Macroautophagy is linked to various diseases in humans, including cancer and neurodegeneration. The morphological hallmark is the formation of the double-membrane autophagosome, which is the most complex aspect of macroautophagy. We demonstrate a role for post-Golgi Sec proteins, Sec2 and Sec4, in autophagosome formation.
Abstract
In eukaryotic cells, autophagy mediates the degradation of cytosolic contents in response to environmental change. Genetic analyses in fungi have identified over 30 autophagy-related (ATG) genes and provide substantial insight into the molecular mechanism of this process. However, one essential issue that has not been resolved is the origin of the lipids that form the autophagosome, the sequestering vesicle that is critical for autophagy. Here, we report that two post-Golgi proteins, Sec2 and Sec4, are required for autophagy. Sec4 is a Rab family GTPase, and Sec2 is its guanine nucleotide exchange factor. In sec2 and sec4 conditional mutant yeast, the anterograde movement of Atg9, a proposed membrane carrier, is impaired during starvation conditions. Similarly, in the sec2 mutant, Atg8 is inefficiently recruited to the phagophore assembly site, which is involved in autophagosome biogenesis, resulting in the generation of fewer autophagosomes. We propose that following autophagy induction the function of Sec2 and Sec4 are diverted to direct membrane flow to autophagosome formation.
INTRODUCTION
In eukaryotic cells, dynamic vesicle trafficking between the membrane-bounded organelles of the secretory pathway makes it possible for them to carry out their function, while a high degree of regulation allows these compartments to maintain their normal and discrete morphology. In contrast to membrane trafficking in the secretory pathway where the cargo-containing vesicle is formed from a preexisting organelle by budding, vesicle formation in autophagy is considered to be de novo; this is critical to autophagic function, which necessitates the use of sequestering vesicles of varying size to accommodate a wide range of cargo. Autophagy is a ubiquitous process conserved in eukaryotes that mediates an adaptive response to environmental change by degrading cytoplasm, including entire organelles, in the lysosome, or the fungal equivalent, the vacuole. Beyond its role as a degradative pathway, recent studies have elucidated the role of autophagy in human pathophysiology (Huang and Klionsky, 2007). Based on the cargo sequestration process, autophagy can be divided into several types. The best-characterized type so far is macroautophagy, which we refer to as autophagy hereafter.
Genetic analyses in yeast have significantly enhanced our understanding of the molecular basis of autophagy. For example, many of the yeast Atg proteins have homologues in higher eukaryotic cells (Xie and Klionsky, 2007). In Saccharomyces cerevisiae, the autophagy pathway can be induced by starvation. In response to nutrient depletion, cytosolic components are enwrapped in a double-membrane compartment termed a phagophore. This structure expands to form a double-membrane vesicle, an autophagosome. The completed autophagosome fuses with the vacuole, releasing the inner vesicle, termed an autophagic body, into the vacuolar lumen. The autophagic body and its contents are then degraded by vacuolar hydrolases. A perivacuolar locus called the phagophore assembly site (PAS) is proposed to be the vesicle formation site during autophagy (Suzuki et al., 2001; Kim et al., 2002). As detected by fluorescence microscopy, most Atg proteins converge at this locus as a single dot, and the non-PAS population is diffuse in the cytosol.
In yeast, autophagy also mediates the cytoplasm-to-vacuole targeting (Cvt) pathway (Klionsky et al., 1992; Harding et al., 1995; Hutchins and Klionsky, 2001). Unlike the nonspecific autophagy induced by starvation, the Cvt pathway operates exclusively as a biosynthetic process, specifically transporting certain vacuolar hydrolases, such as the precursor form of aminopeptidase I (prApe1), into the vacuole; this pathway is constitutively active in growing cells. The Cvt pathway has similar morphological features to starvation-induced autophagy and uses much of the same protein machinery (Scott et al., 1996; Baba et al., 1997). Therefore, the Cvt pathway can be regarded as a specific form of autophagy. Besides the Cvt pathway, other types of specific autophagy that mediate the removal of excess organelles or invasive bacteria have also been reported in fungi as well as higher eukaryotes (Nakagawa et al., 2004; Dunn et al., 2005; Iwata et al., 2006; Kanki et al., 2009).
Ever since the initial observation of autophagy, a central question that remains to be answered is the origin of the membrane used for autophagosome formation. The biggest obstacle in investigating this question is that the autophagosome is almost totally lacking in transmembrane proteins (Hirsimaki et al., 1982; Stromhaug et al., 1998; Fengsrud et al., 2000). As a result, proteomic analyses have failed to reliably identify organelle-specific markers, suggesting that resident proteins are excluded from the source membrane(s) used to generate the autophagosome. Data from yeast indicate that the secretory pathway contributes to the vesicle formation step during autophagy. Sec12, a guanine nucleotide exchange factor (GEF) involved in vesicle budding from the endoplasmic reticulum (ER), and some components of the COPII complex are essential for autophagosome formation (Ishihara et al., 2001). Similarly, strains that are defective in Golgi complex function, such as those with mutations in Sec7 or subunits of the COG complex, also show a block in autophagy (Reggiori et al., 2004b; Yen et al., 2009; van der Vaart et al., 2010). In these studies, however, it is difficult to determine whether the autophagic block is due to a defect in a particular component of the secretory pathway or just reflects an indirect effect when membrane flow through the early secretory pathway is impaired. Along these lines, the defective autophagic phenotype observed in early sec mutants may indicate the involvement of a particular cargo protein that is transported via the secretory pathway. In addition, an impaired secretory pathway leads to cellular dysfunction in processes such as ribosome synthesis, endocytosis, and organization of the nucleus (Mizuta and Warner, 1994; Hicke et al., 1997; Nanduri et al., 1999), which makes it difficult to differentiate which defect directly causes the block in autophagy. Thus, there has been considerable speculation and confusion regarding the role of the secretory pathway in autophagy.
In this report, we analyzed the function of two proteins involved in the late stage of the secretory pathway, Sec2 and Sec4, and show that both play important roles in autophagy. Sec2 is a GEF protein that acts on the yeast Rab protein Sec4, and both participate in the polarized transport of secretory vesicles toward the sites of active growth (bud and mother/daughter neck; Nair et al., 1990; Itzen et al., 2007). As a Rab protein, Sec4 oscillates between the inactive GDP-bound form and the active GTP-bound form and thus functions as a molecular switch to regulate vesicle delivery (Walworth et al., 1992). Activated by Sec2, Sec4, together with Sec2, reversibly associates with secretory vesicles and the exocyst complex on the plasma membrane and thus directs the vesicles toward the secretion site. We propose that during autophagy the secretion machinery at the trans-Golgi network, including Sec2 and Sec4, is diverted to direct membrane flow to the process of autophagosome formation.
MATERIALS AND METHODS
Media and Growth Conditions
Yeast cells were grown in rich (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal (SMD; 0.67% yeast nitrogen base, 2% glucose, and auxotrophic amino acids and vitamins as needed) media. For starvation conditions, SD-N medium (0.17% yeast nitrogen base without ammonium sulfate or amino acids, and 2% glucose) was used. For experiments with temperature-sensitive mutants, cells were grown at 24°C to midlog phase and shifted to a nonpermissive temperature (34 or 37°C) for 30 min to inactivate the mutants. If not otherwise indicated, cells were grown at 30°C.
Plasmids and Strains
pGFP-Atg8(316), pCu416, and pRS413 have been reported previously (Christianson et al., 1992; Labbé and Thiele, 1999; Suzuki et al., 2001). To clone pSec2(413) and pSec4(413), the open reading frames (ORFs) as well as 500 base pairs of the 5′ promoter regions were amplified by PCR from yeast genomic DNA and cloned into the XmaI/ClaI sites of pRS413. pCuSec2(416) and pCuSec4(416) were generated by amplifying the ORFs from pSec2(413) and pSec4(413) and then cloning them into the EcoRI/ClaI sites of pCu416. To generate pCuHASec2(416), pCuHASec2(1-450)(416) and pCuHASec2(1-508)(416), a forward primer with additional sequence encoding a single hemagglutinin (HA) epitope at the 5′ end was used to introduce the HA tag at the N terminus. Corresponding reverse primers were designed depending on the truncation site. PCR fragments amplified from pCuSec2(416) were inserted into the EcoRI/ClaI sites of pCu416. To make pGFP-Atg8(405), the GFP-Atg8 coding sequence from pGFP-AUT7(316) (Suzuki et al., 2001) was moved as a PvuI fragment to pRS405. pCuYpt31(416) was made by inserting the sequence corresponding to the Ypt31 ORF into the XmaI/ClaI sites of pCu416. pVAM3ts(404) was a generous gift from Dr. Zhiping Xie (Nankai University, China).
Yeast strains used in this article are listed in Table 1. Gene deletion, truncation, and C-terminal tagging were carried out using a PCR-based method as described previously (Longtine et al., 1998; Gueldener et al., 2002). For integration of Atg9-3xGFP, the pAtg9-3xGFP(306) plasmid (Monastyrska et al., 2008) was linearized with BglII and integrated into the ATG9 genomic locus. Red fluorescent protein (RFP)-Ape1 was incorporated into the chromosome by integrating AvrII-digested pRFP-Ape1(305) (Stromhaug et al., 2004) into the APE1 locus. To integrate green fluorescent protein (GFP)-Atg8, pGFP-Atg8(405) was linearized with AflII and integrated into the LEU2 locus.
Table 1.
