yMK16 and yMK36 a prediction signal of >10 was set to ensure only the highest quality SNPs (single nucleotide polymorphisms) were called. To detect SNPs that were present in yMK16, but not YMK36, SNPs present in both YMK36 and YMK16 were removed using the compare_strains perl script included with the SNPscanner program.
Western Blot Analysis of eIF2α and Phosphoserine 51 eIF2α
Yeast strains were grown to an OD600 of 0.7 in SCD and divided into 25-ml aliquots. The cells were pelleted, and the media or alcohol treatments indicated in the figure legends were carried out. All cells were lysed, and protein samples were prepared, electrophoretically separated, and subjected to immunoblot analysis as described previously (Ashe et al., 2000). The phosphospecific eIF2α (Invitrogen, Carlsbad, CA) and eIF2α (Dr. T. Dever, NIH, Bethesda. MD) antibodies were used for the detection.
Coimmunoprecipitation of eIF2B Complex
Strains were grown in YPD to an OD600 of 1.0, and 100-ml cultures were either treated with 1% butanol for 10 min at 30°C or left untreated and rapidly pelleted in a clinical centrifuge. Pellets were flash-frozen, and protein extracts were prepared by grinding under liquid nitrogen. Cell powders were resuspended in 250 ml buffer A (30 mM HEPES.KOH, pH 7.5, 100 mM potassium acetate, 2 mM magnesium acetate, 1 mM PMSF), and unlysed cells were pelleted in a clinical centrifuge. Cell extract, 600 μg, was incubated with preconjugated anti-Myc agarose beads and incubated at 4°C for 2 h. Beads were collected by centrifugation at 400 × g, and the supernatant fraction was removed. The beads were then washed three times with buffer A. Finally, the pellet fractions were eluted by boiling in 100 μl SDS-PAGE loading buffer. One-tenth of the pellet and one-thirtieth of the input was used for Western blotting.
Measurement of Ternary Complex
Cell extracts were prepared as described above except buffer B (100 mM Tris-HCl, pH 8.0, 100 mM KCl, 5 mM MgCl2, 5 mM NaF, 2.5 mM PMSF, 7 mM β-mercaptoethanol [BME], 10% (vol/vol) glycerol, 0.1% (vol/vol) Triton-X, protease inhibitor cocktail (Roche, Indianapolis, IN) and RNAsin Plus (Promega, Madison, WI; 80 units per 100 μl) was used instead of buffer A. Protein, 1 mg, in a volume of 200 μl of buffer B was added to 100 μl of Red ANTI-FLAG M2 affinity gel (Sigma, St. Louis, MO) and incubated at 4°C for 2 h. The resin was washed with 200 μl buffer B (with 50 μM GMPP-N-P) and incubated for 20 min. This was repeated, and the sample was split 80:20 (160 μl:40 μl). The 40-μl aliquot was centrifuged, and the resin was resuspended in 40 μl of SDS-PAGE buffer (lacking BME). Samples were boiled for 10 min, and 10 μl of these eluates was loaded onto SDS-PAGE gels for Western blotting. For the input, 3 μl of the original extract was used for SDS-PAGE and Western analysis.
RNA was prepared from both the 160-μl eluate fraction and a 30-μl (made up to 160 μl) sample of the original extract. Briefly, samples were extracted in 320 μl Trizol (Invitrogen, Carlsbad, CA) and 80 μl chloroform. Glycan blue carrier, 3 μl, (Ambion, Austin, TX), 15 μl 3 M NaOAc, pH 5.2, and 140 μl isopropanol were added and mixed. Precipitates were pelleted and resuspended in formaldehyde-loading dye. Samples were boiled and loaded onto 8% polyacrylamide gels that were processed for Northern blotting as described in Singh et al. (2007). Blots were probed using a 5′ end-labeled [32P]oligonucleotide probe (5′-AGCCCTGCGCGCTTCCACTG-3′) specific to the initiator methionyl tRNA.
Microscopy
Real-time 2D deconvolved projections from continuous z-sweep acquisition were generated using a Delta Vision RT microscope (Applied Precision, Issaquah, WA) with an Olympus 100× 1.40 NA DIC oil PlanApo objective (Melville, NY) and Roper CoolSnap HQ camera (Tucson, AZ) using Applied Precision Softworx 1.1 software and 2 × 2 binning at room temperature. Z-sweep acquisition allowed fast visualization of all planes while minimizing fluorescent bleaching. Time-course experiments were performed by acquiring images every 5 s over a 2-min time period. ImageJ (http://rsb.info.nih.gov/ij/; NIH) was used to manually track the movement of the 2B body in the images acquired during the time course. The values calculated for the mean total distance moved by the 2B body were subjected to statistical analysis. A two-sample t test was used to determine if the mean total distance moved by the 2B body in cells treated with either 1% (vol/vol) butanol or 2% (vol/vol) butanol was significantly different from the movement of the 2B body in untreated cells of the same strain background. Images for quantitation of eIF2 in the 2B body were captured on a confocal microscope (SP5; Leica, Bannockburn, IL) using a 63× 0.6–1.40 NA plan Apo oil objective (Leica). Images were acquired using Application Suite 1.6.3 (Leica). For densitometric analysis, a merged 12-image z-series was taken, and the images were measured using ImageJ software (NIH). FRAP analysis was carried out as previously described (Campbell et al., 2005).
Mean Squared Displacement
2B body movement was measured at 5-s intervals over a 2-min time period and this movement was tracked using ImageJ software (NIH). These data were imported into Excel and used to calculate the Mean squared displacement (MSD) for each 2B body for Δt across the time-lapse experiments using the following equation (Platani et al., 2002):
where Δt corresponds to the time interval between images and d(t) to the position of the 2B body at any time t.
RESULTS
eIF2B Mutant Strains Differ in Their Susceptibility to 1-Butanol
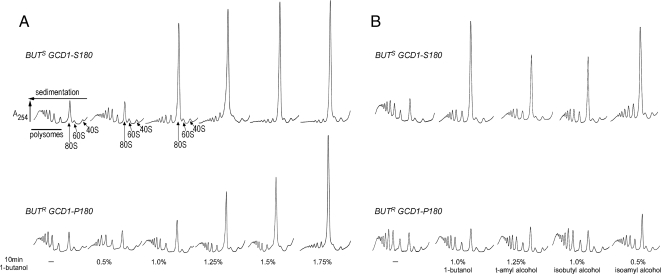
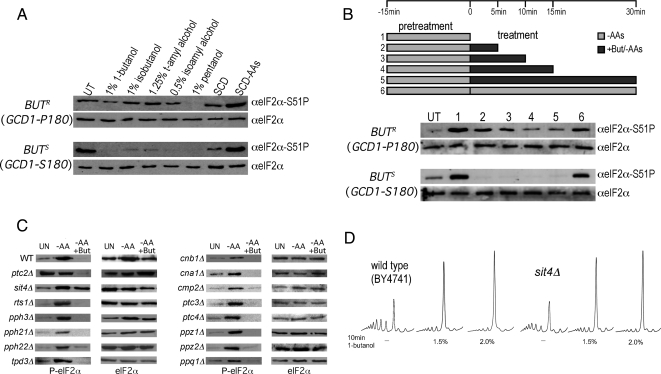
To expand our previous analysis of 1-butanol and its impact on protein synthesis, we made use of BUTS (butanol sensitive) and BUTR (butanol resistant) W303-1A laboratory isolates, where the difference in butanol sensitivity is explained by an allelic variation, P180 (BUTR) or S180 (BUTS), in the GCD1 gene encoding eIF2Bγ (Ashe et al., 2001). In both strains translation initiation is inhibited 10 min after the addition of butanol (Figure 1A). However, there is a fundamental difference in the concentration of butanol required to bring about the inhibitory effect. By plotting the percentage of translationally active ribosomes (i.e., polysome associated ribosomes) across the titration series for both yeast strains (Figure S1A), the median inhibitory concentration (IC50) for the BUTR isolate was found to be ∼1.5% (vol/vol) butanol, whereas for the BUTS isolate the IC50 was ∼0.8% (vol/vol) butanol. Consistent with this, for a whole range of different fusel alcohols a more effective inhibition of translation initiation is observed in the BUTS strain relative to the BUTR strain (Figure 1B). These results show that translation initiation is inhibited by fusel alcohols in both the BUTR and BUTS strains; however, the susceptibility differs dramatically. Interestingly, alcohols such as ethanol, isopropanol, and both d- and l-enantiomers of amino alcohols such as leucinol do inhibit translation initiation at high concentrations, but no difference is observed between the susceptibility of the BUTR and BUTS strains (data not shown).
Figure 1.
