The delivery of proteins and organelles to the vacuole by autophagy involves membrane rearrangements that result in the formation of autophagosomes. We have investigated the role of the Golgi in autophagy and found that, in yeast, this organelle plays a crucial role in supplying lipid bilayers necessary for autophagosome biogenesis.
Abstract
The delivery of proteins and organelles to the vacuole by autophagy involves membrane rearrangements that result in the formation of large vesicles called autophagosomes. The mechanism underlying autophagosome biogenesis and the origin of the membranes composing these vesicles remains largely unclear. We have investigated the role of the Golgi complex in autophagy and have determined that in yeast, activation of ADP-ribosylation factor (Arf)1 and Arf2 GTPases by Sec7, Gea1, and Gea2 is essential for this catabolic process. The two main events catalyzed by these components, the biogenesis of COPI- and clathrin-coated vesicles, do not play a critical role in autophagy. Analysis of the sec7 strain under starvation conditions revealed that the autophagy machinery is correctly assembled and the precursor membrane cisterna of autophagosomes, the phagophore, is normally formed. However, the expansion of the phagophore into an autophagosome is severely impaired. Our data show that the Golgi complex plays a crucial role in supplying the lipid bilayers necessary for the biogenesis of double-membrane vesicles possibly through a new class of transport carriers or a new mechanism.
INTRODUCTION
It is crucial for the cell that unwanted and damaged structures are efficiently eliminated from the cytoplasm to maintain cellular homeostasis. Autophagy plays a major role in this process of removal, because in addition to proteins, it also allows the degradation of multiprotein complexes such as ribosomes or entire organelles such as mitochondria or endoplasmic reticulum (ER) (van der Vaart et al., 2008; Kraft et al., 2009). This highly conserved pathway is characterized by the formation of large double-membrane vesicles called autophagosomes that sequester the cargo destined for degradation (Xie and Klionsky, 2007). Once the autophagosome is completed, it docks and fuses with the vacuole/lysosome, allowing the internal vesicle, called the autophagic body, to be liberated into the interior of this organelle where, together with the cargo, it is degraded by resident hydrolases. This leads to the generation of new building blocks that can be reused for the synthesis of new cellular components. Autophagy was originally identified as a response to starvation, but numerous other functions have been identified more recently. For example, it is now known that this pathway plays a role in immunity, aging and development (Cecconi and Levine, 2008; Vyas et al., 2008; Vellai, 2009). Furthermore, autophagy is also important in a number of pathologies, including neurodegenerative disorders, cancer, and myopathies (Mizushima et al., 2008; van der Vaart et al., 2008).
The use of Saccharomyces cerevisiae as a model system has generated a substantial amount of information about the molecular mechanism of autophagy. Thirty-one autophagy-related (ATG) genes have been identified so far, but the precise molecular function of the Atg proteins remains largely unknown. On autophagy induction, most of the Atg proteins are recruited to a perivacuolar location called the phagophore assembly site (PAS), the place where autophagosomes are generated. Autophagosome biogenesis can be divided into at least four distinct steps: 1) induction of the PAS assembly, which includes Atg protein recruitment; 2) generation and the expansion by lipid bilayer acquisition of an initial cisterna called a phagophore; 3) vesicle completion, leading to the autophagosome formation; and 4) fusion of the autophagosome with the vacuole. The exact molecular mechanism underlying these four steps remains to be fully understood, but it is clear that the delivery of membranes to the PAS is of crucial importance because lipid bilayers are the major component of autophagosomes (Reggiori, 2006). To date, it is unknown how the membranes necessary for the formation of the autophagosome are recruited to the PAS and where they originate from. This is largely due to a lack of specific protein markers that can be used to monitor this event and consequently several contradicting reports about the membrane origin have been published previously (Reggiori, 2006).
The Golgi complex is a highly dynamic organelle, consisting of an intricate network of cisternae, tubules, and associated vesicles, and it is one of the principal places in the cell where the sorting of newly synthesized proteins and lipids occurs. Proteins are mainly directed toward either the plasma membrane and extracellular space or intracellular organelles such as endosomes or lysosomes/vacuoles. There are some indications that the Golgi complex plays an important role in autophagy (Geng et al., 2010). In particular, it has been shown that the yeast guanine nucleotide exchange factor (GEF) Sec7, which localizes to the late Golgi and is indispensable for secretion (Novick et al., 1980; Franzusoff and Schekman, 1989), is essential for autophagy (Reggiori et al., 2004b). It is unknown, however, whether Sec7 has a role in autophagy that is independent from its function at the Golgi. Sec7 is one of the GEFs for the Golgi-localized ADP-ribosylation factors (Arfs), which are small GTPases important for the regulation of membrane dynamics in eukaryotic cells. Arf proteins exist in two forms, an inactive guanosine diphosphate (GDP)-bound and an active guanosine triphosphate (GTP)-bound form. GEF activities facilitate the activation of the Arf GTPases. In particular, binding to GTP triggers a conformational change in the Arf protein that leads to the release of the myristoylated N-terminus from its core. This free myristoyated tail associates with lipid bilayers and as a result, the Arf effectors are recruited to membranes (Casanova, 2007). The most common function of the Arf proteins is to promote the formation of complete, cargo-containing transport vesicles at different sites in the cell. As a consequence, for example, deletion of the Golgi Arf proteins blocks the transport through this organelle (Gaynor et al., 1998). In accordance with the role of Arf proteins in membrane trafficking, vesicle coat complexes such as the coatomer and adaptor proteins are among the Arf effector proteins (Nie et al., 2003).