Yeast strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| BY4742 | MATα his3Δ leu2Δ lys2Δ ura3Δ | Invitrogen |
| FRY143 | SEY6210 vps4Δ::TRP1 pep4Δ::LEU2 | Cheong et al. (2005) |
| HAY572 | TN124 atg1Δ::URA3 | Abeliovich et al. (2003) |
| HCY76 | SEY6210 vps4ΔTRP1 pep4Δ::Kan atg1Δ::URA3 | Cheong et al. (2007) |
| JGY107 | sec2-41 ATG9-3xGFP::URA3 RFP-APE1::LEU2 atg1Δ::ble | This study |
| JGY112 | TN124 sec2-59-PA::Kan | This study |
| JGY113 | SEY6210 sec2-59-3HA::Kan | This study |
| JGY124 | SEY6210 SEC2::TRP1 | This study |
| JGY125 | SEY6210 sec2-78::TRP1 | This study |
| JGY126 | SEY6210 sec2(Δ451-508)::TRP1 | This study |
| JGY127 | TN124 SEC2::TRP1 | This study |
| JGY128 | TN124 sec2-78::TRP1 | This study |
| JGY129 | TN124 sec2(Δ451-508)::TRP1 | This study |
| JGY146 | SEY6210 GFP-ATG8::LEU2 | This study |
| JGY159 | sec4-8 pho8Δ60::URA3 pho13Δ::ble | This study |
| JGY166 | LRB939 pho8Δ60::URA3 pho13Δ::ble | This study |
| JGY168 | LRB932 pho8Δ60::URA3 pho13Δ::ble | This study |
| JGY169 | SEY6210 GFP-ATG8::LEU2 vam3ts::TRP1 | This study |
| JGY171 | SEY6210 GFP-ATG8::LEU2 vam3ts::TRP1 sec2-41-3HA::HIS3 | This study |
| JGY172 | SEY6210 GFP-ATG8::LEU2 vam3ts::TRP1 sec2-59-3HA::HIS3 | This study |
| JGY180 | SEY6210 vps4Δ::TRP1 pep4Δ::LEU2 sec2-59-3HA::Kan | This study |
| JGY183 | SEY6210 GFP-ATG8::LEU2 sec2-59-3HA::Kan | This study |
| JGY184 | SEY6210 GFP-ATG8::LEU2 sec2-41-3HA::Kan | This study |
| JGY192 | NSY340 ATG9-3xGFP::URA3 RFP-APE1::LEU2 atg1Δ::ble | This study |
| JGY197 | BY4742 ATG9-3xGFP::URA3 RFP-APE1::LEU2 atg1Δ::Kan | This study |
| JGY198 | sec4-8 ATG9-3xGFP::URA3 RFP-APE1::LEU2 atg1Δ::ble | This study |
| JGY207 | NSY340 ATG9-3xGFP::URA3 RFP-APE1::LEU2 | This study |
| JGY209 | NSY340 pho8Δ60::URA3 pho13Δ::ble | This study |
| JGY210 | NSY340 GFP-ATG8::LEU2 | This study |
| JGY217 | NSY128 GFP-ATG8::LEU2 | This study |
| JGY220 | NSY128 pho8Δ60::URA3 pho13Δ::ble | This study |
| JGY222 | NSY340 pep4Δ::LEU2 vps4Δ::ble | This study |
| JGY227 | NSY128 trp1Δ::Kan vam3ts::TRP1 GFP-ATG8::LEU2 | This study |
| LRB932 | LRB939 sec4-2 | Babu et al. (2002) |
| LRB939 | MATα his3 leu2 ura3-52 | Babu et al. (2002) |
| NSY128 | MATα ade2 his3Δ200 leu2-3,112 lys2-801 ura3-52 | Jedd et al. (1997) |
| NSY340 | MATα leu2-3,112 lys2-801 ura3-52 ypt31Δ::HIS3 ypt32A141D | Liang et al. (2007) |
| sec2-41 | BY4742 sec2-41 | This study |
| sec4-8 | BY4742 sec4-8 | This study |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | Robinson et al. (1988) |
| TN124 | MATa leu2-3,112 trp1 ura3-52 pho8::pho8Δ60 pho13Δ::LEU2 | Noda et al. (1995) |
| YTS158 | BY4742 pho8::pho8Δ60 pho13Δ::Kan | He et al. (2006) |
To generate the sec2-78 and sec2(Δ451-508) strains, the endogenous copy of SEC2 was replaced with mutated alleles by homologous recombination. First, pCuSec2C483Y(416) was made by PCR-based site-directed mutagenesis using pCuSec2(416) as the template. To construct pCuSec2(Δ451-508)(416), DNA fragments encoding Sec2(1-450) and Sec2(509-759) were amplified from pCuSec2 and annealed together as the template for the second round PCR that amplified the Sec2(Δ451-508) fragment. The resulting fragment was cloned into the EcoRI/ClaI sites of pCu416 to generate pCuSec2(Δ451-508)(416). Next, from these two plasmids and pCuSec2(416), the Sec2, Sec2C483Y and Sec2(Δ451-508) sequences plus the CYC1 terminator were amplified by PCR and inserted into the PstI/BglII sites of pFA6a-TRP1, which are upstream of the TRP1 gene (Longtine et al., 1998). Then, the 700-base pair genomic region downstream of the SEC2 ORF including its terminator was cloned and ligated into the EcoRI/SpeI sites of pFA6a-TRP1, which are downstream of TRP1, to complete the pSec2-TRP1, pSec2C483Y-TRP1, and pSec2(Δ451-508)-TRP1 plasmids. These three plasmids were then digested with PstI/SpeI, and the resulting fragments were transformed into yeast cells to replace the endogenous copy of SEC2. PCR and DNA sequencing were used to confirm incorporation of the mutations.
EM Analysis
Cells were cultured to midlog phase in YPD medium at 24°C and shifted to 37°C for 30 min to inactivate the temperature-sensitive mutants. After 2-h starvation at 37°C, samples were harvested and analyzed by transmission electron microscopy as described previously (Cheong et al., 2007). Images of cells with intact morphology and clear vacuoles were collected. To measure the size of autophagic bodies, vesicles were outlined manually using Adobe Photoshop (San Jose, CA) and the area values of outlined regions were measure by ImageJ (http://rsb.info.nih.gov/ij/). The area value (S) was then converted into diameter by the formula: d = 2(S/π)1/2.
Fluorescence Microscopy
Cells were grown in YPD or SMD media without auxotrophic amino acids to midlog phase. For starvation experiments, cells were incubated in SD-N medium as indicated in the corresponding figure legends, and 1 OD600 unit of cells (equivalent to 1 ml culture at A600 = 1.0) were collected by centrifugation and resuspended in 20 μl of SMD without vitamins to reflect growing conditions or in SD-N for starvation conditions. When indicated in the figure legend, fixation was carried out as described previously (Legakis et al., 2007); otherwise, living cells were used. The samples were then examined by microscopy (DeltaVision Spectris, Applied Precision,) and pictures were captured with a CCD camera (CoolSnap HQ, Photometrics). For each microscopy picture, 12 Z-section images were captured. The distance between two neighboring sections was 0.5 μm and the total depth of each stack was 5.5 μm, which was the approximate diameter of a normal yeast cell. To show the fluorescence signal of the whole cell in one picture, the stack of Z-section images were projected into a 2-D image by sum projection and quantified using softWoRx software (Applied Precision, Issaquah, WA). To label vacuolar membrane, FM 4-64 (Molecular Probes, Eugene, OR) staining was performed as described previously (Cheong et al., 2005).
Radioactive Pulse Chase
To study the kinetics of prApe1 maturation we used a pulse-chase analysis as described previously (Yen et al., 2007). To quantify protein secretion, wild-type cells were grown in SMD to midlog phase at 30°C, and 30 OD600 units of cells (equivalent to a 30 ml culture at A600 = 1.0) were harvested. The cells were labeled in 1 ml SMD with 200 μCi [35S]methionine/cysteine for 15 min. After washing, the cells were subjected to a nonradioactive chase in 4 ml SMD containing 0.2% yeast extract and 2 mM cysteine and methionine with or without 10 ng/ml cycloheximide or 0.8 μg/ml rapamycin, or in SD-N. At the indicated time points, 400-μl samples were collected and precipitated with 10% trichloroacetic acid. Protein samples were analyzed with a scintillation counter or SDS-PAGE. For the sec2-59 mutant, cells were grown at 24°C and incubated at 37°C for 30 min before labeling. The subsequent labeling and chase were done similar to the wild type, but were carried out at 37°C.
Autophagy Assays
GFP-Atg8 processing and Pho8Δ60 assays were performed as described previously (Abeliovich et al., 2003; Shintani and Klionsky, 2004).
RESULTS
Sec2 Is Required for Both Autophagy and the Cvt Pathway
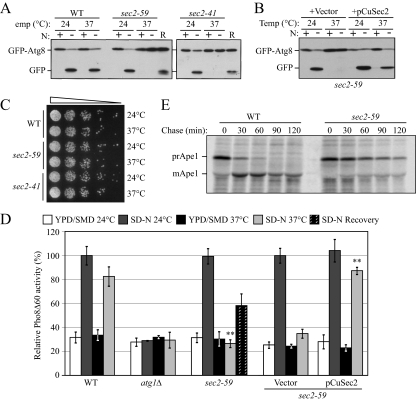
To identify new essential genes involved in autophagy, we screened a collection of S. cerevisiae temperature-sensitive mutants for an autophagic defect, using the GFP-Atg8 processing assay to monitor autophagy. After induction of autophagy, GFP-Atg8 is transported into the vacuole; the GFP moiety is released by proteolysis and it is relatively stable so that free GFP reflects the level of autophagy (Shintani and Klionsky, 2004). Using this method, we found a clear defect in autophagy in the sec2-41 mutant and subsequently confirmed the phenotype in a second allele, sec2-59 (Figure 1A). At permissive temperature (PT), the appearance of free GFP in both sec2-59 and sec2-41 after starvation was comparable to that in wild-type cells. However, at nonpermissive temperature (NPT), no free GFP was detected in either sec2 mutant after starvation (Figure 1A). To confirm that the autophagic defect was due to the dysfunction of Sec2, we cloned the SEC2 gene on a centromeric plasmid. Exogenous expression of Sec2 restored the induction of free GFP at the NPT in both sec2-59 (Figure 1B) and sec2-41 (our unpublished data). To address the possibility that the autophagic defect was due to sec2 mutant cell death at the NPT, we tested the viability of the sec2 mutants at the conditions used in the previous experiments. After 2-h starvation at the NPT, the cell culture was diluted and spotted onto agar plates. Both sec2 alleles showed normal growth similar to the wild-type cells (Figure 1C). In addition, in the GFP-Atg8 processing assay after 2-h starvation at the NPT we shifted the cells back to the PT for another 2-h starvation (recovery period); under these conditions free GFP could be clearly detected (Figure 1A) showing that autophagy induction was recovered. These results suggested that the defect in autophagy in the sec2 mutants was not due to loss of viability at the NPT.