Fusel alcohols differentially inhibit translation initiation in butanol-sensitive (BUTS) and -resistant (BUTR) strains. (A) Polysome traces from yMK23 (BUTR) and yMK36 (BUTS) strains grown in YPD and incubated with added 1-butanol at the concentrations indicated for 10 min at 30°C. Polysomes were analyzed as described in Materials and Methods. The 40S (small ribosomal subunit), 60S (large ribosomal subunit), 80S (monosome), and polysome peaks are labeled. (B) Polysome traces from the yMK23 (BUTR) and yMK36 (BUTS) strains as in A, except the yeast were incubated with the indicated concentrations of tert-amyl alcohol (2-methyl 2-butanol), isobutyl alcohol (2-methyl 1-propanol), and isoamyl alcohol (3-methyl 1-butanol) before polysome analysis.
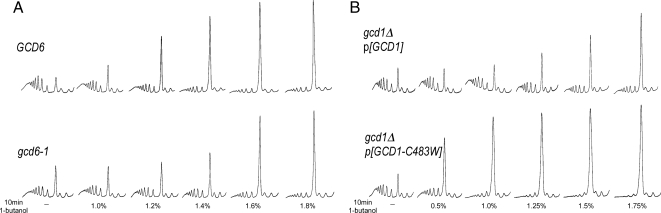
To extend the analysis of eIF2B and its role in the susceptibility of yeast to fusel alcohols, we also analyzed a gcd6-1 mutant strain (Bushman et al., 1993b). Previously, this mutant of the GCD6 gene, coding for the ε subunit of eIF2B exhibited increased resistance in terms of growth to butanol treatment, even though the strain background already harbored the GCD1-P180 (BUTR) allele (Ashe et al., 2001). To confirm and extend these studies, we compared the resistance of the gcd6-1 mutant to the wild-type strain background using polysome analysis as a measure of translational effects. Consistent with the Gcd− phenotype, Figure 2A shows that the gcd6-1 mutant strain has reduced translational activity in the absence of alcohol, as judged by elevated monosome levels and decreased polysome levels relative to the wild-type strain (see Figure S1B). Intriguingly though, the butanol-dependent inhibition of translation in this strain is not observed until a concentration of 1.4% (vol/vol) is reached. The control wild-type strain is inhibited at lower concentrations of 1.2% (vol/vol). This is an interesting result because the Gcd− phenotype and consequent inhibited translation of the gcd6-1 mutant in the absence of fusel alcohols suggest decreased basal eIF2B activity, whereas the resistance to fusel alcohols suggests that at specific alcohol concentrations the eIF2B activity in this strain becomes greater than a wild-type strain. We determined the mutation in GCD6, which was responsible for the Gcd− phenotype of the gcd6-1 strain. A C383W change was identified that was found to cause both the Gcd− phenotype and the butanol resistance phenotype of this strain (data not shown).
Figure 2.
Parallel mutations within the left-handed parallel β-helix (LbH) domains of eIF2Bε and γ lead to opposite phenotypes after exposure to butanol. (A) Polysome traces from yMK658 (GCD6) or yMK644 (gcd6-1) strains, grown in YPD and treated with butanol at the indicated concentrations for 10 min at 30°C. (B) Polysome traces from yMK974 (GCD1) or yMK976 (GCD1-C483W) strains, grown in selective media and treated with butanol at the indicated concentrations for 10 min at 30°C. Polysome analysis was as described in Materials and Methods.
The ε and γ subunits of eIF2B (encoded by GCD6 and GCD1, respectively) show homology across a number of domains. The residue, which is mutant in gcd6-1, lies within the third predicted hexapeptide repeat of the left-handed parallel β-helix (LbH) domain (pfam00132) of eIF2Bε (Mohammad-Qureshi et al., 2007). This residue is conserved in the third hexapeptide repeat present in eIF2Bγ as C483 (see Figure S2). Therefore, we altered the C483 to W in GCD1-P180 and tested the susceptibility of the resulting strain to butanol. Figure 2B and Figure S1C show that this strain is highly sensitive to butanol with concentrations of 0.5% (vol/vol), eliciting a dramatic effect on protein synthesis. This is all the more surprising considering that this C483W mutation is placed in the context of the “resistant” P180 form of GCD1.
Overall, these results show that fusel alcohols can generally inhibit translation initiation, yet various mutations in the eIF2B ε and γ subunits produce subtle yet measurable differences in susceptibility to these alcohols. This can either make strains more sensitive or resistant to these alcohols. However, existing knowledge of the precise makeup of the eIF2B complex is insufficient to allow an accurate prediction as to the finite impact of these mutations or the mechanism by which the subtle differences in alcohol sensitivity are brought about.
eIF2Bα Mutations Confer Butanol Resistance
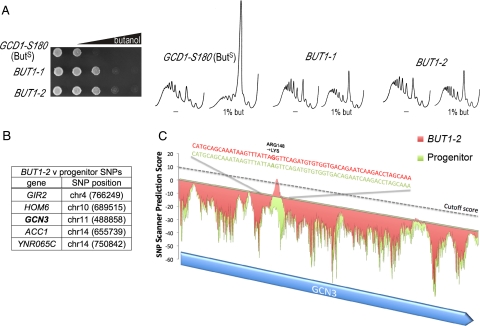
In an attempt to understand more about the inhibition of translation initiation by butanol, we undertook a genetic screen. Starting with the BUTS (GCD1-S180) W303-1A isolate (Ashe et al., 2001), we selected for resistance in terms of growth on 1.25% (vol/vol) butanol plates. Two promising mutants were identified, and these were further tested for translational resistance to the impact of butanol using polysome analysis. Both mutants exhibited increased resistance to butanol in terms of both growth and translation initiation (Figure 3A). PCR amplification from genomic DNA followed by sequencing showed that the mutants were not simply alterations at the GCD1-S180 site. In addition, no linkage to GCD1 was identified in backcrosses to the GCD1-P180 W303-1A strain, demonstrating that the new mutations were unlikely to represent intragenic GCD1 suppressors of the sensitive phenotype (data not shown). The mutations were found to be semidominant in that the heterozygote diploid grows faster on butanol plates than a homozygote BUTS diploid yet slower than the BUTR homozygote diploid (data not shown). In addition, when these mutants were directly crossed and tetrads were dissected, all of the resulting progeny were resistant to butanol, suggesting that these mutations are present within the same gene (data not shown). On this basis, these mutants were called BUT1-1 and BUT1-2 (Figure 3A).
Figure 3.
DNA tiling arrays highlight single nucleotide polymorphisms (SNPs) in the α subunit of eIF2B, which may explain the resistance to butanol. (A) Gradient plate and polysome analysis on yMK54 (BUT1-1), yMK16 (BUT1-2), and yMK36 (progenitor) showing the resistant profiles of the mutants relative to the progenitor BUTS strain. The gradient plates contain a gradient of butanol up to a maximum of 3% vol/vol, and equal numbers of the yeast cells are spotted at every position. (B) The five SNPs identified as present in yMK16 (BUT1-2) strain but not in yMK36 (progenitor) are shown along with the genomic position, i.e., chromosome number and coordinate. These include a SNP in the α subunit of eIF2B (GCN3). (C) A plot showing the SNP prediction signal across the GCN3 gene locus obtained from the tiling microarray analysis. A stringent cutoff value of 10 was used, and a prominent potential SNP was identified for the yMK16 (BUT1-2) strain (red) but not for the yMK36 (progenitor) strain (green). Sequencing analysis of multiple independently PCR-amplified DNA samples across the GCN3 locus identified a single amino acid change of arginine to lysine at position 148 for the yMK16 (BUT1-2) strain relative to the progenitor.
A number of attempts were made to identify the mutant gene using standard complementation cloning techniques both with 2-μm genomic libraries or genomic libraries generated from the mutant strains (Rose and Broach, 1991). In addition, attempts were made to generate transposon-mediated insertion mutants in the same complementation group, as this strategy has been previously used to clone semidominant mutations (Burns et al., 1994). However, all of these cloning attempts were unsuccessful, largely because of the number of false positives that impact on growth on butanol via mechanisms outside of translation initiation, combined with the subtlety of the butanol-resistant and -sensitive phenotypes. More recently, precise identification of SNPs has been achieved in yeast using oligonucleotide tiling arrays (Gresham et al., 2006). Therefore we prepared and fragmented genomic DNA from the BUT1-2 mutant and the GCD1-S180 progenitor strain and hybridized the resulting DNA to the GeneChip S. cerevisiae Tiling 1.0R arrays (Affymetrix, Santa Clara, CA). These tiling arrays were designed to precisely complement the S. cerevisiae genome database (i.e., the S288c background sequence). Therefore, after a software training procedure (Gresham et al., 2006), the SNPscanner software identified many SNPs above a specific threshold across both strains (data not shown). As a result, we selected those SNPs that are only present in BUT1-2 and not in the progenitor strain. Five potential SNPs were identified that might explain the difference between the BUT1-2 mutant strains and the wild-type progenitor (Figure 3B). One of these was particularly conspicuous, in that the potential SNP was identified in the GCN3 gene, which encodes eIF2Bα (Figure 3C). PCR amplification of the GCN3 gene from both the BUT1-1 and BUT1-2 mutant strains, followed by sequencing, revealed a single nucleotide change for both strains. This change would result in an arginine-to-lysine change at position 148 of Gcn3p for BUT1-2 (Figure 3C) and a threonine to lysine change at position 41 of Gcn3p for BUT1-1 (data not shown).