Because the Golgi complex plays a major role in membrane sorting, we have explored the role of this organelle in yeast autophagy. To this end, we have analyzed whether Sec7 and its main downstream effectors are involved in this pathway. We discovered that a block of Golgi transport functions severely impairs autophagy. In particular, Sec7 and Golgi Arf proteins are essential for autophagy. Inactivation of these proteins does not prejudice the correct assembly of the Atg machinery at the PAS, but rather blocks the expansion of the phagophore. Our data thus highlight the relevance of the Golgi system in providing lipid bilayers and possibly proteins necessary for the formation of autophagosomes.
MATERIALS AND METHODS
Strains, Plasmids, and Media
The S. cerevisiae strains used in this study are listed in Table 1. For gene disruptions, the entire PEP4, ATG1, and PHO13 coding regions were replaced with either the Escherichia coli KANr, the Saccharomyces pombe HIS5, or the Kluyveromyces lactis LEU2 gene by using polymerase chain reaction (PCR) primers containing ∼60 bases of identity to the regions flanking the open reading frame. The vector to integrate pho8Δ60 at the PHO8 locus was a kind gift of Dr. Takahiro Shintani (Laboratory of Bioindustrial Genomics, Tohoku University, Sendai, Japan), whereas the plasmid allowing the integration of RFP-APE1 (pPS130) has been described previously (Stromhaug et al., 2004). Integrations of the dsRED and GFP genes at the 3′ end of SEC7 and ATG9, respectively, were performed as described previously (Longtine et al., 1998; Losev et al., 2006). Western blot analyses using specific antibodies or inspection of the loci by PCR were used to confirm all deletions and integrations.
Table 1.
Strains used in this study
| Name | Genotype | Reference/origin |
|---|---|---|
| AFM69-1A | MATa sec7-4 his3,11-15 leu2-3,112 ura3-1 | Franzusoff and Schekman (1989) |
| APY022 | MATα ura3-52 leu2-3,112 his3-200 lys2-801 ade2-101 | Peyroche et al. (2001) |
| gea2Δ::HIS3 gea1-6 | ||
| AVY004 | AFM69-1A pep4Δ::LEU2 | This study |
| AVY006 | SEY6210 SEC7-dsRED::URA3 | This study |
| AVY007 | SSY146 pep4Δ::KAN | This study |
| AVY012 | AFM69-1A SEC7ts-dsRED::URA3 | This study |
| AVY037 | FLY89 pep4Δ::LEU2 | This study |
| AVY042 | FLY89 pho8Δ60::HIS5 pho13Δ::KAN | This study |
| AVY046 | AFM69-1A SEC7ts-dsRED::URA3 ATG9-GFP::HIS5 | This study |
| AVY047 | SEY6210 RFP-APE1::LEU2 | This study |
| AVY048 | AFM69-1A RFP-APE1::LEU2 | This study |
| AVY065 | AFM69-1A ATG9-GFP::HIS5 atg1Δ::URA3 | This study |
| erg6Δ | BY4742 erg6Δ::KAN | Euroscarf |
| FLY89 | MATa ura3 leu2 his3 trp1 suc2Δ9 ret3-1 | Cosson et al. (1996) |
| FRY162 | SEY6210 ATG9-GFP::HIS5 | Reggiori et al. (2005) |
| FRY170 | SEY6210 ATG9-GFP::HIS5 atg1Δ::URA3 pep4Δ::LEU2 | This study |
| FRY326 | AFM69-1A ATG9GFP::HIS5 | This study |
| FRY341 | SEY6210 SEC7-dsRED::URA3 ATG9-GFP::TRP1 | This study |
| PC130 | MATa ura3 leu2 his4 lys, suc2Δ9 ret2-1 | Cosson et al. (1996) |
| SEY6210 | MATa ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 | Robinson et al. (1988) |
| suc2-Δ9 mel GAL | ||
| SSY146 | MATa arf1-4::HIS3 arf2Δ::URA3 ura3 leu2 lys2 bar1 his3 | Lay et al. (2005) |
| TVY1 | SEY6210 pep4Δ::LEU2 | Gerhardt et al. (1998) |
| YTS113 | SEY6210 atg1Δ::HIS5 pep4::LEU2 | Shintani et al. (2002) |
| WHY001 | SEY6210 atg1Δ::HIS5 | Shintani et al. (2002) |
The pCuGFPATG8414 and pCuGFPATG8416 plasmids that express the GFP-Atg8 fusion under the control of the CUP1 promoter have been described previously (Kim et al., 2002), as well as the pTS112 vector expressing GFP-Atg2 under control of the ATG2 authentic promoter and the one carrying the ATG1ts allele (Shintani et al., 2001; Suzuki et al., 2001).
Yeast cells were grown in rich (YPD; 1% yeast extract, 2% peptone, 2% glucose) or synthetic minimal media (SMD; 0.67% yeast nitrogen base, 2% glucose, amino acids and vitamins as needed). Starvation experiments were conducted in synthetic medium lacking nitrogen (SD-N; 0.17% yeast nitrogen base without amino acids, 2% glucose) or by addition of 0.2 μg/ml rapamycin to the culture (Suzuki et al., 2007). For the experiment with brefeldin A (BFA), the erg6Δ cells were grown to an early logarithmic (log) phase at 30°C, and 75 μg/ml BFA diluted in methanol was added to the growth medium (Graham et al., 1993).
Fluorescence Microscopy
Cells were grown in SMD medium lacking the appropriate auxotrophic substance to an early log phase or treated with rapamycin for 1 h. Fluorescence signals were visualized with a DeltaVision RT fluorescence microscope (Applied Precision, Issaquah, WA). The images were captured with a CoolSNAP camera and deconvolved using SoftWoRx software (Applied Precision).