Figure 1.
Sec2 is involved in both autophagy and the Cvt pathway. (A) GFP-Atg8 processing is blocked in sec2 mutants. GFP-Atg8 was expressed by chromosomal integration in wild-type (JGY146), sec2-59 (JGY183), and sec2-41 (JGY184) strains. Cells were cultured in YPD at 24°C to midlog phase. For each strain, the culture was divided into two parts. One-half was incubated at 37°C for 30 min to inactivate the sec2 mutant, whereas the other one remained at 24°C. Then cells were shifted to SD-N and incubated for 2 h at the same temperature. Samples were taken before and after starvation. For recovery (R), cells starved at 37°C for 2 h were shifted back to 24°C and starved for another 2 h. Immunoblotting was done with anti-YFP antibody (that recognizes GFP) and the positions of full-length GFP-Atg8 and free GFP are indicated. (B) Exogenous expression of Sec2 restores autophagy in the sec2-59 mutant. sec2-59 GFP-Atg8 cells (JGY183) harboring an empty vector or pCuSec2(416) were cultured in SMD medium and examined as described in A. (C) sec2 mutants are viable after 2-h starvation at 37°C. The same amount of starved cells from A were diluted and plated on a YPD plate, followed by 2-d incubation at room temperature. Cells were diluted 1:5 in each step from left to right. (D) Pho8Δ60 activity in the sec2-59 mutant. Wild-type (TN124), atg1Δ (HAY572), and sec2-59 (JGY112) cells as well as sec2-59 cells expressing an empty vector or pCuSec2(416) were cultured as in A. The Pho8Δ60 assay was performed as described in Materials and Methods. Error bar, SD of three independent experiments. Significant difference compared with wild-type or vector alone, **p < 0.01. (E) The Cvt pathway is defective in the sec2-59 mutant. Wild-type (SEY6210) and sec2-59 (JGY113) cells were grown in SMD at 24°C. After a 30-min inactivation at 37°C, cells were subjected to pulse-chase labeling as described in Materials and Methods.
To extend our analysis, we used a more quantitative method, the Pho8Δ60 assay, to confirm the defect in autophagy in the sec2 mutants. PHO8 encodes an alkaline phosphatase that is transported to the vacuole via the ALP pathway; removal of a propeptide activates the zymogen in the vacuole lumen (Kaneko et al., 1987; Klionsky and Emr, 1989). In contrast, Pho8Δ60, a mutated version without its transmembrane domain, can only be delivered to the vacuole via autophagy (Noda et al., 1995). Thus, in a strain with a deletion of PHO13, which encodes the other yeast alkaline phosphatase, and where pho8Δ60 replaces PHO8, the activity of alkaline phosphatase represents the magnitude of autophagy. At both the PT and NPT, wild-type cells showed a strong increase in Pho8Δ60 activity after incubation in starvation medium, whereas the activity did not increase in atg1Δ cells in which autophagy is completely blocked (Figure 1D). In sec2-59, although the induction of Pho8Δ60-dependent alkaline phosphatase activity was normal at the PT, there was no significant increase when the cells were starved at the NPT. Similar to the GFP-Atg8 processing results, the Pho8Δ60 activity of sec2-59 was partially recovered when the cells were shifted back to the PT (recovery period) or in the presence of plasmid-expressed SEC2 (Figure 1D).
In yeast, the majority of molecular components are shared between the Cvt pathway and nonspecific autophagy. The precursor form of Ape1 is transported to the vacuole through the Cvt pathway in nutrient-rich conditions and via autophagy in starvation conditions (Baba et al., 1997). On delivery to the vacuole, the N-terminal propeptide is removed to generate the mature form of the enzyme (mApe1; Klionsky et al., 1992). The molecular-weight difference between mApe1 and prApe1 make it a useful marker to monitor protein sorting through the Cvt pathway. Therefore, we tested the kinetics of prApe1 maturation in the sec2 mutant using pulse-chase experiments. Wild-type and sec2-59 cells were shifted from 24 to 37°C for 30 min to inactivate Sec2 function. The cells were then subjected to a radioactive pulse chase; samples at each time point were analyzed by immunoprecipitation. In wild-type cells, most prApe1 was converted to mApe1 after a 60-min chase, and no precursor was detected after 90 min (Figure 1E). In contrast, in sec2-59 cells only a portion of prApe1 was processed into mApe1 even after a 120-min chase (Figure 1E), suggesting the Cvt pathway was severely blocked in the sec2 mutant. As a control, we examined the CPY pathway to confirm that the sec2 mutation does not generally affect vacuolar protein sorting. As reported previously, in the sec2-59 mutant most carboxypeptidase Y was processed into its mature form, although the kinetics were slightly slower than seen in the wild type (our unpublished data; Stevens et al., 1982). Therefore, sec2 mutants specifically affected autophagy and the Cvt pathway in starvation and growing conditions, respectively.
Mutations in Amino Acids 451-508 of Sec2 Result in a Temperature-sensitive Phenotype in Autophagy
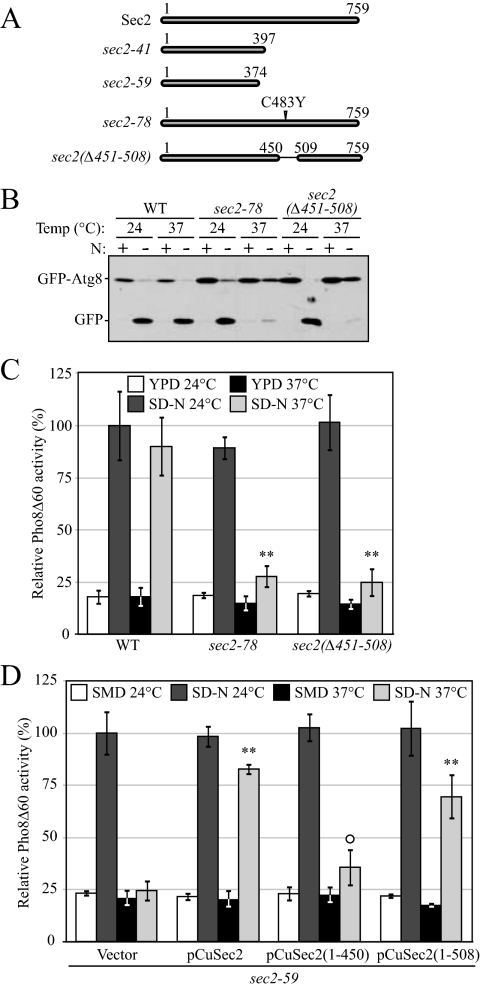
Comprehensive studies have been performed on the role of Sec2 in the secretory pathway. The N-terminal half of Sec2 is required and sufficient for Sec2-Sec4 interaction and its GEF activity (Nair et al., 1990). In contrast, the C-terminal half of Sec2 is dispensable for exchange activity, but truncation mutants display a temperature-sensitive phenotype (Elkind et al., 2000). As reported previously, sec2-59 (which contains amino acids 1-374) and sec2-41 are truncated alleles (Nair et al., 1990). Sequencing data show that a TGG-to-TGA mutation in sec2-41 results in a premature protein product with 397 amino acids (Figure 2A). Within the C-terminal half of Sec2, a stretch of 58 amino acids (451-508) are critical for its proper localization and function. A point mutation within this domain (sec2-78, C483Y) results in a temperature-sensitive phenotype (Walch-Solimena et al., 1997; Elkind et al., 2000). To investigate whether this domain is also involved in autophagy, we next examined the induction of autophagy in strains expressing Sec2 proteins lacking this 58-amino acid domain or carrying the C483Y mutation.
Figure 2.
A 58-amino acid domain on Sec2 is essential for its role in autophagy. (A) Schematic representation of Sec2 alleles. (B) Mutations within the 58-amino acid domain result in defective GFP-Atg8 processing. Wild-type (JGY124), sec2-78 (JGY125) and sec2(Δ451-508) (JGY126) cells were transformed with the pGFP-Atg8(316) plasmid and examined as in Figure 1A. (C) An autophagic defect in sec2-78 and sec2(Δ451-508) is confirmed by the Pho8Δ60 assay. The corresponding Pho8Δ60 strains (JGY127, JGY128, and JGY129) to those used in B were examined by the Pho8Δ60 assay. Error bar, SD of three independent experiments. Significant difference compared with wild-type, **p < 0.01. (D) The extreme C terminus of Sec2 is dispensable to complement the sec2-59 mutant. sec2-59 (JGY112) cells were transformed with plasmids expressing various truncated forms of Sec2 and examined by the Pho8Δ60 assay. Error bar, SD of three independent experiments. Significant difference compared with vector alone, **p < 0.01; no significant difference, ○p > 0.1.
Sec2 mutant alleles with amino acids 451-508 deleted (sec2(Δ451-508)) or containing the C483Y point mutation (sec2-78) were used to replace the wild-type SEC2 gene in the chromosome. Autophagy activity was examined by the GFP-Atg8 processing and Pho8Δ60 assays. Both the sec2(Δ451-508) and sec2-78 strains showed a clear temperature-sensitive phenotype for autophagy. After 2-h starvation at 37°C, there was severely reduced cleavage of GFP-Atg8 with either sec2(Δ451-508) or sec2-78 (Figure 2B), and this result was confirmed with the Pho8Δ60 assay. The increase of Pho8Δ60-dependent alkaline phosphatase activity after starvation was normal at the PT in both mutant strains, whereas it was barely above the background level at the NPT (Figure 2C). Therefore, at the NPT, amino acids 451-508 of Sec2 were required for autophagy. In contrast to the previously published temperature-sensitive phenotype of the sec2-78 mutant (Walch-Solimena et al., 1997), cell growth was not completely blocked in the sec2-78 allele that we constructed, although the growth rate was slower than in the corresponding wild-type cells (our unpublished data). This result meant that in this mutant the late stage of the secretory pathway necessary for cell growth was not completely blocked at 37°C, even though autophagy under this condition was substantially diminished. Therefore, these findings suggested that it was not protein secretion per se that was required for autophagy, but rather Sec2 itself.