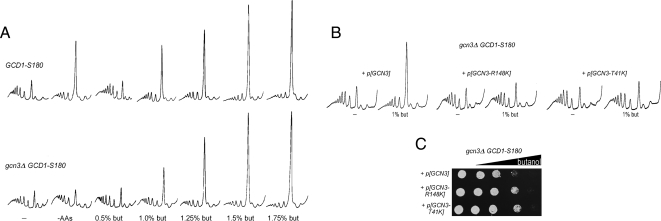
To establish whether these mutations were responsible for the observed alterations in butanol sensitivity, we performed a number of genetic tests. We first investigated impact of the deletion of the GCN3 gene. Figure 4A shows that this deletion has a relatively minor effect on the level of butanol sensitivity or resistance. However, the mutations identified are semidominant; therefore, if these represent gain of function alleles, a deletion mutant might not necessarily exhibit a robust phenotype. Therefore, we next evaluated the genetic linkage of these mutations to GCN3 via meiotic mapping. As such, we integrated URA3 at a position proximal to the GCN3 gene (Chromosome XI: 493505—between the BCH2 and SAP190 genes) and assessed linkage of the phenotype conferred by the BUT1-1 and BUT1-2 alleles to this locus. Table 2 shows an analysis of the resulting tetrads where only four from the BUT1-2 cross and three from the BUT1-1 cross gave a pattern indicative of a single meiotic crossover, and a double meiotic crossover was never observed in either cross. This suggests that the gene responsible for the phenotype in the BUT1-1 and BUT1-2 mutants lies in very close proximity to the GCN3 gene. Finally, we tested whether the mutations described above explain the phenotype by generating these mutations in a plasmid copy of GCN3. Figure 4, B and C, shows that expression of either GCN3-R148K or GCN3-T41K in a gcn3Δ GCD1-S180 strain produced a strain with a butanol-resistant phenotype both in terms of growth and translation initiation. In contrast, expression of wild-type GCN3 gave a sensitive phenotype (due to the GCD1-S180 allele present in the strain genome). Incidentally, these mutants also fail to inhibit translation initiation in response to amino acid starvation (i.e., they exhibit a Gcn− phenotype; data not shown). It is intriguing that these mutants phenocopy the null allele with regard to amino acid starvation yet with regard to the butanol-resistant phenotype they are clearly different from the null allele.
Figure 4.
Specific point mutations but not deletions of the eIF2Bα gene are resistant to butanol treatment. (A) Polysome traces from yMK36 (GCD1-S180) or yMK1262 (GCD1-S180 gcn3Δ) strains, grown in selective media and incubated either in amino acid–free media for 15 min or in media containing butanol at the indicated concentrations for 10 min at 30°C. (B) Polysome traces from gcn3Δ strains bearing various plasmids, yMK1442 with p[GCN3 URA3 CEN], yMK1441 with p[GCN3-R148K URA3 CEN], and yMK1451 with p[GCN3-T41K URA3 CEN]. Strains were grown in selective media and incubated in fresh media or media containing butanol at the indicated concentrations for 10 min at 30°C. Polysome analysis was carried out as described in Materials and Methods. (C) Gradient plate analysis on the above strains showing the resistant profiles of the GCN3 mutant strains relative to the wild type. The gradient plates contain a gradient of butanol up to a maximum of 3% vol/vol, and equal levels of yeast are spotted at every position.
Table 2.
Results of meiotic mapping from diploid strains
| (a) BUTS/BUTR (BUT1-2) v ura3/GCN3::URA3 |
(b) BUTS/BUTR (BUT1-1) v ura3/GCN3::URA3 |
||||
|---|---|---|---|---|---|
| PDa | TTb | NPDc | PDa | TTb | NPDc |
| 37 | 4 | 0 | 42 | 3 | 0 |
Diploid strains were constructed by crossing either (a) yMK1465 (BUT1-2 GCN3::URA3) and yMK39 (ura3) or (b) yMK53( BUT1-1 ura3) and yMK1475 (GCN3::URA3).
aThe number of tetrads that are parental ditype (PD); i.e., if the parents are AB × ab then a PD tetrad pattern consists of two AB spores and two ab spores.
bThe number of tetrads which are tetratype (TT), i.e., if the parents are AB × ab then a TT tetrad pattern consists of AB, ab, Ab and aB spores.
cThe number of tetrads which are non-parental ditype (NPD), i.e., if the parents are AB × ab then a NPD tetrad pattern consists of two Ab spores and two aB spores.
Overall, where other mutant cloning techniques have failed, we have used oligonucleotide tiling arrays to identify two alleles of GCN3 that can both suppress the butanol-sensitive phenotype. Thus we have now identified mutations in at least three eIF2B subunits that alter the sensitivity of yeast strains to fusel alcohols. Intriguingly, the identified Gcn3p or eIF2Bα subunit is required as part of a regulatory subcomplex for the regulation of translation initiation via interaction with phosphorylated eIF2.
1-Butanol Elicits a Dephosphorylation of eIF2α
It is well established that starvation for amino acids generates an increase in eIF2α phosphorylation via activation of the only S. cerevisiae eIF2α kinase Gcn2p and that this inhibits eIF2B guanine nucleotide exchange activity in an eIF2Bα-dependent manner (Dever et al., 1992; Yang and Hinnebusch, 1996; Pavitt et al., 1997). Given that eIF2Bα mutants are resistant to fusel alcohols, it is perhaps surprising that either deletion of GCN2 or a specific mutation at the site of phosphorylation on eIF2α (SUI2 S51-A) has no impact on the ability of fusel alcohols to inhibit translation initiation (Ashe et al., 2001). We were equally surprised to discover that exposure to 1% (vol/vol) butanol causes eIF2α to become very rapidly dephosphorylated in the BUTS strain (Figure 5A). A similar effect is noted for other fusel alcohols that inhibit translation initiation. Markedly, little or no dephosphorylation of eIF2α was observed for the BUTR strain under these conditions. Therefore even though we have demonstrated genetically that eIF2α phosphorylation (or dephosphorylation) is not the root cause of the butanol-dependent inhibition of translation initiation, there is a striking correlation between the effects of butanol on translation initiation and its impact on eIF2α dephosphorylation (cf. Figure 5A with Figure 1B).
Figure 5.
Butanol causes a Sit4p-dependent dephosphorylation of eIF2α. (A) Western blots from the yMK23 [GCD1-P180 (BUTR); top panels] and yMK36 [GCD1-S180 (BUTS); bottom panels] strains after growth in SCD. Cells were either untreated (UT) or treated for 15 min in the indicated concentrations of alcohol. Cells were also pelleted and resuspended in complete media (SCD) or media lacking amino acids (SCD-AAs) for 15 min. Protein extracts were blotted and probed with antibodies to eIF2α and phospho-specific antibodies to phosphoserine 51 on eIF2α. (B) As in A, except the strains were pretreated in amino acid starvation media for 15 min to induce eIF2α phosphorylation before 1% (vol/vol) butanol addition. UT, untreated cells; 1, the pretreatment alone, 2 through 5, 5, 10, 15, and 30 min of butanol addition under continuing amino acid starvation conditions; 6, a control of 30-min amino acid starvation alone after the pretreatment. (C) Protein extracts were made from yMK472 (BY4741 wild type) and a range of phosphatase subunit mutants (yMK1235-49) to screen for mutants that were defective in the butanol-induced dephosphorylation of eIF2α. After growth on SCD media, extracts were made from untreated cells (UT), cells starved for amino acids for 15 min (−AAs), and cells treated with 2% (vol/vol) butanol after the amino acid starvation (−AAs +But). (D) Polysome traces from yMK472 (BY4741) and yMK1246 (BY4741 sit4Δ) strains, grown in rich media and incubated in media containing butanol at the indicated concentrations for 10 min at 30°C.