Electron Microscopy
Preparations for electron microscopy (EM) analysis were made as described previously (Griffith et al., 2008). In brief, early log phase cells grown in YPD medium at 24°C were either shifted to 37°C for 2 h or transferred to the SD-N medium for 2 h at either 24 or 37°C. Cells were collected by centrifugation at the beginning and at the end of each treatment before being fixed with potassium permanganate, dehydrated with acetone, and embedded in Spurr's resin. Subsequently, 55- to 65-nm sections were cut with a diamond knife mounted on an Ultracut E ultramicrotome (Leica Microsystems, Vienna, Austria). Sections were finally contrasted with uranyl acetate and lead citrate before being viewed with a 1200 electron microscope (JEOL, Tokyo, Japan). The number of autophagic bodies per vacuole was determined as follows. Two different grids with sections obtained from the same preparation were evaluated. For every grid, the number of autophagic bodies in 60 cells with apparent vacuoles was counted. Error bars represent the SD from the counting of the two grids.
For immunoelectron microscopy (IEM) analysis, the sec7ts cells were grown to an early log phase at 24°C before being transferred to SD-N medium for 1 h at 37°C and processed as described previously (Griffith et al., 2008). A polyclonal anti-Ape1 antiserum was used to localize the prApe1 oligomer (a kind gift from Maria Mazón and Ignacio Sandoval, Universidad Autónoma de Madrid).
Miscellaneous Procedures
The Pho8Δ60 assay, protein extraction, and Western blot analyses were conducted as described previously (Reggiori et al., 2003). A monoclonal anti-green fluorescent protein (GFP) antibody (Roche, Basel, Switzerland) was used for the examination of GFP by Western blot, and the results were quantified using an Odyssey infrared imaging system (LI-COR Biosciences, Lincoln, NE).
RESULTS
GEF Activity of Sec7 Is Required for Autophagy
The Golgi complex is vital for eukaryotic cells and deletions of genes carrying out essential functions for this organelle are consequently lethal. Yeast genetics has permitted the generation of numerous thermosensitive alleles (ts), which allowed studying the role of several essential genes. For example, sec7ts cells grow normally at the permissive temperature of 24°C, but when transferred to the restrictive temperature of 37°C, the secretory pathway is blocked (Franzusoff and Schekman, 1989). The same cells also have been exploited to assess the role of Sec7 in autophagy (Reggiori et al., 2004b). To study autophagy, we opted for the use of the GFP-Atg8 processing assay, which simply needs the transformation of a plasmid in the strain that has to be analyzed (Shintani and Klionsky, 2004; Klionsky et al., 2008). This assay exploits the fact that part of the Atg8 pool is present in the interior of autophagosomes and together with the autophagic body, this pool is transported into the vacuole lumen where it becomes accessible to resident hydrolases. As a result, when Atg8 is N-terminally tagged with GFP, vacuolar hydrolases cleave this fusion protein releasing GFP. Free GFP has a long half-life in the vacuole interior and therefore it can easily be detected by Western blot using specific antibodies. Because the rate of autophagosome formation and fusion with the vacuole massively increases when autophagy is induced, augmentation in the amount of free GFP over time allows monitoring the progression of this pathway.
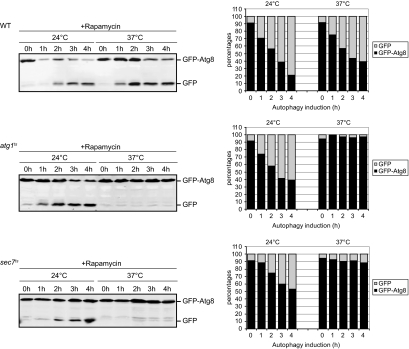
First, we tested whether we could observe the autophagy defect documented previously for sec7ts cells by using the GFP-Atg8 processing assay. Accordingly, a plasmid expressing GFP-Atg8 was transformed into the sec7ts mutant but also in a wild type (WT) strain and cells carrying a thermosensitive allele of ATG1 (atg1ts). Transformants were grown at 24°C, and after induction of autophagy by addition of rapamycin, cells were either kept at this temperature or transferred to 37°C. Culture aliquots were then collected at intervals of 1 h during a period of 4 h, and GFP-Atg8 cleavage was determined by Western blot analysis of the cell extracts. As expected, the appearance of a band corresponding to the 25-kDa free GFP was observed in WT cells after autophagy induction at both 24 and 37°C (Figure 1). Quantification of this band showed an increase in the amount of GFP over time, accompanied with a concomitant decrease of GFP-Atg8 levels, which reflects the normal progression of autophagy (Figure 1). Atg1 is a protein essential for autophagy and when the GFP-Atg8 processing assay was performed in the atg1ts mutant at permissive temperature, free GFP appeared in a time-dependent manner as in the WT strain. In contrast, no increase in the amount of GFP was detected at 37°C, revealing the block in autophagy of these cells. Importantly, a similar temperature-dependent block was obtained in sec7ts cells (Figure 1). Identical results were obtained when the same strains were nitrogen starved in SD-N medium, indicating no major difference between the autophagy induction by rapamycin treatment or by nitrogen deprivation (Supplemental Figure S1). Together, these data confirm that the autophagy impairment in the sec7ts mutant can be monitored using the GFP-Atg8 processing assay.
Figure 1.