Next, we examined whether the amino acids at the extreme C terminus (downstream of amino acids 451-508) played a role in autophagy. Plasmids expressing full-length or truncated Sec2 were transformed into the sec2-59 strain and the Pho8Δ60 activity was measured. As expected, full-length Sec2 complemented the autophagic defect at 37°C. The expression of Sec2(1-508) also restored the Pho8Δ60 activity to ∼69% the level of full-length Sec2 (Figure 2D). A similar result was observed in sec2-59 cells expressing Sec2(1-541) (data not shown). As negative controls, when empty vector or Sec2(1-450) lacking the 58 amino acid domain were expressed, the Pho8Δ60 activity was barely rescued. Thus, the C-terminal 509-759 amino acids of Sec2 were not required to complement the autophagic defect seen in sec2-59 cells.
Fewer Autophagic Bodies Are Formed in sec2-59 at the Nonpermissive Temperature
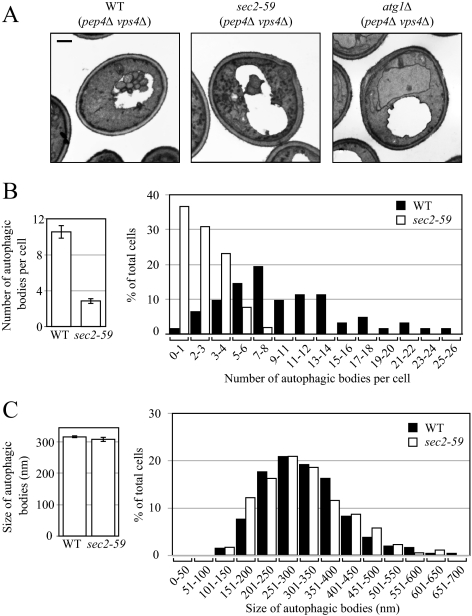
To further understand how autophagy is affected in the sec2-59 mutant, we performed a morphological analysis using transmission electron microscopy. After being delivered into the vacuole, autophagic bodies are degraded in a Pep4-dependent manner (Takeshige et al., 1992). To visualize intact autophagic bodies, the PEP4 gene was deleted, which allows the accumulation of autophagic bodies inside the vacuole. In addition to PEP4, the VPS4 gene was also knocked out to avoid the presence of small vesicles derived from the Mvb pathway (Babst et al., 1998). Wild-type and sec2-59 cells (both harboring pep4Δ vps4Δ double mutations) cultured in YPD medium were shifted from 24 to 37°C and incubated for 30 min. The cells were then shifted to SD-N medium and starved at 37°C for another 2 h, and samples were examined by electron microscopy as described in Materials and Methods.
In wild-type cells, a large number of autophagic bodies (ABs) accumulated in the vacuole after 2-h starvation (Figure 3A). The average number was 10.55 ± 0.73 ABs per vacuole (mean ± SEM, n = 62 vacuoles; Figure 3B). Under the same conditions, however, in sec2-59 cells fewer ABs were observed (Figure 3A); the average number was 2.88 ± 0.31 ABs per vacuole (mean ± SEM, n = 56 vacuoles; Figure 3B), substantially lower than the number in wild-type cells. As a negative control, in atg1Δ cell there were no ABs visible in the vacuole (Figure 3A). A histogram showed that more than 65% of the sec2-59 vacuoles had fewer than four ABs, whereas in wild-type cells <10% fell into that category (Figure 3B). Statistical analysis using a t test showed that the difference of AB number between wild-type and sec2-59 cells was significant (p < 0.01). Although the disruption of Sec2 function at the NPT resulted in the formation of fewer autophagic bodies, it had little effect on their size. In wild-type cells, the average diameter of ABs was 315.42 ± 3.78 nm (mean ± SEM, n = 651 ABs) and that of sec2-59 cells was 307.77 ± 7.73 nm (mean ± SEM, n = 172 ABs; Figure 3C); this difference between wild-type and sec2-59 cells was not statistically significant (p = 0.36). Therefore, fewer autophagic bodies with normal size were formed in the sec2 mutant.
Figure 3.
Sec2 mutation results in a reduced number of autophagic bodies, but has no effect on their size. After inactivation, the wild-type (pep4Δ vps4Δ, FRY143), atg1Δ (atg1Δ pep4Δ vps4Δ, HCY76), and sec2-59 (sec2-59 pep4Δ vps4Δ, JGY180) strains were starved for 2 h at 37°C and then analyzed by EM as described in Materials and Methods. (A) Representative images of wild-type, atg1Δ, and sec2-59 strains. Scale bar, 500 nm. (B and C) Number and size of autophagic bodies. Quantification was done as described in Materials and Methods. Error bars, SEM.
Atg8 Recruitment to the PAS Is Affected in sec2 Mutants
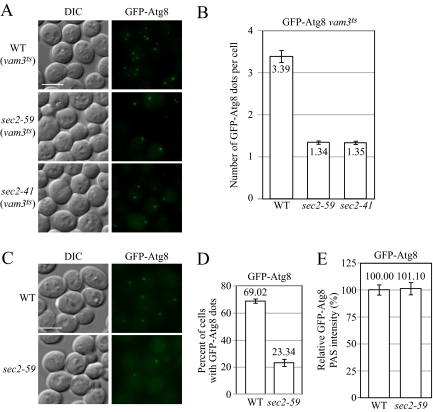
EM analysis showed that fewer autophagic bodies accumulated in a sec2 mutant. However, this analysis could not differentiate the step at which autophagy was affected. In particular, the substantially reduced number of ABs could be due to a defect in autophagosome formation or a block in autophagosome fusion with the vacuole. The apparent absence of accumulated autophagosomes in the cytosol (Figure 3A) favored the former explanation. However, to further address this question, we took advantage of a unique property of Atg8: Unlike most Atg proteins that dissociate from the completed autophagosome, a portion of Atg8 remains conjugated to the inner membrane of the autophagosome and is then delivered into the vacuole lumen following fusion (Geng and Klionsky, 2008b). Completed autophagosomes fuse with the vacuole in a Vam3-dependent manner (Darsow et al., 1997). In the vam3ts (temperature sensitive) mutant, completed autophagosomes cannot fuse with the vacuole and accumulate in the cytosol (Darsow et al., 1997); Atg8 is associated with completed autophagosomes and serves as a marker for these compartments (Kirisako et al., 1999). We observed multiple GFP-Atg8 puncta in starved vam3ts cells at the NPT (Figure 4A), whereas most starved wild-type cells only displayed one GFP-Atg8 dot per cell (Figure 4C). Therefore, examining the accumulation of GFP-Atg8 puncta in the vam3 mutant background allows us to trace the formation of autophagosomes. On the basis of this strategy, we expressed GFP-Atg8 in either a vam3ts mutant or a sec2 vam3ts double mutant by genomic integration and measured the accumulation of GFP-Atg8 puncta after starvation at the NPT. Instead of the 2-h starvation used in previous experiments, in this assay we used a relatively short period of incubation in SD-N to avoid the appearance of too many GFP-Atg8 dots in the cytosol, which would make quantification more difficult.
Figure 4.
Recruitment of Atg8 to the PAS is affected in the sec2-59 mutant. (A and B) In the vam3ts background, sec2-59 cells accumulate fewer GFP-Atg8 dots in the cytosol. After preinactivation, vam3ts (JGY169) and vam3ts sec2 mutants (JGY171, JGY172) expressing chromosomally integrated GFP-Atg8 were starved for 20 min at 37°C. Cells were fixed to avoid any movement of GFP-Atg8, and fluorescence microscopy was performed as described in Materials and Methods. Twelve Z-section images were projected to visualize all dots present throughout the entire cell. Representative images are shown in A, and quantification of the number of GFP-Atg8 puncta is shown in B. Error bar, SEM; n = 100. (C–E) PAS intensity of GFP-Atg8 is not affected in the sec2-59 mutant although fewer cells have GFP-Atg8 dots. The wild-type (JGY146) and sec2-59 (JGY183) GFP-Atg8 strains were incubated in SD-N for 20 min at 37°C, and cells were examined by microscopy. (C) Representative images after projection of 12 Z-section pictures. (D) The percentage of cells that had at least one GFP-Atg8 dot was determined. Error bar, SD from three independent experiments; n = 300. (E) Relative PAS intensity of GFP-Atg8. The intensity in wild-type cells was set to 100%, and the other value was normalized. Error bar, SEM; n = 50. Scale bar, 5 μm.
After 20-min starvation at the NPT, 98% of control cells (vam3ts; WT) showed GFP-Atg8 dots, whereas only 48 and 46% of sec2-59 vam3ts and sec2-41 vam3ts cells, respectively, had at least one GFP-Atg8 dot (Figure 4A). To normalize for the different percentages of punctate cells, we quantified the number of GFP-Atg8 puncta per cell, counting only those cells with at least one GFP-Atg8 dot. In wild-type (vam3ts) cells, there were 3.39 ± 0.16 (mean ± SEM, n = 100 cells) GFP-Atg8 puncta per cell (Figure 4B); with a long incubation time in starvation conditions, the number of GFP-Atg8 puncta elevated significantly and became impossible to count due to overlapping dots. In contrast, the sec2-59 vam3ts and sec2-41 vam3ts cells had only 1.34 ± 0.06 and 1.35 ± 0.06 (mean ± SEM, n = 100 cells) GFP-Atg8 puncta per cell, respectively (Figure 4B). At longer time points of starvation, the number of GFP-Atg8 puncta in these mutants barely increased (Figure S1), suggesting that the reduced number of GFP-Atg8 dots in the sec2 mutants was not due to a minor kinetic delay but rather reflected a severe reduction in autophagosome formation.