To further characterize the influence of butanol on the levels of phosphorylated eIF2α, we prestarved cells for amino acids to induce high levels of phospho-eIF2α and then added 1% (vol/vol) butanol. Figure 5B shows that under these conditions butanol addition causes robust dephosphorylation of eIF2α such that even after persistent amino acid starvation, butanol leads to either a complete dephosphorylation in the BUTS background or a return to basal levels in the BUTR background. These results are striking in their similarity to observations using volatile anesthetics, where exposure of cells to such compounds causes an inhibition of translation initiation and also leads to the dephosphorylation of eIF2α (Palmer et al., 2005). These data are also striking in their contrast with observations after amino acid starvation, where translation initiation is inhibited as a direct consequence of eIF2α phosphorylation (Hinnebusch, 2005).
Sit4p Is Required for eIF2α Dephosphorylation But Not for Translational Regulation by Fusel Alcohols
To explore the mechanism of eIF2α dephosphorylation in response to fusel alcohols, we tested a panel of phosphatase mutants from the EUROSCARF deletion collection (Frankfurt, Germany; in the BY4741 background;). Figure 5C shows that in the BY4741 wild-type strain, amino acid starvation elicits the expected increase in phospho-eIF2α levels and that as a strain with the GCD1-P180 (BUTR) genotype, addition of a higher level of butanol (2%; vol/vol) is required in order to decrease phospho-eIF2α to undetectable levels. Most of the mutants in the various phosphatase subunits generate a similar pattern of phosphorylation and dephosphorylation in response to amino acid starvation and butanol addition, respectively (Figure 5C). However, two mutant strains exhibited a different pattern. First, the ptc2Δ strain, which is mutant in the type 2C protein phosphatase Ptc2p, exhibited high basal levels of phospho-eIF2α that were not further increased by amino acid starvation yet were reduced normally by butanol addition. The high basal level and lack of further induction after amino acid starvation is suggestive that the Gcn2p kinase is somehow active under nonstarvation conditions. Gcn2p itself is subject to regulation by phosphorylation and dephosphorylation (Cherkasova and Hinnebusch, 2003), and it is entirely possible that Ptc2p is somehow involved in repressing Gcn2p under nonstarvation conditions.
In contrast, the sit4Δ strain, which is mutant in the type 2A–related phosphatase Sit4p, exhibited normal induction of phospho-eIF2α by amino acid starvation, suggesting that Sit4p does not impact on either basal or activated Gcn2p kinase activity. Strikingly, however, there was little or no reduction in phospho-eIF2 after butanol treatment (Figure 5C). The simplest interpretation of these results is that fusel alcohols activate Sit4p, and this activation is required for the reduction in levels of phosphorylated eIF2α. This potential role for Sit4p in promoting eIF2α dephosphorylation where translation is inhibited by butanol is almost exactly opposite to the situation where translation initiation is inhibited by rapamycin treatment. Here inhibition of Sit4p has been implicated in an increase in phosphorylated eIF2α, because of suggested functions in the dephosphorylation of either the Gcn2p kinase or eIF2α itself (Cherkasova and Hinnebusch, 2003; Rohde et al., 2004).
Serine/threonine phosphatases are relatively promiscuous in terms of their substrate specificity (Gallego and Virshup, 2005). It is therefore plausible that a phosphatase such as Sit4p that is being activated toward eIF2α would also act on other sites in the eIF2B/eIF2 complex. Therefore, even though the dephosphorylation of eIF2α is not a requirement for the inhibition of translation initiation, this change may be serving as a marker for other phosphorylation changes that are important for the regulation of protein synthesis by fusel alcohols. Therefore to test whether activation of Sit4p is a requirement for the inhibition of translation initiation in response to fusel alcohols, we assessed this regulatory response in a SIT4-deleted strain. Figure 5C shows that the BY4741 wild-type strain is relatively resistant to butanol, and that at least 1.5% (vol/vol) butanol is required to generate a dramatic change in polysome profiles. For the sit4Δ strain, translation initiation is inhibited by butanol in a manner similar to that of the wild type. Therefore Sit4p activation is not required for the inhibition of translation initiation in response to fusel alcohols. In addition, this result also confirms that the capacity to rapidly dephosphorylate eIF2α is not required for this butanol-dependent regulation of protein synthesis.
Butanol Does Not Alter eIF2B Levels, Integrity, or Localization But Does Alter Ternary Complex Levels
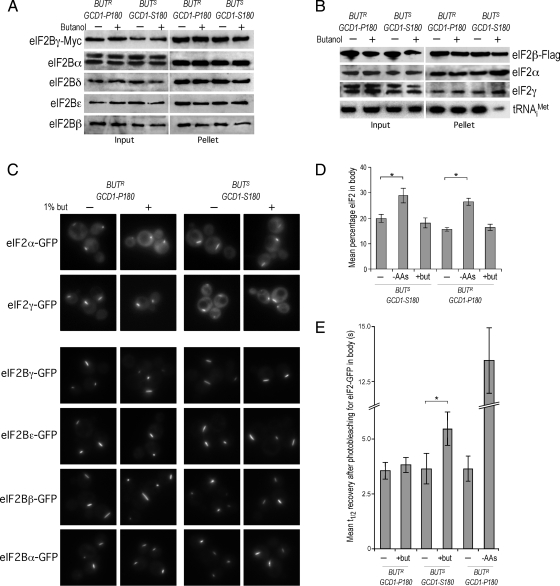
A variety of different mutants in the eIF2B subunit genes that cause the human disease VWM have been shown to impact on either the stoichiometry or the integrity of the eIF2B complex (Li et al., 2004; Richardson et al., 2004). Given that yeast mutations analogous to the human VWM mutations have also been shown to cause increased sensitivity to fusel alcohols (Richardson et al., 2004), we assessed the impact of fusel alcohols on the eIF2B complex. We generated strains in both the BUTS (GCD1-S180) and BUTR (GCD1-P180) backgrounds where the genomic GCD1 gene is c-terminally tagged with the c-myc epitope (Janke et al., 2004). Hence, in these strains Gcd1p-myc serves as the sole source of Gcd1p, yet no obvious phenotype is apparent and levels of Gcd1p are not significantly altered (data not shown). Critically, the GCD1-MYC S180 (BUTS) and P180 (BUTR) strains exhibit the expected translational sensitivity and resistance to butanol, respectively (data not shown). Figure 6A shows that neither the levels of each of the eIF2B subunits (Input lanes) or the complex integrity, as judged by the level of each of the eIF2B subunits immunoprecipitated with Gcd1p-Myc (Pellet lanes) is altered in response to butanol.
Figure 6.
Butanol reduces ternary complex levels and modestly reduces the transit of eIF2 through the 2B body. (A) Myc-immunoprecipitation experiments using extracts prepared from the yMK1125 [bearing GCD1-P180-MYC (BUTR)] and yMK1126 [bearing GCD1-S180-MYC (BUTS)] strains grown in rich media and then either incubated in media (−) or media with 1% (vol/vol) butanol (+) for 15 min. Samples were separated into input and pellet fractions, as indicated, and were used for Western blots using antibodies against eIF2Bα to ε. (B) Flag-immunoprecipitation experiments using extracts prepared from strains yMK1613 [GCD1-P180 (BUTR)] and yMK1615 [GCD1-S180 (BUTS)] (both bearing a eIF2β-Flag–expressing plasmid) grown in rich media and then either incubated in media (−) or media with 1% (vol/vol) butanol (+) for 15 min. Samples were separated into input and pellet fractions, as indicated, and 20% was used for Western blots using antibodies against Flag, eIF2α to eIF2γ. The remaining 80% was used to prepare RNA that was used to perform Northern blots using an oligonucleotide probe to tRNAiMet. (C) A panel of live-cell images from the BUTR (GCD1-P180) strains: yMK883 (eIF2α-GFP), yMK1211 (eIF2γ-GFP), yMK880 (eIF2Bγ-GFP), yMK882 (eIF2Bε-GFP), yMK1347 (eIF2Bβ-GFP), yMK1356 (eIF2Bα-GFP) and the BUTS (GCD1-S180) strains: yMK914 (eIF2α-GFP), yMK1212 (eIF2γ-GFP), yMK876 (eIF2Bγ-GFP), yMK878 (eIF2Bε-GFP), yMK1363 (eIF2Bβ-GFP), and yMK1355 (eIF2Bα-GFP) either treated with 1% (vol/vol) butanol for 15 min (+) or untreated (−). (D) Bar chart depicting the mean percentage eIF2 in the 2B body for the strains yMK883 (SUI2-GFP GCD1-P180 BUTR) and yMK914 (SUI2-GFP GCD1-S180 BUTS) after 15-min treatments in complete media (−), media with l% (vol/vol) butanol (+but), or media lacking amino acids (−AAs). eIF2 levels in the 2B body were measured using confocal microscopy and densitometry. Fifteen image-merged z-stacks from at least 20 single yeast cells were quantified using ImageJ to obtain the plotted values. Error bars, ±1 SEM; *p < 0.01 from an analysis of variance (ANOVA). (E) Bar chart showing half times of FRAP derived from experiments performed on eIF2α-GFP–bearing strains yMK883 (SUI2-GFP GCD1-P180 BUTR) and yMK914 (SUI2-GFP GCD1-S180 BUTS). Strains were grown in SCD media then incubated for 15 min in either complete media (−), media with l% (vol/vol) butanol (+but), or media lacking amino acids (−AAs) before single-cell FRAP studies. Error bars, ±1 SEM (where n is at least 18). *p < 0.05 from analysis of variance (ANOVA).