Processing of GFP-Atg8 is blocked in the sec7 mutant. WT (SEY6210), atg1ts (WHY1 transformed with the plasmid expressing Atg1ts), and sec7ts (AFM69-1A) cells carrying the pCuGFPAtg8416 plasmid were grown at 24°C to an early log phase. Autophagy was then induced by addition of rapamycin, and cells were placed either at 24 or 37°C. Culture aliquots were collected at intervals of 1 h during a period of 4 h, and GFP-Atg8 cleavage was determined by Western blot analysis of the cell extracts. Bands were quantified using the Odyssey software, and the percentages of GFP-Atg8 (black) and GFP (gray) were plotted. Data for the sec7ts mutant graph represent the average of four experiments.
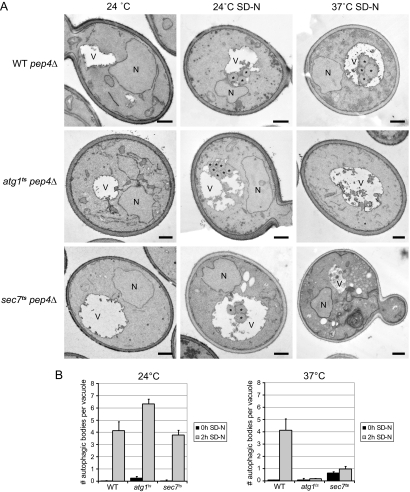
Next, we analyzed the sec7ts cells by EM to determine which step of autophagy is blocked. For example, the accumulation of complete autophagosomes in the cytoplasm indicates a block in the fusion of vesicles with the vacuole (Darsow et al., 1997). This approach also provides an alternative way to quantify autophagy, because it allows determining the number of autophagic bodies produced during a precise period of time (Bugnicourt et al., 2008). Autophagic bodies are rapidly degraded and therefore the gene coding for the main vacuolar protease PEP4 has to be deleted to make them detectable (Takeshige et al., 1992). WT, atg1ts, and sec7ts strains were grown in rich medium before being nitrogen starved for 2 h at permissive and restrictive temperature. Cells collected before and after nutrient deprivation were processed for EM, and autophagic bodies were counted as described in Materials and Methods. No autophagic bodies were observed when the three strains were grown in the presence of nitrogen (Figure 2A). In contrast, an average of four of these vesicles per vacuole was found when cells were starved for 2 h at 24°C. As expected, an identical result was obtained in WT cells at 37°C, whereas no autophagic bodies were counted in the atg1ts mutant under the same conditions, reflecting the autophagy block at restrictive temperature typical of this strain. The sec7ts cells also showed a temperature-dependent defect in autophagic body accumulation similar to the one observed in the atg1ts mutant (Figure 2B). This result is in complete agreement with the data obtained with the GFP-Atg8 processing assay. That is, the autophagic activity in the sec7ts strain is comparable to that of WT cells at 24°C, but it is blocked at restrictive temperature. Further investigation of the EM sections of sec7ts cells starved at 37°C revealed that there are no autophagosomes in the cytoplasm, indicating that the impairment of Sec7 function blocks their biogenesis rather than their fusion with the vacuole or their breakdown. The EM sections of the sec7ts strain revealed the presence of two cell populations with different morphologies. The first included cells with a previously defined ultrastructure of accumulating aberrant Golgi structures and ER proliferation, the reported phenotypes of the sec7 mutant (Novick et al., 1980). These were the cells used to count the number of autophagic bodies in the vacuole. The second population displayed an abnormal cytoplasmic density, lipid depositions, large aberrant membranous conformations and an overall altered yeast morphology. This phenotype has previously been observed in dying cells (Ludovico et al., 2001). The percentage of these cells strongly increased if the cultures were kept at 37°C under starvation conditions for longer periods (data not shown), suggesting that those are indeed dying cells.
Figure 2.
Electron microscopy analysis of WT, atg1ts, and sec7ts cells in nutrient rich and starvation conditions. (A) WT pep4Δ (TVY1), atg1ts pep4Δ (the YTS113 strain transformed with the plasmid expressing Atg1ts), and sec7ts pep4Δ (AVY004) cells were grown in rich medium at 24°C to an early log phase and then transferred to the SD-N medium for 2h at either 24 or 37°C. In parallel, cells were also placed at 37°C for 2 h. Culture aliquots were collected at the beginning and at the end of each incubation and processed for EM as described in Materials and Methods. (B) Quantification of the accumulated autophagic bodies. The results are expressed as the average number of autophagic bodies per vacuole. Error bars represent the SD in the counting of two different grids. N, nucleus; V, vacuole; *, autophagic body. Bar, 500 nm.