Recently, it has been shown that the amount of Atg8 at the PAS controls the level of autophagy by regulating the size of autophagosomes (Xie et al., 2008). On the basis of this result, we examined whether the amount of Atg8 at the PAS was affected in the sec2 mutant. Similar to the results in the vam3ts background, fewer sec2-59 cells harbored at least one GFP-Atg8 punctum in an otherwise wild-type background (Figure 4C). After a 20-min incubation in SD-N at the NPT, 69% of the wild-type cells had at least one GFP-Atg8 dot. In contrast, only 23% of sec2-59 cells had GFP-Atg8 puncta (Figure 4D). To measure the amount of Atg8 at the PAS, we took advantage of a recent finding that within a certain range the fluorescence signal intensity correlates with the protein amount (Wu and Pollard, 2005; Geng et al., 2008), which means we can measure the PAS fluorescence intensity of GFP-Atg8, and the corresponding value indicates the level of GFP-Atg8 at this site. Quantification of the data showed that after 20-min starvation at the NPT, the PAS intensity of GFP-Atg8 was the same in wild-type and sec2-59 cells (Figure 4E). This result suggested that although fewer sec2-59 cells have Atg8 puncta corresponding to the PAS, once Atg8 is recruited to this site it is ultimately recruited at a level similar to that in wild-type cells, suggesting a substantial kinetic delay rather than a complete block in Atg8 delivery to the site of autophagosome formation. Combined with the EM data indicating that the reduced number of autophagosomes that were generated in the sec2-59 mutant attained the normal size, this finding further supports the model that Atg8 plays a role in the regulation of autophagosome size during nonspecific autophagy (Geng and Klionsky, 2008a; Xie et al., 2009).
Sec2 Affects Atg9 Anterograde Movement
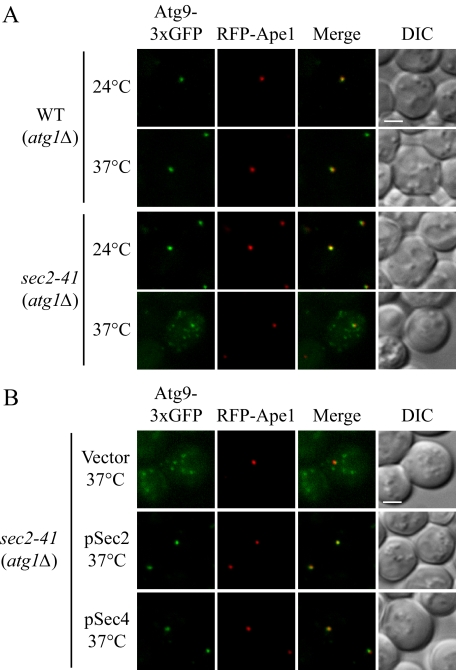
Recently, it was reported that Atg8 localization to the PAS is dependent on the presence of Atg9 (Suzuki et al., 2007). Of the core machinery required for autophagosome formation, Atg9 is the only transmembrane protein, and it shuttles between the PAS and other peripheral membrane structures (Noda et al., 2000; Reggiori et al., 2005). The anterograde movement of Atg9, from peripheral sites to the PAS, requires the presence of Atg27 (Yen et al., 2007). In an atg27Δ mutant the number of autophagosomes is reduced compared with wild-type cells upon the induction of autophagy, but the size of the autophagosomes is not affected. This phenotype is similar to what we observed in the sec2 mutant at the NPT. Therefore, we examined whether the anterograde transport of Atg9 was affected in the absence of functional Sec2.
To examine the cycling of Atg9 between the PAS and peripheral sites, we used the TAKA assay (Transport of Atg9 after Knocking out ATG1; Shintani and Klionsky, 2004). This epistasis analysis is based on a previous finding that the retrieval of Atg9 from the PAS to peripheral sites is dependent on the Atg1–Atg13 complex (Reggiori et al., 2004a). In an atg1Δ mutant, Atg9 is restricted at the PAS as a single dot in contrast to the multiple dots detected in wild-type cells. If the introduction of a second mutation in the atg1Δ background results in multiple Atg9 puncta, the second mutation is epistatic to atg1Δ, suggesting that the corresponding protein is required for the anterograde movement of Atg9. Using this strategy, we looked at Atg9 localization in an atg1Δ sec2-41 double mutant in starvation conditions. Atg9 was tagged with triple GFP at the C terminus, which does not affect the normal localization and functionality of Atg9 (He et al., 2008; Monastyrska et al., 2008), and the chimera appears to be stable based on the absence of a detectable free GFP band when examined by Western blot analysis; furthermore, GFP expressed by itself does not display the type of punctate distribution pattern seen with Atg9-3xGFP (our unpublished data). As expected, in the atg1Δ mutant, essentially 100% of the Atg9-3xGFP accumulated at the PAS (marked by RFP-Ape1) at both 24 and 37°C (Figure 5A). At the PT, the Atg9-3xGFP signal in the atg1Δ sec2-41 cells was localized to a single dot that colocalized with RFP-Ape1. However, when the cells were starved at the NPT, Atg9-3xGFP was localized to multiple dots (Figure 5A). In some cells, one of these dots colocalized with RFP-Ape1. These results suggested that in the sec2-41 mutant the anterograde transport of Atg9 was reduced, but not completely blocked. To extend our analysis, we analyzed the localization of Atg9 in the sec2-41 mutant in the presence of Atg1. After 2-h starvation at both 24 and 37°C, we observed the localization of Atg9 as multiple punctate structures in sec2-41 cells. Among all the cells at 24°C, 49% had one of the Atg9 dots colocalized with RFP-Ape1. In contrast, at 37°C only 26% of the cells showed any Atg9 colocalization at the PAS. This result was in agreement with the TAKA assay that Atg9 trafficking toward the PAS is partially defective in the sec2 mutant.
Figure 5.
Atg9 anterograde movement is less efficient in the sec2 mutant. (A) In the TAKA assay, Atg9 is localized to multiple dots in the sec2-41 mutant at the NPT. Wild-type (Atg9-3xGFP RFP-Ape1 atg1Δ, JGY197) and sec2-41 mutant (Atg9-3xGFP RFP-Ape1 atg1Δ, JGY107) cells were starved at the indicated temperature for 2 h. Cells were fixed and then analyzed by microscopy. 12 Z-section images were stacked, and representative images are shown. (B) Exogenous expression of Sec4 can restore the Atg9 anterograde movement defect in sec2-41 cells as shown by the TAKA assay. sec2-41 (Atg9-3xGFP RFP-Ape1 atg1Δ) cells were transformed with empty vector, pSec2(413) or pSec4(413). Microscopy samples were prepared, and pictures were taken as described in A. Scale bar, 2 μm.
It was reported that a second copy of SEC4 suppresses the secretion defect in the sec2-41 and sec2-59 mutants (Nair et al., 1990). There are other examples where overexpression of a GTPase complements, to some extent, the absence of the corresponding GEF. For instance, the autophagic activity in the sec12 mutant can be restored by the overproduction of Sar1 (Ishihara et al., 2001), and the expression of the constitutively active form of Ypt1 suppresses the Cvt pathway defect resulting from the absence of Trs85 (Lynch-Day et al., 2010). Accordingly, we examined whether an extra copy of Sec4 could restore the proper localization of Atg9 based on the TAKA assay. When empty vector was expressed in atg1Δ sec2-41 cells, there was no effect on Atg9 localization; Atg9-3xGFP was still detected in multiple dots (Figure 5B). As expected, plasmid-based Sec2 restored the Atg9 localization to a single dot at the PAS. When Sec4 was expressed on a plasmid under the control of its native promoter, the Atg9 signal again accumulated at the PAS. Therefore, the defect in anterograde Atg9 transport in sec2 cells could be rescued by the overexpression of Sec4.
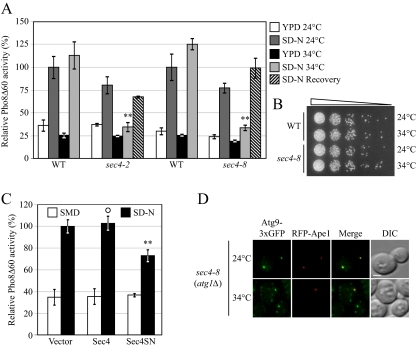
Autophagy Is Severely Compromised in sec4 Mutants
So far, Sec4 is the only Rab protein for which Sec2 has exchange activity. Because Sec4 overexpression complemented the sec2 phenotype in the TAKA assay, we hypothesized that Sec2 functions through Sec4 during autophagosome formation. To test this hypothesis, we studied the occurrence of autophagy in sec4 mutants. Previously it was published that sec4-2 shows a block in Pho8Δ60 activity when starved at 37°C, but the alkaline phosphatase activity could not be recovered when the cells were shifted back to 24°C. At a restrictive temperature of 34°C, sec4-2 shows both secretion and growth defects; however, at this temperature, autophagy is not affected (Ishihara et al., 2001). To examine this result, we constructed Pho8Δ60 strains bearing two sec4 alleles, sec4-2 and sec4-8. Both sec4 alleles were defective for autophagy induction based on the level of Pho8Δ60-dependent alkaline phosphatase activity at 34°C, and the defect could be mostly recovered when the cells were starved again at permissive temperature, indicating that the cells were not dead (Figure 6, A and B). To confirm that Sec4 participates in autophagy as a GTPase, we examined the function of Sec4S34N, a GDP-bound mutant of Sec4, in autophagy. When Sec4S34N was expressed in sec4-8 cells, it could not restore the Pho8Δ60 activity (our unpublished data). Furthermore, when Sec4S34N was overexpressed in wild-type cells, it showed a dominant-negative effect on the magnitude of autophagy. In Sec4S34N-overexpressing cells, the Pho8Δ60 activity was reduced to 73% compared with cells expressing an empty vector, whereas overexpression of wild-type Sec4 did not have a clear effect (Figure 6C). Thus, the activation of Sec4 is necessary for autophagy and the constitutively inactive mutant of Sec4 has a dominant-negative effect on the magnitude of autophagy.
Figure 6.