We also decided to directly test whether the reduced protein synthesis observed in response to fusel alcohols occurs as a result of reduced ternary complex levels, as would be predicted if eIF2B activity has been inhibited. Therefore we generated BUTR (GCD1-P180) and BUTS (GCD1-S180) strains bearing a Flag-tagged version of the SUI3 gene that encodes the β subunit of eIF2. Immunoprecipitation of eIF2 from extracts prepared from these strains (either treated with 1% butanol or untreated) resulted in equivalent levels of the eIF2 subunits (Figure 6B). Therefore, the integrity of the eIF2 complex is not significantly altered after butanol addition. However, it is clear that treatment with butanol leads to a dramatic reduction in the level of initiator methionyl tRNA associated with eIF2 for the BUTS strain but not the BUTR strain. This directly shows that butanol reduces the level of the eIF2-GTP Met-tRNAi ternary complex; a result that corresponds well with indirect measurements of ternary complex levels via GCN4-lacZ translational reporter assays (Ashe et al., 2001).
Recently, we have shown that eIF2B resides in a single focus within yeast cells that we have termed the “2B body.” Experiments using FRAP suggest guanine nucleotide exchange occurs in this body (Campbell et al., 2005). These previous studies were performed in the BUTR (GCD1-P180) W303-1A background. To evaluate whether the 2B body is also present in the BUTS (GCD1-S180) strain and to establish whether butanol impacts on the body, we GFP-tagged a number of eIF2B and eIF2 subunit genes in both the BUTS (GCD1-S180) and BUTR (GCD1-P180) backgrounds using a strategy where the tagging cassette is integrated downstream of the endogenous gene (Janke et al., 2004). Neither the strain background nor treatment with butanol appeared to impact on the localization of eIF2B or eIF2 subunits to the 2B body (Figure 6C). Similarly no difference was observed across the two strain backgrounds in the presence or absence of butanol when the total fraction of fluorescence from eIF2B (data not shown) or eIF2 subunits (Figure 6D) in the 2B body was assessed. The result with eIF2 is intriguing because after amino acid starvation, we observed (both previously and in this study) that the percentage of total eIF2 associated with the eIF2B focus increases by 50–100% (Figure 6D; Campbell et al., 2005). This suggests that the fundamental mechanism by which these alcohols impact on eIF2B is different from that of amino acid starvation. Therefore, it appears fusel alcohols do not cause increased residence of eIF2 on eIF2B to competitively inhibit exchange of other eIF2 molecules.
Butanol Inhibits the Dynamics of the 2B Body
The dynamic cycling of eIF2 through the 2B body can be assessed using FRAP (Campbell et al., 2005; Campbell and Ashe, 2006). Figure 6E shows that amino acid starvation leads to a significant increase in the half-time of recovery after photobleaching for eIF2. This is entirely consistent with the model where the phosphorylated eIF2 that is produced as a result of amino acid starvation exhibits increased occupancy on eIF2B and thus inhibits further exchange events. Butanol treatment also causes an increase in the half-time of recovery after eIF2 photobleaching; however, the magnitude of this change is very much smaller than that seen after amino acid starvation (Figure 6E). Given that both amino acid starvation and fusel alcohol treatment have a similar impact on translation initiation (Ashe et al., 2001), then it seems inconceivable that this small change in the rate of eIF2 cycling through the 2B body is sufficient to explain the inhibition of translation.
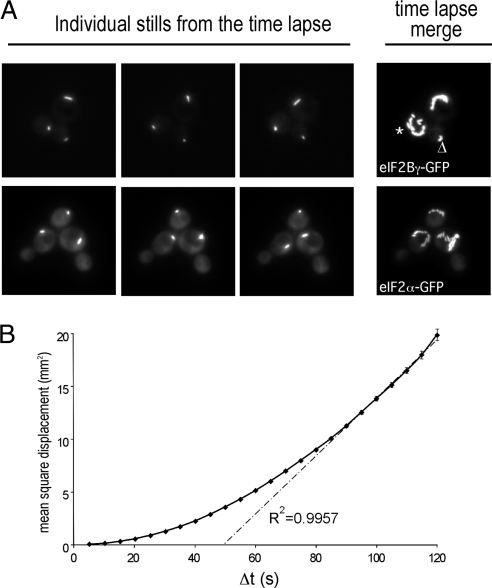
Another property of the 2B body is that it traverses almost all regions of the cytoplasm. For instance, it can be observed to transit around the vacuole and also in rare cases it can be seen to enter the daughter cell transiently before rapidly reentering the mother cell (data not shown). Time-lapse microscopy was used to investigate the dynamics of the 2B body, where 25 images were collected over a 2-min period. For each strain depicted in Figure 7A, three individual images from this set are shown, as well as an overlay of all 25 images, which gives a representation of the extent of movement over the time course. From these and many other examples, we observe two phases in the movement of the 2B body. First, the body can move around the cytoplasm in what appears a relatively haphazard manner (e.g., Figure 7A, Time-Lapse Merge, the body marked *). The second type of movement is where the 2B body appears tethered to a specific site in the cell (e.g., Figure 7A, Time-Lapse Merge, the body marked ▵). Here although very localized movement is observed (for instance the body can turn or switch orientation), there is very little overall alteration in position. To further analyze these types of movement a standard MSD plot was generated using the amassed time-lapse data (Figure 7B; Platani et al., 2002). This plot shows that across short time intervals (e.g., Δt values of 5–20 s) most 2B bodies do not move significantly, whereas over longer time intervals (Δt > 20 s) there is a finite chance of movement, and this movement appears to occur via random diffusion (because the R2 value of the latter part of the MSD plot approximates to 1; Platani et al., 2002). Therefore the 2B body appears to cycle between two phases: a tethered nonmobile phase and a mobile diffusing phase.
Figure 7.
The 2B body rapidly moves throughout the cell. (A) Images from time-lapse microscopy studies using the yMK880 (eIF2Bγ-GFP) and yMK883 (eIF2α-GFP) strains in the top and bottom rows, respectively. Each row contains three stills from a series of 25 images over a period of 2 min as well as a merged image of all 25 stills, which serves to depict the total extent of 2B body movement. (B) A plot of the mean square displacement (MSD) as a function of time interval (Δt) from >25 single particle–tracking experiments using the yMK880 (eIF2Bγ-GFP) strain. For short time intervals the 2B body does not become displaced linearly, suggesting that there is some form of tethering in operation. At progressively longer time intervals the MSD approaches a linear relationship, which is indicative of movement by diffusion.
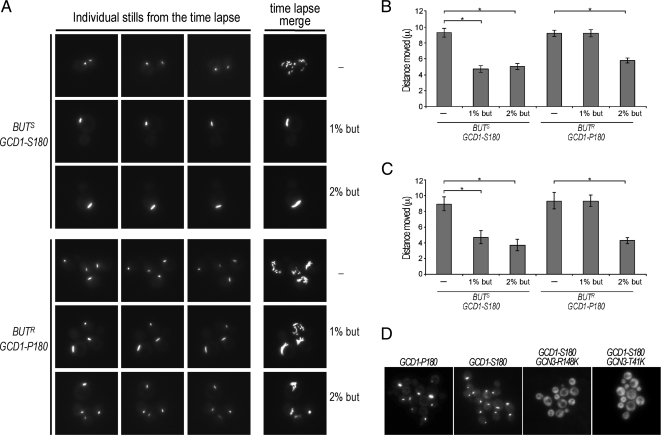
To investigate the impact of fusel alcohols on these phases of 2B body movement, we performed similar time-lapse studies in the presence and absence of butanol using markers for both eIF2B and eIF2 (Figure 8). In both the BUTS (GCD1-S180) and BUTR (GCD1-P180) strain backgrounds, the degree of movement of the 2B body was similar in the unstressed situation (Figure 8A). Quantitation of the average displacement over 120 s shows that the 2B body (assessed using strains bearing either GFP-tagged eIF2 or eIF2B subunits) moves ∼9 μm in either strain background (Figure 8, B and C). For the BUTS (GCD1-S180) strain, a 10-min incubation with either 1 or 2% (vol/vol) butanol causes the level of movement to drop substantially (∼4–5 μm over 120 s; Figure 8, A–C). For the BUTR strain (GCD1-P180), although 2% (vol/vol) butanol similarly inhibits movement of the 2B body, 1% (vol/vol) butanol has little impact (Figure 8, A–C). The difference between these strains correlates exactly with the difference in sensitivity observed at the level of translation initiation and growth (Figure 1). As the 2B body moves less after exposure to concentrations of fusel alcohols that also inhibit translation initiation, it seems possible that at least part of the inhibition of translation initiation is linked to increased tethering of the 2B body. By inference, then the capacity of the 2B body to move rapidly around the cell may allow sustained and robust levels of translation initiation.