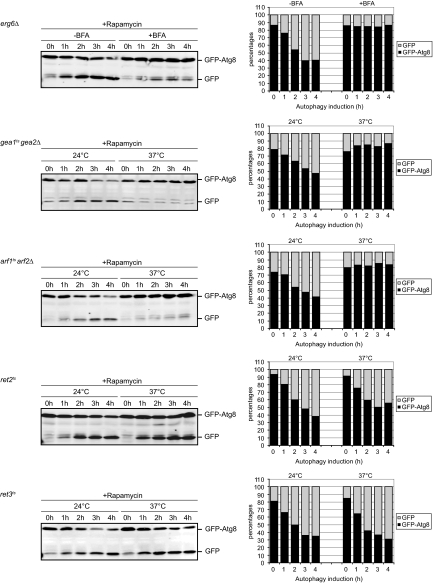
Sec7 is a large protein of ∼230 kDa, and so far all its cellular functions have been correlated to the GEF activity of its Sec7 domain. Consequently, we explored the role of the Sec7 domain in autophagy. The temperature sensitive allele of sec7 used in our experiments, e.g., sec7-4, codes for a protein with a single amino acid substitution in the Sec7 domain. Using an in vitro nucleotide exchange assay, it was determined that this mutation causes a loss of GEF activity at restrictive temperature (Jones et al., 1999). This suggests that the autophagy impairment observed in the sec7ts cells is probably due to a defect in GEF activity. However, because the point mutation is not in the catalytic site (Mossessova et al., 1998), it cannot be excluded that it provokes a more general conformational defect in the protein affecting other domains. To prove that the GEF activity of Sec7 is essential for autophagy, we used BFA. This fungal toxin stabilizes the interaction between Arf-GDP and the Sec7 domain and consequently inhibits the catalytic GEF activity (Peyroche et al., 1999; Jackson and Casanova, 2000). Because WT cells are impermeable to BFA, we made use of the erg6Δ strain for this experiment. The erg6Δ mutant has a defect in the biosynthesis of ergosterol, which renders the plasma membrane more permeable to chemicals (Graham et al., 1993). We triggered autophagy with rapamycin in erg6Δ cells carrying the plasmid expressing GFP-Atg8 in the absence or presence of BFA and GFP-Atg8 processing was determined by Western blot. As shown in Figure 3, autophagy was normal in the absence of BFA. In contrast, in the presence of BFA autophagy was blocked. Identical data were obtained when autophagy was induced by nitrogen starvation (data not shown). This result demonstrates that the GEF activity of Sec7 is necessary for autophagy.
Figure 3.
Activation of the Golgi Arf proteins by the GEF activity of Sec7, Gea1, and Gea2 is essential for autophagy. The erg6Δ, gea1ts gea2Δ (APY022), arf1ts arf2Δ (SSY146), ret2ts (PC130), and ret3ts (FLY89) cells carrying the pCuGFPAtg8416 or the pCuGFPAtg8414 plasmid were grown and processed as described in Figure 1, with the exception of erg6Δ, which was analyzed at 30°C. To inhibit all the Sec7 GEF activity, BFA was added to the erg6Δ mutant. Quantification of the GFP-Atg8 processing was done using an Odyssey system. Graphs represent an average of two experiments. Identical results were obtained when the same experiments were repeated, and autophagy was induced by nitrogen starvation in the SD-N medium (data not shown).
BFA, however, is not a specific inhibitor of Sec7. S. cerevisiae also expresses two uncharacterized Sec7-domain containing proteins and the nonessential GEF Syt1, which are sensitive to BFA. Interestingly, it was shown that the major targets of BFA in the secretory pathway of yeast, besides Sec7, are the two Gea proteins (Peyroche et al., 1999). Gea1 and Gea2 are 50% identical and are at least partially functionally redundant (Peyroche et al., 2001). They localize to the early Golgi cisternae, and they are involved in maintaining Golgi structure and function (Peyroche et al., 2001; Chantalat et al., 2003). Because BFA also inhibits the GEF activity of the Gea proteins, we could not rule out that this contributes to the autophagy block in BFA-treated cells. To investigate this, we used the thermosensitive gea1ts gea2Δ mutant (Peyroche et al., 2001) and performed the GFP-Atg8 processing assay. This revealed that the gea1ts gea2Δ strain also had a block in autophagy at restrictive temperature (Figure 3). These data indicate that Gea1 and Gea2 are essential for autophagy and suggest that the autophagy defect observed in BFA-treated cells is caused by a combined impairment of the GEF activity of Sec7, Gea1, and Gea2.
Role of Sec7 Downstream Effectors in Autophagy
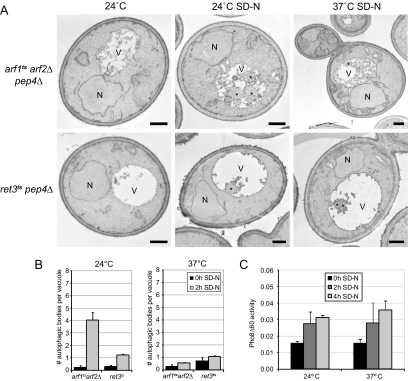
Interestingly, Sec7, Gea1, and Gea2 share at least one common downstream effector; the Arf GTPases (Casanova, 2007; Anders and Jurgens, 2008). Yeast possesses 3 Arf GTPases: Arf1, Arf2, and Arf3 (Gaynor et al., 1998). Arf1 and Arf2 are 96% similar in their amino acid sequence and have redundant functions in membrane transport through the Golgi (Gaynor et al., 1998; Mossessova et al., 1998), whereas Arf3 seems to be exclusively devoted to the establishment of cell polarity, actin organization, and endocytosis (Smaczynska-de Rooij et al., 2008; Tsai et al., 2008). Simultaneous deletion of ARF1 and ARF2 is lethal (Gaynor et al., 1998). To assess the role of the Golgi Arf proteins in autophagy, we made use of the thermosensitive arf1ts arf2Δ strain. We first analyzed the delivery of GFP-Atg8 into the vacuole lumen. As shown in Figure 3, normal processing of GFP-Atg8 was observed when autophagy was examined at permissive temperature. Alternatively, the appearance of free GFP was almost completely blocked when the same assay was performed at 37°C. We also analyzed the efficiency of autophagy in these cells by counting the number of autophagic bodies. This approach also showed that this process is severely impaired in arf1ts arf2Δ cells at restrictive temperature but not at 24°C (Figure 4, A and B). Together, these results reveal that the Golgi-associated Arf GTPases are necessary for autophagy.
Figure 4.