The sec4 mutant displays an autophagic defect. (A) Two sec4 alleles show a defect in Pho8Δ60 activity. The sec4-2 (JGY168) and sec4-8 (JGY159) mutants and the corresponding wild-type (JGY166, YTS158) strains were cultured in YPD medium at 24°C to midlog phase. Half of the culture was inactivated for Sec4 function at 34°C for 30 min, whereas the other half was kept growing at 24°C. Then cells were shifted to SD-N, and the temperature was maintained. Samples were collected before and after 2-h starvation. For recovery, cells starved at 34°C were shifted back to 24°C and incubated for another 2 h. Error bar, SD from three independent assays. Significant difference compared with corresponding wild-type cells, **p < 0.01. (B) The sec4 mutant is viable after 2-h starvation at 34°C. The same amount of wild-type (YTS158) and sec4-8 (JGY159) cells after 2-h starvation at the indicated temperature were diluted and spotted on a YPD plate. Cells were diluted 1:5 in each step from left to right. The plate was incubated at room temperature for 2 d. (C) Sec4S34N has a dominant-negative effect on autophagy. Wild-type cells (TN124) were transformed with an empty vector or a plasmid expressing either Sec4 or Sec4S34N under the control of the CUP1 promoter. Transformants were cultured in SMD medium at 30°C and shifted to SD-N for 2 h. Before and after starvation, samples were collected and tested by the Pho8Δ60 assay. Error bar, SD from three independent assays. Significant difference compared with vector alone, **p < 0.01; no significant difference, ○p > 0.1. (D) Atg9 movement to the PAS is defective in the sec4-8 mutant. sec4-8 (Atg9-3xGFP RFP-Ape1 atg1Δ, JGY198) cells were used for microscopy and the procedure was similar to that described in Figure 5A. Scale bar, 2 μm.
Next, we examined Atg9 trafficking in the sec4 mutant by the TAKA assay. At the PT, starved atg1Δ sec4-8 cells showed one single dot of Atg9-3xGFP, which was localized at the PAS. In contrast, when atg1Δ sec4-8 cells were starved at 34°C, Atg9-3xGFP was localized to multiple dots, although in some cells one of the dots colocalized with RFP-Ape1 at the PAS (Figure 6D). This phenotype was very similar to what was observed in sec2. Considering the complementation experiment using Sec4 overexpression (Figure 5D), this result suggested that Sec2 and Sec4 function as a GTPase–GEF complex in Atg9 anterograde movement. We further tested whether the overexpression of Sec4 can restore the autophagy activity in sec2 mutants. However, the overexpression of neither wild-type Sec4 nor a constitutively active mutant of Sec4 (Walworth et al., 1992) had a substantial effect on Pho8Δ60 activity in the sec2 mutant (our unpublished data). These results suggested that although a higher dosage of Sec4 could complement Atg9 movement in the sec2 mutant, there is another defect(s) in sec2 mutants that could not be suppressed by Sec4 overexpression.
Regulation between Protein Secretion and Autophagy
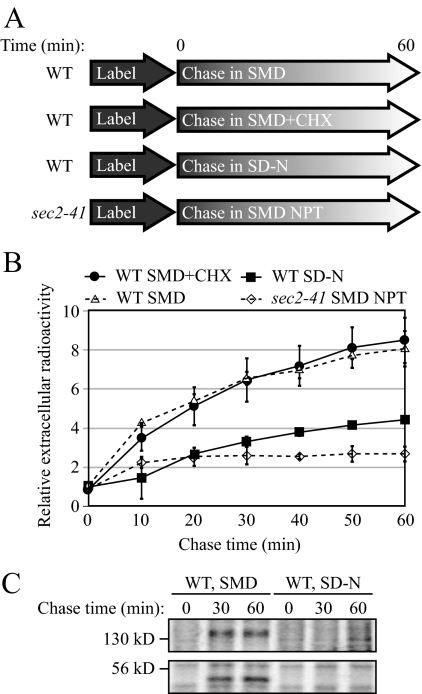
We have identified several genes involved in the late-stage secretory pathway that also play a role in autophagy. One hypothesis is that there is a regulatory mechanism between exocytosis and autophagy when the nutrient condition is changed. If so, similar post-Golgi machinery being used by two different pathways makes it possible to quickly switch the membrane flow from one pathway to the other in response to environmental changes. To investigate this hypothesis, we tested whether the level of protein secretion is down-regulated under starvation conditions. To measure the quantity of secreted proteins in either nutrient rich or starvation conditions, wild-type cells were labeled with [35S]methionine/cysteine and chased in either SMD or SD-N medium (Figure 7A). At each time point, the extracellular media was harvested, and the secreted proteins were collected by TCA precipitation. The pellet was then analyzed for radioactivity to determine the magnitude of protein secretion within a certain period of time.
Figure 7.
Protein secretion is down-regulated during starvation conditions. (A) Schematic representation of the procedure used for the pulse-chase experiments. (B) Relative radioactivity of the extracellular media. Cells were chased in the indicated media at 30°C, except for the sec2-41 cells, which were chased at 37°C. Samples were collected at the indicated time points. Extracellular proteins were precipitated and the amount of radioactivity quantified using a scintillation counter. The values at time zero were set to 1.0, and other values were normalized. Error bar, SD from three independent experiments. (C) Secretion of extracellular protein was examined by SDS-PAGE. Samples collected at 0, 30, and 60 min from B were separated by SDS-PAGE and visualized by autoradiography.
When wild-type cells were chased in nutrient-rich medium, the amount of secreted proteins increased dramatically over time, and radioactivity was elevated by eightfold after a 60-min chase (Figure 7B). As a negative control, sec2-41 cells chased at the NPT showed much less of an increase in the amount of secreted protein. When wild-type cells were chased in starvation medium, a very small amount of protein was secreted into the medium, and the secretion kinetics was comparable to that of sec2-59 at the NPT (Figure 7B). Protein synthesis is generally halted in starved cells. To exclude the possibility that the loss of protein secretion is because some newly synthesized proteins are necessary for exocytosis, we chased the wild-type cells in nutrient-rich medium in the presence of cycloheximide. Cycloheximide efficiently blocked protein synthesis (Figure S2). In nutrient-rich medium with cycloheximide, the protein secretion kinetics was not affected (Figure 7B) suggesting newly synthesized proteins were not required for exocytosis over the time course we utilized. To further confirm this result, we examined the protein pellet from the extracellular media by SDS-PAGE. After autoradiography, secreted proteins with molecular mass of ∼50 and 140 kDa were visible in wild-type cells chased in nutrient-rich medium (Figure 7C). In cells chased in starvation conditions, however, these bands were completely absent, indicating a block in protein secretion. Accordingly, these results demonstrated that protein secretion was down-regulated when cells encountered a shortage of nutrient.
Exit from the trans-Golgi Is Essential for Autophagy
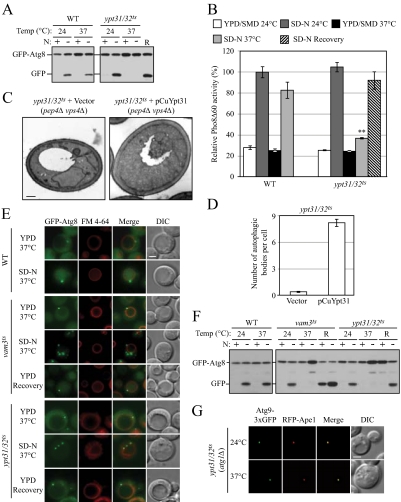
To further test the hypothesis that membrane flow from the Golgi complex is diverted into the autophagy pathway during starvation, we investigated whether vesicle exit from the Golgi complex is required for autophagy. Studies show that Ypt31/32 are required for the exit of secretory vesicles from the trans-Golgi and that these proteins are involved in the recruitment of Sec2-Sec4 to secretory vesicles (Jedd et al., 1997; Ortiz et al., 2002). Ypt32 also interacts with Sec2, although Sec2 does not have exchange activity for Ypt32 (Ortiz et al., 2002). Ypt32 shares a very similar amino acid sequence with Ypt31, and deletion of both genes is lethal (Benli et al., 1996). Thus, we investigated whether there was an autophagic defect in cells with ypt31Δ and a conditional ypt32 mutation (ypt31/32ts) and whether the phenotype was similar to that of sec2 and sec4 cells, based on the GFP-Atg8 processing assay. The ypt31/32ts mutant displayed a clear temperature-sensitive autophagy phenotype. At the NPT, the absence of free GFP after 2-h starvation indicated a defect of autophagy induction, and this defect was reversible when the cells were shifted back to the PT (Figure 8A). The failure of autophagy induction in ypt31/32ts was also observed by the Pho8Δ60 assay (Figure 8B). In addition, morphological analysis also confirmed the autophagic defect when the function of Ypt31/32 was disrupted. In the ypt31/32ts mutant, ABs could hardly been seen by transmission electron microscopy after 2-h starvation at the NPT (Figure 8C); the average number of ABs was 0.40 ± 0.12 (mean ± SEM, n = 75 vacuoles; Figure 8D). When plasmid-based Ypt31 was overexpressed in this mutant, the accumulation of ABs within the vacuole was restored; under the same conditions, there were 8.22 ± 0.46 ABs per vacuole (mean ± SEM, n = 79 vacuoles; Figure 8, C and D). The complementation of autophagy activity by exogenous Ypt31 was further demonstrated by GFP-Atg8 processing (Figure S3A), suggesting the autophagic defect was a result of Ypt31/32 dysfunction.
Figure 8.