Figure 8.
Butanol inhibits 2B body movement and butanol-resistant GCN3 mutants lack the 2B body. (A) Images from time-lapse microscopy studies using the eIF2Bγ-GFP bearing strains yMK876 [GCD1-S180-GFP (BUTS)] and yMK880 [GCD1-P180-GFP (BUTR)] strains, top three rows and bottom three rows, respectively. The strains were incubated in media (−) or media with 1% (vol/vol) or 2% (vol/vol) butanol for 10 min as indicated. Each row contains three stills from a series of 25 images over a period of 2 min, as well as a merged image of all 25 stills, which serves to depict the total extent of 2B body movement. (B) Bar chart depicting the mean distance moved in μ over a 2-min period from at least 40 time-lapse experiments (as in A) using eIF2α-GFP to mark the 2B body in the strains yMK914 [SUI2-GFP GCD1-S180 (BUTS)] and yMK883 [SUI2-GFP GCD1-P180 (BUTR)] Error bars, ±1 SEM; *p < 0.01 from an analysis of variance (ANOVA). (C) As in B, except eIF2Bγ is GFP-tagged to follow the 2B body in the strains yMK876 [GCD1-S180-GFP (BUTS)] and yMK880 [GCD1-P180-GFP (BUTR)]. (D) Images showing the absence of the eIF2B body in the yMK1567 [BUT1-1 (GCN3-R148K)] and yMK1595 [(BUT1-2 (GCN3-T41A)] strains relative to wild-type controls yMK1596 (GCD1-P180 BUTR) and yMK1597 (GCD1-S180 BUTS) using eIF2Bε-GFP (GCD6-GFP) to mark the eIF2B body.
As part of the mutational analysis described earlier two suppressors of the BUTS butanol-sensitive phenotype were characterized that are mutated in the eIF2Bα gene GCN3. Therefore, we generated strains where the endogenous GCD6 (eIF2Bε) gene is GFP tagged in these mutants and both the BUTS and BUTR wild-type strains. For both mutant strains the eIF2B body is not present (Figure 8D). The fact that the eIF2B body is not present in these mutants, where sensitivity to these alcohols is suppressed, suggests that the eIF2B body is important for the translational inhibition. It is also possible that in these mutants lacking the eIF2B body, the free eIF2B no longer has the potential to become tethered after exposure to fusel alcohols and that this suppresses the effect of butanol on translation initiation.
DISCUSSION
Eukaryotic cells utilize multiple eIF2B targeting pathways (Proud, 2005). Previously, we have characterized S180 and P180 allelic variants in the yeast eIF2Bγ (GCD1 gene), which confer sensitivity and resistance to fusel alcohols (e.g., 1-butanol), respectively (Ashe et al., 2001). Here, we find that both strains are translationally inhibited by butanol, yet the butanol concentration required to achieve inhibition differs dramatically. Such differential sensitivity is not observed for ethanol (data not shown) but is seen for other alcohols, including well-characterized products of branched-chain amino acid catabolism (isoamyl alcohol and isobutyl alcohol; Hazelwood et al., 2008). Therefore, it seems likely that there is some general mechanism where alcohols of a certain carbon chain length elicit effects on translation.
In our initial study, a panel of eIF2B mutants exhibited butanol-dependent phenotypes (Ashe et al., 2001). Furthermore, butanol-sensitive phenotypes have since been observed for specific eIF2B VWM mutants in yeast (Richardson et al., 2004). In this current study, we have characterized an eIF2Bε mutant, gcd6-1 (Bushman et al., 1993a). As a Gcd− mutant, this strain is slow growing and has reduced basal translation rates. Somewhat surprisingly, we found that this mutant is butanol resistant, and we identified C383W as the mutated residue. This mutation impacts on one of the eIF2Bε hexapeptide repeats that constitute the left-handed parallel LbH domain (Aravind and Koonin, 2000). This domain is also present in eIF2Bγ, so we generated the analogous eIF2Bγ-C483W mutant strain, which exhibited extreme sensitivity to fusel alcohols. Therefore, although the LbH domains of both eIF2Bε and γ are important for the fusel alcohol response, it is unlikely that they function similarly. The LbH domain is found in a variety of acyltransferases and is generally involved in the formation of trimeric complexes (Johnson et al., 2005), although other conformations are possible (Gorman and Shapiro, 2004). However, the relevance and functions of this domain and its structure in the context of the eIF2B heteropentamer are currently unknown.
Two butanol-resistant eIF2Bα mutations (GCN3-R148K and GCN3-T41K) were identified using an oligonucleotide array strategy. Mutation at T41 has previously been described as a Gcn− mutant (Pavitt et al., 1997), whereas mutation of R148 has not been characterized before. Both residues are well conserved, and both R148 and T41 are present in human eIF2B1 (data not shown). Both mutations phenocopy the gcn3Δ strain after amino acid starvation, because they fail to inhibit translation initiation. In stark contrast, the butanol response is unaltered in a gcn3Δ mutant, whereas the two GCN3 mutants are butanol resistant. These genetic intricacies probably reflect the complicated subunit interactions and regulation of this factor. Overall, eIF2Bα, which is required for the inhibition of translation initiation as a result of eIF2α phosphorylation, is also a key player in the fusel alcohol response.
In contrast to other translational inhibitory mechanisms, eIF2α becomes dephosphorylated in response to fusel alcohols. This was unexpected given that the kinetics and scale of translational inhibition are virtually identical after either fusel alcohol addition or amino acid starvation (which induces eIF2α phosphorylation; Smirnova et al., 2005). Two phosphatase deletion mutants, ptc2Δ and sit4Δ, exhibited unusual eIF2α phosphorylation profiles. The ptc2Δ mutant exhibited high basal phosphorylated eIF2α levels that were not further elevated upon amino acid starvation. Ptc2p is a type 2C phosphatase that dephosphorylates Hog1p and Ire1p, influencing the osmotic stress and unfolded protein responses, respectively (Welihinda et al., 1998; Young et al., 2002). Ptc2p also influences the yeast cell cycle via Cdc28p (Cheng et al., 1999) and the DNA checkpoint pathway (Guillemain et al., 2007). The fact that a ptc2Δ mutant has constitutively high phospho-eIF2 levels that are unresponsive to amino acid starvation suggests that this strain harbors nonregulatable Gcn2p kinase. Therefore, it is possible that in a wild-type strain, the Ptc2p phosphatase acts on Gcn2p to inhibit or dampen the basal kinase activity. However, Ptc2p is clearly not required for the fusel alcohol–induced dephosphorylation of eIF2α.
In the case of the type 2A–related phosphatase Sit4p, a deletion mutant exhibits virtually no eIF2α dephosphorylation after exposure of yeast cells to fusel alcohols. Therefore, it seems likely that Sit4p is activated after fusel alcohol treatment and either directly or indirectly leads to a decrease in phosphorylated eIF2. However, in the sit4Δ mutant, translation initiation is still inhibited after fusel alcohol treatment; therefore, neither the activation of Sit4p nor the dephosphorylation of eIF2 is involved in the effects of fusel alcohols on translation initiation. However, in a physiological sense the dephosphorylation of eIF2α observed after fusel alcohol treatment may prevent the inappropriate activation of an amino acid starvation cellular response. This might be especially important given that fusel alcohol stress can be viewed as metabolically opposed to amino acid starvation; i.e., fusel alcohols signal the catabolism of amino acids as cells desperately attempt to find nitrogen sources, whereas amino acid starvation signals the up-regulation of anabolic pathways involved in channelling nitrogen reserves toward the biosynthesis of amino acids. The activation of Sit4p in response to fusel alcohols is also compatible with other responses that are activated by this stress, such as the pseudophyphal response (Lorenz et al., 2000). Indeed, a sit4Δ mutant is incapable of undergoing pseudohyphal growth after nitrogen starvation (Cutler et al., 2001).