Analysis of autophagy in arf1tsarf2Δ and ret3ts cells. (A) arf1ts arf2Δ pep4Δ (AVY007) and ret3ts pep4Δ (AVY037) cells were grown in rich medium at 24°C to an early log phase and processed for EM after been transferred at 37°C for 2 h or starved in SD-N medium for 2 h at either 24 or 37°C. (B) Quantification of the accumulated autophagic bodies. The results are expressed as the average number of autophagic bodies per vacuole. Error bars represent the SD in the counting of two different grids. N, nucleus; V, vacuole; *, autophagic body. Bar, 500 nm. (C) Ret3 is not essential for autophagy. The ret3ts pho8Δ60 pho13Δ (AVY042) cells were grown at 24°C in rich medium to an early log phase before transfer into SD-N medium to induce autophagy. Cells were then incubated at either 24 or 37°C and culture aliquots were taken after 0, 2, and 4 h. The Pho8Δ60 assay was performed as described in Materials and Methods. Error bars represent the SD of three experiments.
To identify which downstream effectors of the Golgi Arf GTPases are involved in autophagy, we directed our attention to the best described interactors of the Arf proteins: The COPI coat. This oligomer forms a proteinaceous coat on vesicles and is involved in retrograde transport between Golgi cisternae and from the Golgi to the ER (Bethune et al., 2006). The coatomer subunits are recruited to the Golgi membranes and assemble into the COPI vesicle coat by activated, GTP-bound Arf proteins (Nie et al., 2003). The COPI coat is composed of seven subunits, and six of them are essential for cell viability. We tested the involvement of the coatomer in autophagy by analyzing the role of Ret2/δ-COP and Ret3/ξ-COP by using thermosensitive strains. Surprisingly, the GFP-Atg8 processing was normal in both ret2ts and ret3ts cells at permissive and restrictive temperature (Figure 3). The GFP cleavage was not caused by a secondary effect due to these mutations, but by the proteolytical activity in the vacuole because free GFP was not detected in the ret3ts strain when PEP4 was absent (Supplemental Figure 2). Consequently, this result indicates that Ret2 and Ret3, and by extension the coatomer, do not play a role in autophagy.
We then studied autophagy by EM in ret3ts cells, but we found that this process does not reach WT activity levels at permissive temperature; e.g., we counted an average of one autophagic body per vacuole instead of four after starvation induction (Figure 4B). The different genetic background of the analyzed strains is probably causing this discrepancy. Nevertheless, we also found an average of one autophagic body per vacuole when cells were nitrogen starved at restrictive temperature (Figure 4B). This observation indicates that Ret3 is not essential for autophagy. To confirm these data we next performed a more sensitive test to monitor autophagy: The Pho8Δ60 assay (Noda et al., 1995). Pho8 is a vacuolar alkaline phosphatase that, when N-terminally truncated (Pho8Δ60), resides in the cytoplasm in an unprocessed, enzymatically inactive form. Under starvation conditions, part of the Pho8Δ60 pool is delivered by bulk autophagy into the vacuole lumen where resident proteases process it into its active form (Noda et al., 1995). Pho8Δ60 activity can be colorimetrically measured and reflects the level of autophagy. As shown in Figure 4C and Supplemental Figure 3, autophagy is not impaired in ret3ts cells because the Pho8Δ60 enzymatic activity reaches the same levels after autophagy induction at permissive and restrictive temperatures by either nitrogen starvation or rapamycin treatment. This result further suggests that Ret3 and by extension the COPI coat are not required for autophagy.
Rud3, the homologue of GMAP-210, is so far the only other protein shown to interact with the activated Arf1 in yeast (Gillingham and Munro, 2007). We explored whether this factor is involved in autophagy by performing a GFP-Atg8 processing assay and by counting the number of autophagic bodies accumulated in absence of PEP4 in the rud3Δ deletant. We also carried out the same analyses in strains lacking the Golgi-localized γ-ear–containing Arf-binding proteins Gga1 and Gga2; the phospholipase D Spo14; and clathrin. The mammalian counterparts of these proteins are downstream effectors of the Golgi Arf proteins in higher eukaryotes (Nie et al., 2003). These assays revealed that none of the tested proteins are essential for autophagy (Supplemental Figure 4), indicating that a so far unidentified downstream effector of Golgi-localized Arf proteins plays a crucial role in this catabolic process.
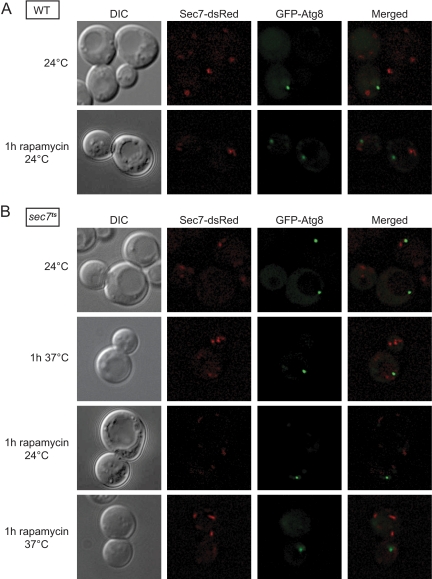
Sec7 Maintains Its Subcellular Distribution during Autophagy
Next, we asked whether Sec7 executes its function(s) in autophagy at the PAS or at a different location. We genomically tagged SEC7 with the gene coding for dsRed to obtain endogenous expression levels of the fusion protein (Losev et al., 2006). Then, we transformed the strain with a plasmid expressing GFP-Atg8, a protein marker for the PAS (Suzuki et al., 2007), and we performed fluorescence microscopy-based localization studies. As expected, when WT cells carrying GFP-Atg8 were grown in rich medium, Sec7-dsRed was distributed to several cytoplasmic puncta, which were previously shown to be trans-Golgi compartments (Figure 5A). Importantly, no colocalization between Sec7-dsRed and GFP-Atg8 was observed. We repeated the same experiment in conditions triggering autophagy by adding rapamycin for 1h and we obtained an identical result. Based on these observations, we concluded that Sec7 does not relocalize when autophagy is induced and that it does not associate with the PAS under any of the tested conditions.