Autophagy defect and abnormal Atg8 localization in the ypt31/32ts mutant. (A) GFP-Atg8 processing is blocked in the ypt31/32ts mutant as shown by GFP-Atg8 processing. Wild-type (NSY128) and ypt31/32ts (NSY340) cells transformed with the pGFP-Atg8(316) plasmid were grown in SMD at 24°C. Samples were collected after incubation at 24 or 37°C and examined as described in Figure 1A. Recovery (R) refers to cells shifted back to 24°C after the 37°C incubation. (B) Pho8Δ60 assay in the ypt31/32ts mutant. Wild-type (JGY220) and ypt31/32ts (JGY209) cells were examined by the Pho8Δ60 assay. Error bar, SD of three independent experiments. Significant difference compared with wild-type, **p < 0.01. (C and D) On starvation, no autophagic bodies were accumulated in ypt31/32ts cells. ypt31/32ts pep4Δ vps4Δ (JGY222) cells harboring pCu416 or pCuYpt31(416) were cultured in SMD at 24°C. After inactivation for 30 min, cells were starved for 2 h at the NPT. EM samples were prepared, and EM data were analyzed as described in Figure 3. (C) Representative EM images. Scale bar, 500 nm. The number of autophagic bodies was quantified and shown in D. Error bars, SEM. (E and F) ypt31/32ts is defective in Atg8 localization. Wild-type (JGY217), vam3ts (JGY227), and ypt31/32ts (JGY210) cells were cultured in YPD medium at 24°C, shifted to 37°C for 30 min, and subsequently starved at 37°C. To recover the temperature-sensitive phenotype, vam3ts and ypt31/32ts cells were shifted back to 24°C and incubated in YPD or SD-N medium for another 2 h. (E) Representative fluorescence microscopy images. The vacuolar membrane was stained with FM 4-64 as described in Materials and Methods. For each condition, only one Z-section with a clearly defined vacuole was shown. Scale bar, 2 μm. (F) Samples taken at the indicated conditions were examined by immunoblotting using anti-YFP antibody. (G) Ypt31/32 are not involved in movement of Atg9 toward the PAS. ypt31/32ts (Atg9-3xGFP RFP-Ape1 atg1Δ, JGY192) mutant cells were examined using the TAKA assay and the procedure was similar to that described in Figure 5A. Scale bar, 2 μm.
Next we examined the localization of Atg8 in the ypt31/32ts mutant. In both wild-type and ypt31/32ts cells, GFP-Atg8 was mostly diffuse in the cytosol with a PAS punctum in nutrient-rich conditions (Figure 8E). After starvation at the NPT, in addition to a prominent PAS dot, wild-type cells had GFP signal accumulated within the vacuolar lumen. However, when yp31/32ts cells were starved at the NPT, the vacuole was relatively empty of GFP signal, suggesting a substantial reduction of autophagic flux. Furthermore, there were multiple GFP-Atg8 puncta in the cytosol of ypt31/32ts cells under starvation conditions (Figure 8E). Overexpression of Ypt31 in the ypt31/32ts mutant restored the distribution of GFP-Atg8 as a single PAS dot and also allowed vacuolar accumulation (Figure S3B). There are two possibilities to explain why Atg8 was localized to multiple dots in the ypt31/32ts strain: 1) Ypt31/32 is required for the fusion between formed autophagosomes and the vacuole. Thus, the multiple GFP-Atg8 dots in the ypt31/32ts mutant indicate the accumulation of autophagosomes in the cytosol; 2) Ypt31/32 is involved in double-membrane vesicle formation, and in the ypt31/32ts mutant Atg8 is mislocalized to aberrant membrane structures. To further characterize the property of the multiple GFP-Atg8 puncta in the ypt31/32ts mutant, cells were recovered in YPD medium at the PT. In this nutrient-rich condition no more autophagosomes would be formed. If there are completed autophagosomes accumulated in the cytosol, once cells are shifted back to the PT, even in nutrient-rich conditions, GFP-Atg8 dots will be delivered to the vacuole, resulting in a GFP signal within the vacuole. For example, in vam3ts cells GFP-Atg8 dots were accumulated in the cytosol after starvation at the NPT. When cells were recovered in YPD medium at the PT, the GFP signal was mostly enriched in the vacuolar lumen (Figure 8E), indicating that the completed autophagosomes rapidly fused with the vacuole (Figure S3C). In contrast, in ypt31/32ts mutant cells, after the recovery period GFP-Atg8 regained its localization of one PAS dot, but there was no vacuolar GFP signal (Figure 8E, S3C), suggesting that the GFP-ATG8 puncta detected at the NPT did not correspond to completed autophagosomes, but rather were incomplete/aberrant structures that were able to coalesce into a PAS at the PT. The presence or absence of the vacuolar GFP signal was confirmed by the analysis of GFP-Atg8 processing by immunoblot. After recovery in YPD, free GFP was detected in vam3ts but not in ypt31/32ts cells; when the recovery was carried out in starvation conditions, both vam3ts and ypt31/32ts cells showed cleavage of GFP-Atg8, but the intensity of the GFP band in the vam3ts cells was stronger than that in the ypt31/32ts mutant cells (Figure 8F). Therefore, the ypt31/32ts mutation may result in disruption of autophagosome formation.
In contrast to the sec2 or sec4 mutants, the ypt31/32ts mutant appeared similar to the wild type with regard to Atg9 localization. In the TAKA assay, at both the PT and NPT, ypt31/32ts atg1Δ showed a single Atg9-3xGFP punctum that colocalized with the PAS marker RFP-Ape1 (Figure 8G). This result indicates that in the ypt31/32ts mutant the Atg9 movement from peripheral sites to the PAS was not affected. In addition, we tested whether Atg9 was properly localized to its peripheral sites in ypt31/32ts mutant cells. In starvation conditions, Atg9 was localized to multiple dots in the ypt31/32ts mutant, and there was no clear difference between the localization patterns at the PT and NPT (Figure S3D). Therefore, the transport of Atg9 to its peripheral sites does not require Ypt31/32-dependent vesicle exit from the trans-Golgi complex.
DISCUSSION
In vesicle trafficking, Rab proteins and their downstream effectors coordinate consecutive steps of transport. In this article we reported that a post-Golgi Rab protein (Sec4) and its GEF (Sec2) both participate in the autophagy pathway. Our data suggest that the Sec2-mediated nucleotide exchange on Sec4 (from the GDP- to GTP-bound form) is essential for autophagy. Dysfunction of either protein resulted in a substantial reduction in autophagy. In addition, the GDP-bound mutant of Sec4 showed a dominant-negative effect on autophagy, which further confirmed its involvement in autophagy as a GTPase.
Sec4 and Sec2 as well as their downstream effectors during exocytosis have been studied extensively. By interacting with one of the exocyst complex components, Sec15, Sec2 and Sec4 direct the polarized transport of secretory vesicles toward the plasma membrane (Guo et al., 1999). In addition to Sec15, another effector protein of Sec4, Sro7, has also been identified (Grosshans et al., 2006). However, it remains unknown whether there is an autophagy-specific effector of Sec4. Sec15 can be excluded because it is dispensable for autophagy (Ishihara et al., 2001), and we also confirmed this result. The effector-binding domain on Sec4, which is essential for Sec4–Sec15 interaction, is required for autophagy (a sec4 allele harboring a mutation in this domain could not complement the autophagy defect in sec4-8; our unpublished data). On the basis of this finding, we speculate that an autophagy-specific effector can compete with Sec15 by binding the same region on Sec4 and switch the function of Sec2-Sec4 to the autophagy pathway. sec2-59 and sec2-78 show a stronger interaction with Sec15 compared with the wild-type form of Sec2 (Medkova et al., 2006); thus, it is possible that the secretion and autophagy defect in these two mutants is due to an insufficient pool of free Sec2.
We observed a down-regulation of protein secretion during starvation conditions. Because there are several molecular factors shared between secretion and autophagy, one simple explanation is that starvation-induced autophagy competes for machinery that it uses in common with the secretory pathway. If the “competition” model is correct, it can be predicted that protein secretion will also be reduced when autophagy is triggered by other stimuli besides starvation. The Tor inhibitor rapamycin can clearly activate the autophagy pathway, although its magnitude is lower than starvation-induced autophagy (Figure S4A). According to the “competition” model, the level of protein secretion in rapamycin-treated cells will be lower than control cells, but higher than starved cells. However, when cells were treated with rapamycin, the kinetics of protein secretion was not affected (Figure S4B). Therefore, the inhibition of protein secretion during starvation is not due to simple competition between two parallel pathways. In mammalian cells secretion is also inhibited under conditions of amino acid depletion (Shorer et al., 2005), suggesting a conserved mechanism in various species that turns off secretion in response to nutrient shortage.
Atg9 movement to the PAS is speculated to represent the membrane flow from potential membrane sources toward the autophagosome formation site (Reggiori et al., 2004a), and the amount of Atg9 affects the number of autophagosomes. On the other hand, the amount of Atg8 at the PAS is proposed to regulate membrane extension and determine the size of autophagosomes (Xie et al., 2008). In sec2 cells, Atg9 trafficking toward the PAS is defective, whereas the Atg8 protein level at the PAS ultimately attains that of the wild type (although the total number of cells with Atg8 puncta is substantially reduced). The combination of these two phenotypes results in the formation of fewer autophagosomes with normal size, as indicated by our EM analysis. During PAS assembly, Atg8 recruitment occurs at a relatively late stage (Suzuki et al., 2007). Thus, the lower percentage of Atg8 puncta-positive cells in the sec2 mutant may be a consequence of the defect in Atg9 transport to the PAS. Although the overexpression of Sec4 rescues Atg9 movement in the TAKA assay, it only increases the percentage of Atg8 puncta-positive cells slightly (our unpublished data). Because the TAKA assay mainly examines the anterograde movement of Atg9, it is possible that the proper cycling of Atg9 is not fully restored by Sec4 overexpression. This may explain why autophagy activity in sec2 mutants is not restored by Sec4 overexpression.
A surprising result in our analysis was the finding that the ypt31/32ts double mutant did not show an Atg9 localization defect. This result suggests that Atg9 may not exit from the post-Golgi en route to the PAS. Although the transport of Atg9 to the PAS is less efficient in sec2 and sec4 mutants, we still observed part of the Atg9 population colocalizing with, or in proximity to, mitochondria based on fluorescence microscopy. In contrast, in the sec2 and sec4 mutants there was no clear colocalization between Atg9 and Vrg4, a marker protein of the early Golgi. These results suggest that Sec2-Sec4 function is only required for Atg9 movement from peripheral sites toward the PAS during autophagy, but is not necessary for its transport through the late secretory pathway and localization to the peripheral sites. Nonetheless, it is not clear how Sec2 and Sec4 facilitate Atg9 movement.