Intriguingly, volatile anesthetics, like fusel alcohols, lead to the dephosphorylation of eIF2α and GCN3 mutants are resistant to these compounds (Palmer et al., 2005). This suggests that fusel alcohols and volatile anesthetics impact on translation initiation in a remarkably similar manner. However, subtle differences are apparent. For instance, gcn3Δ strains are more resistant to volatile anesthetics, whereas we show that deletion of GCN3 produces little difference in the response to fusel alcohols. Similarly, a gcn2Δ mutant is resistant to volatile anesthetics, whereas GCN2 deletion does not affect the fusel alcohol response. In response to volatile anesthetics, the impact of the GCN2 deletion was dependent on the strains' auxotrophic status. We observe no correlation between auxotrophy/prototrophy and the effects of fusel alcohols. Therefore, these two reasonably diverse sets of chemicals target protein synthesis via very similar but not entirely identical mechanisms (Ashe et al., 2001; Palmer et al., 2005).
Recently we have found that eIF2B and eIF2 reside in a large cytoplasmic body that we have termed the “2B body” and that this body represents a site where guanine nucleotide exchange of eIF2-GDP to eIF2-GTP can occur (Campbell et al., 2005). Fusel alcohols alter a variety of properties with regard to this 2B body. For instance, they impact on the rate of eIF2 transit through the body. This rate has previously been shown to vary in a manner that equates to the level of guanine nucleotide exchange activity after stress or mutation (Campbell et al., 2005). However, fusel alcohols cause much less pronounced changes than other stresses that inhibit eIF2B and translation initiation. The explanation for this is not clear, but we favor the hypothesis that nonproductive eIF2 transit may occur where the rate of exchange is regulated by mechanisms other than increased eIF2α phosphorylation. Thus the rate of eIF2 transit through the 2B body would be an overestimate of the guanine nucleotide exchange activity. Fusel alcohols also increase the length of time that the body remains in a tethered state, thus reducing the movement of the body around the cell. This effect is more exacerbated in a BUTS strain relative to a BUTR strain. The GCN3 mutants that suppress a BUTS mutation do not possess eIF2B bodies and therefore in these strains eIF2B is unlikely to be present in a tethered state. Interestingly, these data have implications for the function of the eIF2B body. The GCN3 mutant strains that lack the eIF2B body show no decrease in growth or protein synthetic rates, suggesting that the eIF2B body is not required for maximal protein synthesis. However, these strains are resistant to the effects of both amino acid starvation and fusel alcohols on translation initiation. Therefore, it is possible that the concentration of eIF2B into a large body permits a greater degree of regulation and, in the absence of this body, regulatory pathways are compromised.
In terms of the mechanism by which fusel alcohols inhibit eIF2B to generate reduced ternary complex, it is not clear whether the regulation is direct or indirect. Direct regulation of enzymes by alcohols has been described previously. For example, alcohol-binding sites exist in the cysteine-rich domains of various isoforms of protein kinase C, and depending on the precise nature of the bound molecule, they have been shown to both stimulate and inhibit kinase activity (Stubbs and Slater, 1999). Intriguingly, these sites overlap with the anesthetic-binding sites (Das et al., 2006). Therefore, it is possible that the eIF2B guanine nucleotide exchange factor represents a point of regulation for alcohols and volatile anesthetics as a result of such a direct binding reaction. Alternatively, the alcohols could interact and impact on a component of a signaling pathway, and one output of this pathway would be the posttranslational modification of eIF2B. In both of these cases, either the direct binding of alcohols or the posttranslational modification of eIF2B would alter the guanine nucleotide exchange activity of this complex, reducing translation initiation. Another consequence of this inhibition of the guanine nucleotide exchange activity would be increased tethering of the eIF2B body. If true, this would implicate the exchange of guanine nucleotides as important in the release of the eIF2B body from the tethered state.
Alternatively, it is possible that fusel alcohols either directly or indirectly impact on the process by which the eIF2B body becomes untethered. In this model, the increased tethering would lead to reduced guanine nucleotide exchange activity even though the intrinsic activity of eIF2B would remain unaltered. By preventing the movement of the eIF2B, fusel alcohols could locally generate increased eIF2-GTP and decreased eIF2-GDP levels. The decrease in substrate and increases in product could potentially lead to the inhibition observed here.
Overall, this study illustrates that eIF2B is targeted by fusel alcohols to generate reduced ternary complex levels and translation initiation rates in a manner that is similar to the effects of volatile anesthetics. This work also emphasizes the 2B body as an element of the cell biology of protein synthesis that might be fine-tuned as a consequence of cellular stress.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Ashe (University of Manchester) for critical reading of the manuscript and T. Dever (National Institutes of Health), A. Hinnebusch (National Institutes of Health), K. Asano (Kansas State University), C. Singh (Kansas State University), and R. O'Keefe (University of Manchester) for advice and reagents. This work was supported by the Wellcome Trust project Grants 067328/Z/02/Z (G.D.P. and M.P.A.) and 080349/Z/06/Z (M.P.A.). E.J.T., C.D.G., and R.J.H. were supported by Biotechnology and Biological Sciences Research Council studentships, and D.D. is supported by a National Environmental Research Council advanced fellowship.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0962) on May 5, 2010.
REFERENCES
- Aravind L., Koonin E. V. Eukaryote-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 2000;10:1172–1184. doi: 10.1101/gr.10.8.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M. P., De Long S. K., Sachs A. B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe M. P., Slaven J. W., De Long S. K., Ibrahimo S., Sachs A. B. A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J. 2001;20:6464–6474. doi: 10.1093/emboj/20.22.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N., Grimwade B., Rossmacdonald P. B., Choi E. Y., Finberg K., Roeder G. S., Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption Saccharomyces cerevisiae. Gene Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Bushman J. L., Asuru A. I., Matts R. L., Hinnebusch A. G. Evidence that GCD6 and GCD7, translational regulators of GCN4, are subunits of the guanine nucleotide exchange factor for eIF-2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993a;13:1920–1932. doi: 10.1128/mcb.13.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman J. L., Foiani M., Cigan A. M., Paddon C. J., Hinnebusch A. G. Guanine nucleotide exchange factor for eukaryotic translation initiation factor 2 in Saccharomyces cerevisiae: interactions between the essential subunits GCD2, GCD6, and GCD7 and the regulatory subunit GCN3. Mol. Cell. Biol. 1993b;13:4618–4631. doi: 10.1128/mcb.13.8.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell S. G., Ashe M. P. Localization of the translational guanine nucleotide exchange factor eIF2B: a common theme for GEFs? Cell Cycle. 2006;5:678–680. doi: 10.4161/cc.5.7.2607. [DOI] [PubMed] [Google Scholar]
- Campbell S. G., Hoyle N. P., Ashe M. P. Dynamic cycling of eIF2 through a large eIF2B-containing cytoplasmic body: implications for translation control. J. Cell Biol. 2005;170:925–934. doi: 10.1083/jcb.200503162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. Y., Ross K. E., Kaldis P., Solomon M. J. Dephosphorylation of cyclin-dependent kinases by type 2C protein phosphatases. Gene Dev. 1999;13:2946–2957. doi: 10.1101/gad.13.22.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V. A., Hinnebusch A. G. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–872. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler N. S., Pan X., Heitman J., Cardenas M. E. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol. Biol. Cell. 2001;12:4103–4113. doi: 10.1091/mbc.12.12.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., Zhou X. J., Miller K. W. Identification of an alcohol binding site in the first cysteine-rich domain of protein kinase C delta. Protein Sci. 2006;15:2107–2119. doi: 10.1110/ps.062237606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T. E., Feng L., Wek R. C., Cigan A. M., Donahue T. F., Hinnebusch A. G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell. 1992;68:585–596. doi: 10.1016/0092-8674(92)90193-g. [DOI] [PubMed] [Google Scholar]
- Di Como C. J., Arndt K. T. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- Gallego M., Virshup D. M. Protein serine/threonine phosphatases: life, death, and sleeping. Curr. Opin. Cell Biol. 2005;17:197–202. doi: 10.1016/j.ceb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Gorman J., Shapiro L. Structure of serine acetyltransferase from Haemophilus influenzae Rd. Acta Crystallogr. D. 2004;60:1600–1605. doi: 10.1107/S0907444904015240. [DOI] [PubMed] [Google Scholar]