Figure 5.
Sec7 does not localize to the PAS. (A) WT (AVY006) and (B) sec7ts (AVY012) cells expressing endogenous Sec7-dsRed and carrying the pCuGFPATG8416 plasmid were grown at 24°C to an early log phase. Rapamycin was then added and cultures were transferred to the indicated temperatures for 1 h. Alternatively, cells were incubated for 1 h at 37°C in absence of rapamycin. Cells were finally visualized by fluorescence microscopy. DIC, differential interference contrast.
However, the interaction between Sec7 and the PAS components could be transient. Therefore, we thought that the localization analysis of the thermosensitive form of Sec7 could permit us to observe its accumulation on a specific autophagosome intermediate. Accordingly, we genomically tagged sec7-4, the thermosensitive allele used in our experiments, with dsRed. When these cells were grown at permissive temperature, Sec7ts-dsRed has a subcellular distribution identical to that of the WT protein (Figure 5B). The intensity of the fluorescence signal, however, was less pronounced. This difference is caused by the mutation carried by the sec7-4 allele, which renders the encoded protein partially instable even at 24°C, leading to reduced cellular levels of mutant Sec7 (Deitz et al., 1996). When the cells were transferred to 37°C for 1h, no obvious change in Sec7ts-dsRed distribution was observed and this result indicates that the inactive mutant protein is still membrane bound and localizes to late Golgi compartments (Figure 5B). Autophagy induction by the addition of rapamycin for 1 h also did not change the subcellular distribution of the mutant Sec7 both at 24 and at 37°C (Figure 5B). This observation confirms that Sec7 does not associate with the PAS and maintains its normal localization under autophagy-inducing conditions. In addition, the fact that the inactive Sec7 mutant has a normal distribution strongly suggests that the contribution(s) of Sec7 to autophagy occurs at the Golgi.
Assembly of the Atg Machinery Is Not Affected in sec7ts Cells
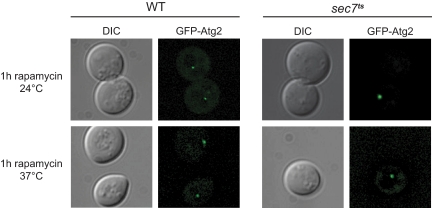
Because Sec7 does not localize to the PAS, it seems that this protein fulfils a crucial function in autophagosome biogenesis at the trans-Golgi compartments. Sec7 is generally involved in membrane sorting; consequently, a possibility is that this protein is carrying out a similar role during autophagy. Membranes are probably necessary for the Atg machinery assembly at the PAS but also for the successive expansion of the phagophore into a double-membrane vesicle. Thus, we first explored whether all the Atg components are correctly recruited to the PAS in the sec7ts cells. The organization of the Atg proteins at the PAS occurs in a hierarchical order, e.g., their recruitment depends on the presence of at least one other Atg factor (Suzuki et al., 2007). As a consequence, by studying the localization of a subset of Atg proteins, it is possible to draw a conclusion about the recruitment of all the Atg proteins to the PAS. Our data indicated that Atg8 retains its proper subcellular distribution in the sec7ts strain at restrictive temperature (Figure 5). Based on the studies of the hierarchical association of Atg proteins to the PAS, this observation does not exclude the possibility that other Atg proteins essential for autophagy are mislocalized in sec7ts cells. In particular, we could not rule out a defect in the recruitment of the Atg2–Atg18 complex (Suzuki et al., 2007); consequently, we analyzed Atg2 subcellular distribution in the sec7ts mutant. WT and sec7ts cells were transformed with a plasmid expressing Atg2 under its endogenous promoter and Atg2 localization was studied in cells treated for 1h with rapamycin both at 24 and 37°C. In the WT strain, Atg2 was present at the PAS, both before (data not shown) and after autophagy induction (Figure 6). This localization was not altered in sec7ts cells, indicating that there is a normal recruitment of Atg2 and by extension of Atg18 to the PAS (Obara et al., 2008). We concluded that Sec7 is not involved in the recruitment and the assembly of the Atg machinery at the PAS.
Figure 6.
Atg2 is correctly recruited to the PAS in the sec7ts mutant. The sec7ts strain (AFM69-1A) and the WT strain (SEY6210) transformed with the pTS112 vector (GFP-Atg2 under control of the ATG2 authentic promoter) were grown at permissive temperature to an early log phase before rapamycin was added and the cells were either kept at 24°C or transferred to 37°C for 1 h. Fluorescence microscopy was used to image the cells. DIC, differential interference contrast.