During autophagosome formation, a continuous supply of lipid is necessary, and multiple membrane sources may be used. In this study, our results suggest that the Golgi apparatus may be, at least one of, the membrane source(s) for autophagosome formation through the action of Sec2-Sec4. Ypt31/32, involved in exit from the post-Golgi, were also required for autophagy, and other independent lines of evidence strengthen this hypothesis. For example, the COG complex and Sec7, which are essential for Golgi function, both participate in autophagy (Reggiori et al., 2004b; Yen et al., 2009; van der Vaart et al., 2010). In mammalian cells, a Golgi Rab protein, Rab33B, interacts with Atg16L and shows some colocalization with Atg16L and LC3 (the mammalian homolog of Atg8; Itoh et al., 2008). All these data suggest membrane flow from the Golgi to the PAS. There are at least two possibilities as to how the Golgi provides lipid flow to the PAS: 1) The Golgi-derived membrane first reaches an Atg9-positive structure and then is delivered to the PAS in an Atg9-dependent manner, or 2) the Golgi directly supplies membrane to the PAS. In any case, the emergence of vesicles from the post-Golgi may occur in a Ypt31/32-dependent manner, with the movement of membrane containing Atg9 dependent upon Sec2-Sec4. It is possible that multiple membrane sources are required for different stages of autophagy and/or that the various sites of membrane mobilization involve distinct sets of proteins. Further analysis needs to be carried out to elucidate the contributions of each organelle to autophagosome formation.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Dr. Lucy C. Robinson (Louisiana State University Health Sciences Center) and Dr. Charlie Boone (University of Toronto) for providing strains and Dr. Zhiping Xie (Nankai University, China) for constructing the vam3ts integrating plasmid. This work was supported by National Institutes of Health Public Health Service Grant GM53396 to D.J.K.
Abbreviations used:
- Ape1
aminopeptidase I
- Atg
autophagy-related
- Cvt
cytoplasm-to-vacuole targeting
- mApe1
mature aminopeptidase I
- NPT
nonpermissive temperature
- ORF
open reading frame
- PAS
phagophore assembly site
- prApe1
precursor aminopeptidase I
- PT
permissive temperature
- ts
temperature sensitive.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0969) on May 5, 2010.
REFERENCES
- Abeliovich H., Zhang C., Dunn W. A., Jr, Shokat K. M., Klionsky D. J. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell. 2003;14:477–490. doi: 10.1091/mbc.E02-07-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Wendland B., Estepa E. J., Emr S. D. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu P., Bryan J. D., Panek H. R., Jordan S. L., Forbrich B. M., Kelley S. C., Colvin R. T., Robinson L. C. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 2002;115:4957–4968. doi: 10.1242/jcs.00203. [DOI] [PubMed] [Google Scholar]
- Benli M., Doring F., Robinson D. G., Yang X., Gallwitz D. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 1996;15:6460–6475. [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Nair U., Geng J., Klionsky D. J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;19:668–681. doi: 10.1091/mbc.E07-08-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C.-W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Darsow T., Rieder S. E., Emr S. D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J.A.K.W., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Elkind N. B., Walch-Solimena C., Novick P. J. The role of the COOH terminus of Sec2p in the transport of post-Golgi vesicles. J. Cell Biol. 2000;149:95–110. doi: 10.1083/jcb.149.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengsrud M., Erichsen E. S., Berg T. O., Raiborg C., Seglen P. O. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur. J. Cell Biol. 2000;79:871–882. doi: 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- Geng J., Baba M., Nair U., Klionsky D. J. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J. Cell Biol. 2008;182:129–140. doi: 10.1083/jcb.200711112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Klionsky D. J. Quantitative regulation of vesicle formation in yeast non-specific autophagy. Autophagy. 2008a;4:955–957. doi: 10.4161/auto.6791. [DOI] [PubMed] [Google Scholar]
- Geng J., Klionsky D. J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 2008b;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans B. L., Andreeva A., Gangar A., Niessen S., Yates J. R., Brennwald P., Novick P. J. The yeast lgl family member Sro7p is an effector of the secretory Rab GTPase Sec4p. J. Cell Biol. 2006;172:55–66. doi: 10.1083/jcb.200510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., Novick P. J. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T. M., Morano K. A., Scott S. V., Klionsky D. J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Baba M., Cao Y., Klionsky D. J. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol. Biol. Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W.-L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicke L., Zanolari B., Pypaert M., Rohrer J., Riezman H. Transport through the yeast endocytic pathway occurs through morphologically distinct compartments and requires an active secretory pathway and Sec18p/N-ethylmaleimide-sensitive fusion protein. Mol. Biol. Cell. 1997;8:13–31. doi: 10.1091/mbc.8.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsimaki Y., Hirsimaki P., Lounatmaa K. Vinblastine-induced autophagic vacuoles in mouse liver and Ehrlich ascites tumor cells as assessed by freeze-fracture electron microscopy. Eur J. Cell Biol. 1982;27:298–301. [PubMed] [Google Scholar]
- Huang J., Klionsky D. J. Autophagy and human disease. Cell Cycle. 2007;6:1837–1849. doi: 10.4161/cc.6.15.4511. [DOI] [PubMed] [Google Scholar]
- Hutchins M. U., Klionsky D. J. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:20491–20498. doi: 10.1074/jbc.M101150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Fujita N., Kanno E., Yamamoto A., Yoshimori T., Fukuda M. Golgi-resident small GTPase Rab33B Interacts with Atg16L and modulates autophagosome formation. Mol. Biol. Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itzen A., Rak A., Goody R. S. Sec2 is a highly efficient exchange factor for the Rab protein Sec4. J. Mol. Biol. 2007;365:1359–1367. doi: 10.1016/j.jmb.2006.10.096. [DOI] [PubMed] [Google Scholar]
- Iwata J., Ezaki J., Komatsu M., Yokota S., Ueno T., Tanida I., Chiba T., Tanaka K., Kominami E. Excess peroxisomes are degraded by autophagic machinery in mammals. J. Biol. Chem. 2006;281:4035–4041. doi: 10.1074/jbc.M512283200. [DOI] [PubMed] [Google Scholar]
- Jedd G., Mulholland J., Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J. Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Hayashi N., Toh-e A., Banno I., Oshima Y. Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. Gene. 1987;58:137–148. doi: 10.1016/0378-1119(87)90036-9. [DOI] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W.-P., Stromhaug P. E., Klionsky D. J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Cueva R., Yaver D. S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992;119:287–299. doi: 10.1083/jcb.119.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Emr S. D. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989;8:2241–2250. doi: 10.1002/j.1460-2075.1989.tb08348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé S., Thiele D. J. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–153. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- Legakis J. E., Yen W.-L., Klionsky D. J. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- Liang Y., Morozova N., Tokarev A. A., Mulholland J. W., Segev N. The role of Trs65 in the Ypt/Rab guanine nucleotide exchange factor function of the TRAPP II complex. Mol. Biol. Cell. 2007;18:2533–2541. doi: 10.1091/mbc.E07-03-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lynch-Day M. A., Bhandari D., Menon S., Huang J., Cai H., Bartholomew C. R., Brumell J. H., Ferro-Novick S., Klionsky D. J. Trs85 directs a Ypt1 GEF, TRAPP III, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA. 2010;107:7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medkova M., France Y. E., Coleman J., Novick P. The rab exchange factor Sec2p reversibly associates with the exocyst. Mol. Biol. Cell. 2006;17:2757–2769. doi: 10.1091/mbc.E05-10-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Warner J. R. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A. D., Li Z., Klionsky D. J. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 2008;19:1962–1975. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J., Müller H., Peterson M., Novick P. J. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J. Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I., et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nanduri J., Mitra S., Andrei C., Liu Y., Yu Y., Hitomi M., Tartakoff A. M. An unexpected link between the secretory path and the organization of the nucleus. J. Biol. Chem. 1999;274:33785–33789. doi: 10.1074/jbc.274.47.33785. [DOI] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W.-P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D. J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Ortiz D., Medkova M., Walch-Solimena C., Novick P. J. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D. J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.-W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. V., Hefner-Gravink A., Morano K. A., Noda T., Ohsumi Y., Klionsky D. J. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl. Acad. Sci. USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorer H., Amar N., Meerson A., Elazar Z. Modulation of N-ethylmaleimide-sensitive factor activity upon amino acid deprivation. J. Biol. Chem. 2005;280:16219–16226. doi: 10.1074/jbc.M500554200. [DOI] [PubMed] [Google Scholar]
- Stevens T., Esmon B., Schekman R. Early stages in the yeast secretory pathway are required for transport of carboxypeptidase Y to the vacuole. Cell. 1982;30:439–448. doi: 10.1016/0092-8674(82)90241-0. [DOI] [PubMed] [Google Scholar]
- Stromhaug P. E., Berg T. O., Fengsrud M., Seglen P. O. Purification and characterization of autophagosomes from rat hepatocytes. Biochem. J. 1998;335(Pt 2):217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Reggiori F., Guan J., Wang C.-W., Klionsky D. J. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vaart A., Griffith J., Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2010;21:2270–2284. doi: 10.1091/mbc.E09-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C., Collins R. N., Novick P. J. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J. Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth N. C., Brennwald P., Kabcenell A. K., Garrett M., Novick P. J. Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:2017–2028. doi: 10.1128/mcb.12.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-Q., Pollard T. D. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Xie Z., Nair U., Geng J., Szefler M. B., Rothman E. D., Klionsky D. J. Indirect estimation of the area density of Atg8 on the phagophore. Autophagy. 2009;5:217–220. doi: 10.4161/auto.5.2.7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Nair U., Klionsky D. J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Legakis J. E., Nair U., Klionsky D. J. Atg27 is required for autophagy-dependent cycling of Atg9. Mol. Biol. Cell. 2007;18:581–593. doi: 10.1091/mbc.E06-07-0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen W.-L., Shintani T., Nair U., Cao Y., Richardson B. C., Li Z., Hughson F. M., Baba M., Klionsky D. J. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J. Cell Biol. 2009;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.