- Gresham D., Ruderfer D. M., Pratt S. C., Schacherer J., Dunham M. J., Botstein D., Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Guillemain G., Ma E., Mauger S., Miron S., Thai R., Guerois R., Ochsenbein F., Marsolier-Kergoat M. C. Mechanisms of checkpoint kinase Rad53 inactivation after a double-strand break in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:3378–3389. doi: 10.1128/MCB.00863-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Fink G. R. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. [Google Scholar]
- Hannig E. M., Hinnebusch A. G. Molecular analysis of Gcn3, a translational activator of Gcn4—evidence for posttranslational control of Gcn3 regulatory function. Mol. Cell. Biol. 1988;8:4808–4820. doi: 10.1128/mcb.8.11.4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hazelwood L. A., Daran J. M., van Maris A. J., Pronk J. T., Dickinson J. R. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 2008;74:2259–2266. doi: 10.1128/AEM.02625-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Janke C., et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Johnson C. M., Roderick S. L., Cook P. F. The serine acetyltransferase reaction: acetyl transfer from an acylpantothenyl donor to an alcohol. Arch Biochem. Biophys. 2005;433:85–95. doi: 10.1016/j.abb.2004.08.014. [DOI] [PubMed] [Google Scholar]
- Jousse C., Oyadomari S., Novoa I., Lu P., Zhang Y. H., Harding H. P., Ron D. Inhibition of a constitutive translation initiation factor 2 alpha phosphatase CReP, promotes survival of stressed cells. J. Cell Biol. 2003;163:767–775. doi: 10.1083/jcb.200308075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern K., Nunn C. D., Pichova A., Dickinson J. R. Isoamyl alcohol-induced morphological change in Saccharomyces cerevisiae involves increases in mitochondria and cell wall chitin content. FEMS Yeast Res. 2004;5:43–49. doi: 10.1016/j.femsyr.2004.06.011. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy T., Pavitt G. D., Zhang F., Dever T. E., Hinnebusch A. G. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2alpha) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 2001;21:5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Valle R., Wittenberg C. A role for the Swe1 checkpoint kinase during filamentous growth of Saccharomyces cerevisiae. Genetics. 2001;158:549–562. doi: 10.1093/genetics/158.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Wang X., Van Der Knaap M. S., Proud C. G. Mutations linked to leukoencephalopathy with vanishing white matter impair the function of the eukaryotic initiation factor 2B complex in diverse ways. Mol. Cell. Biol. 2004;24:3295–3306. doi: 10.1128/MCB.24.8.3295-3306.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Cutler N. S., Heitman J. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell. 2000;11:183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P. D., Harding H. P., Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Anaya C., Dickinson J. R., Sudbery P. E. In yeast, the pseudohyphal phenotype induced by isoamyl alcohol results from the operation of the morphogenesis checkpoint. J. Cell Sci. 2003;116:3423–3431. doi: 10.1242/jcs.00634. [DOI] [PubMed] [Google Scholar]
- Mohammad-Qureshi S. S., Haddad R., Hemingway E. J., Richardson J. P., Pavitt G. D. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2 beta and -2 gamma mediate guanine nucleotide exchange. Mol. Cell. Biol. 2007;27:5225–5234. doi: 10.1128/MCB.00495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero-Lomeli M., Morais B.L.B., Figueiredo D. L., Neto D.C.S., Martins J.R.P., Masuda C. A. The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:21542–21548. doi: 10.1074/jbc.M201977200. [DOI] [PubMed] [Google Scholar]
- Novoa I., Zeng H. Q., Harding H. P., Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2 alpha. J. Cell Biol. 2001;153:1011–1021. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer L. K., Rannels S. L., Kimball S. R., Jefferson L. S., Keil R. L. Inhibition of mammalian translation initiation by volatile anesthetics. Am. J. Physiol. Endocrinol. Metab. 2006;290:E1267–E1275. doi: 10.1152/ajpendo.00463.2005. [DOI] [PubMed] [Google Scholar]
- Palmer L. K., Shoemaker J. L., Baptiste B. A., Wolfe D., Keil R. L. Inhibition of translation initiation by volatile anesthetics involves nutrient-sensitive GCN-independent and -dependent processes in yeast. Mol. Biol. Cell. 2005;16:3727–3739. doi: 10.1091/mbc.E05-02-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt G. D. eIF2B, a mediator of general and gene-specific translational control. Biochem. Soc. Trans. 2005;33:1487–1492. doi: 10.1042/BST0331487. [DOI] [PubMed] [Google Scholar]
- Pavitt G. D., Ramaiah K. V., Kimball S. R., Hinnebusch A. G. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt G. D., Yang W., Hinnebusch A. G. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platani M., Goldberg I., Lamond A. I., Swedlow J. R. Cajal Body dynamics and association with chromatin are ATP-dependent. Nat. Cell Biol. 2002;4:502–508. doi: 10.1038/ncb809. [DOI] [PubMed] [Google Scholar]
- Proud C. G. eIF2 and the control of cell physiology. Semin. Cell Dev. Biol. 2005;16:3–12. doi: 10.1016/j.semcdb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Richardson J. P., Mohammad S. S., Pavitt G. D. Mutations causing childhood ataxia with central nervous system hypomyelination reduce eukaryotic initiation factor 2B complex formation and activity. Mol. Cell. Biol. 2004;24:2352–2363. doi: 10.1128/MCB.24.6.2352-2363.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde J. R., Campbell S., Zurita-Martinez S. A., Cutler N. S., Ashe M., Cardenas M. E. TOR controls transcriptional and translational programs via Sap-Sit4 protein phosphatase signaling effectors. Mol. Cell. Biol. 2004;24:8332–8341. doi: 10.1128/MCB.24.19.8332-8341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Broach J. R. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Rowlands A. G., Panniers R., Henshaw E. C. The catalytic mechanism of guanine-nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J. Biol. Chem. 1988;263:5526–5533. [PubMed] [Google Scholar]
- Scali O., Di Perri C., Federico A. The spectrum of mutations for the diagnosis of vanishing white matter disease. Neurol. Sci. 2006;27:271–277. doi: 10.1007/s10072-006-0683-y. [DOI] [PubMed] [Google Scholar]
- Singh C. R., Udagawa T., Lee B., Wassink S., He H., Yamamoto Y., Anderson J. T., Pavitt G. D., Asano K. Change in nutritional status modulates the abundance of critical pre-initiation intermediate complexes during translation initiation in vivo. J. Mol. Biol. 2007;370:315–330. doi: 10.1016/j.jmb.2007.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova J. B., Selley J. N., Sanchez-Cabo F., Carroll K., Eddy A. A., McCarthy J. E., Hubbard S. J., Pavitt G. D., Grant C. M., Ashe M. P. Global gene expression profiling reveals widespread yet distinctive translational responses to different eukaryotic translation initiation factor 2B-targeting stress pathways. Mol. Cell. Biol. 2005;25:9340–9349. doi: 10.1128/MCB.25.21.9340-9349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs C. D., Slater S. J. Ethanol and protein kinase C. Alcohol Clin. Exp. Res. 1999;23:1552–1560. [PubMed] [Google Scholar]
- Vattem K. M., Wek R. C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. M., Proud C. G. A novel mechanism for the control of translation initiation by amino acids, mediated by phosphorylation of eukaryotic initiation factor 2B. Mol. Cell. Biol. 2008;28:1429–1442. doi: 10.1128/MCB.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Cannon J. F., Dever T. E., Hinnebusch A. G. Truncated protein phosphatase GLC7 restores translational activation of GCN4 expression in yeast mutants defective for the eIF-2 alpha kinase GCN2. Mol. Cell. Biol. 1992;12:5700–5710. doi: 10.1128/mcb.12.12.5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R. C., Jiang H. Y., Anthony T. G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Welihinda A. A., Tirasophon W., Green S. R., Kaufman R. J. Protein serine/threonine phosphatase Ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol. Cell. Biol. 1998;18:1967–1977. doi: 10.1128/mcb.18.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods Y. L., Cohen P., Becker W., Jakes R., Goedert M., Wang X. M., Proud C. G. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2B epsilon at Ser(539) and the microtubule-associated protein tau at Thr(212): potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem. J. 2001;355:609–615. doi: 10.1042/bj3550609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Hinnebusch A. G. Identification of a regulatory subcomplex in the guanine nucleotide exchange factor eIF2B that mediates inhibition by phosphorylated eIF2. Mol. Cell. Biol. 1996;16:6603–6616. doi: 10.1128/mcb.16.11.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young C., Mapes J., Hanneman J., Al-Zarban S., Ota I. Role of Ptc2 type 2C Ser/Thr phosphatase in yeast high-osmolarity glycerol pathway inactivation. Eukaroyt. Cell. 2002;1:1032–1040. doi: 10.1128/EC.1.6.1032-1040.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.