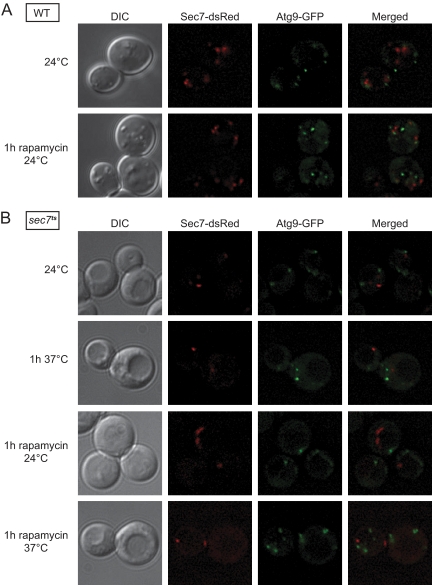
Atg9 Trafficking to the PAS Is Impaired in the sec7ts Strain
Atg9 is the only conserved transmembrane protein involved in double-membrane vesicle formation and plays an important role in generating the PAS (Suzuki et al., 2007; Mari, Griffith, Rieter, Krishnappa, Klionsky, and Reggiori, unpublished data). This protein concentrates into numerous cytoplasmic punctate structures and the relocalization of one or more of these compartments in proximity of the vacuole triggers the recruitment of the rest of the Atg proteins leading to the formation of the PAS (Shintani and Klionsky, 2004; Mari, Griffith, Rieter, Krishnappa, Klionsky, and Reggiori, unpublished data). Newly synthesized Atg9 passes through the Golgi complex before reaching its final destination (Mari, Griffith, Rieter, Krishnappa, Klionsky, and Reggiori, unpublished data). The Atg9-containing structures are thus directly or indirectly connected with the Golgi and consequently the inactivation of Sec7 and Arf proteins could cause an alteration of this compartment homeostasis. We genomically tagged ATG9 with GFP in the strains expressing WT and mutated Sec7-dsRed to investigate whether there is a major alteration in the Atg9 subcellular distribution after Sec7 inactivation. The fluorescence microscopy analyses were performed in growing conditions or under autophagy-induced conditions at both permissive and restrictive temperatures. As reported previously, Atg9-GFP was distributed into several punctate structures (Reggiori et al., 2004a). This localization pattern was identical in both strains and under all the tested conditions (Figure 7, A and B). We concluded that the inactivation of Sec7 does not affect Atg9 distribution.
Figure 7.
The assembly of the Atg machinery is not impaired in the sec7ts mutant. (A) WT (FRY341) and (B) sec7ts (AVY046) cells expressing Sec7-dsRed and Atg9-GFP under the control of their authentic promoters were grown at 24°C to an early log phase. Rapamycin was then added and cultures were transferred to the indicated temperatures for 1 h. Alternatively, cells were incubated for 1h at 37°C in the absence of this drug.
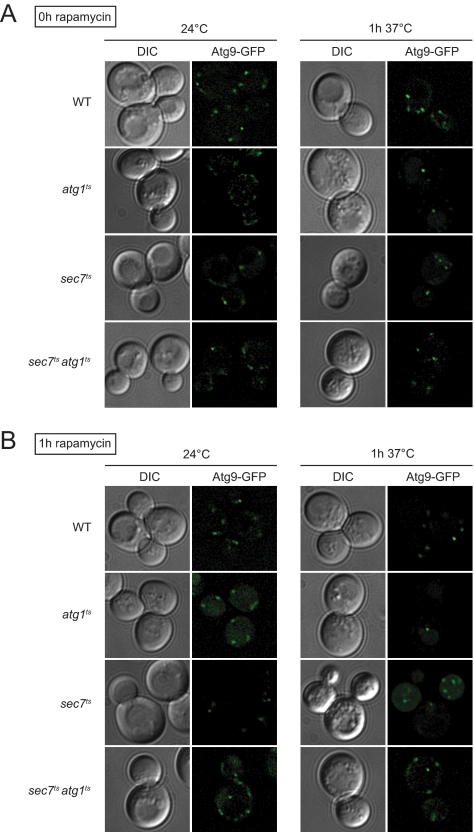
Once the biogenesis of a double-membrane vesicle is complete, Atg9 is retrieved from the PAS back to the cytoplasmic punctate structures (Noda et al., 2000; Reggiori et al., 2004a). It remains unclear whether Atg9 continuously shuttles between these two locations during the formation of a double-membrane vesicle or just at the beginning and the end of this process. That the Atg proteins are correctly assembled at the PAS in sec7ts cells (Figures 5 and 6) shows that at least one of the Atg9-containing compartments correctly relocates from the cytoplasm to the perivacuolar location that will become the PAS. Absence of Atg9 concentration in a single punctum (Figures 5 and 6) suggests that there is not a recycling defect of this protein from the PAS as well (Reggiori et al., 2004a). Our observations, however, could not exclude that additional Atg9 fails to reach the PAS after the initial formation of this specialized site because Atg9 distribution in several punctate structures also can be the result of an impairment of Atg9 anterograde transport to the PAS (Shintani and Klionsky, 2004; Reggiori et al., 2005b; Cheong et al., 2005; He et al., 2006; Reggiori and Klionsky, 2006; Monastyrska et al., 2008). To explore whether this Atg9 trafficking step is disrupted in the sec7ts strain, we decided to use the Transport of Atg9 after Knocking-out Atg1 (TAKA) assay that allows us to analyze the transport of this protein from the cytoplasmic punctate structures to the PAS (Shintani and Klionsky, 2004). In brief, this is an epistasis assay based on the observation that atg1Δ cells accumulate Atg9 almost exclusively at the PAS (Reggiori et al., 2004a). The atg1Δ ATG9-GFP strain carrying the atg1ts-expressing plasmid was grown at permissive temperature and then transferred to 37°C for 1 h in absence or presence of rapamycin. As reported previously (Reggiori and Klionsky, 2006; Monastyrska et al., 2008), Atg9-GFP was dispersed in several punctate structures when the strain was grown at 24°C, whereas the temperature shift caused this fusion protein to accumulate at the PAS as expected in both growing and autophagy conditions (Figure 8). In contrast, Atg9-GFP did not relocalized in the same way when the same experiment was performed in the sec7ts atg1ts double mutant, e.g., Atg9-GFP was still distributed to multiple punctate structures (Figure 8). This result indicates that a possible explanation for the autophagy defect observed in sec7 cells is the failure of additional Atg9 to reach the PAS.
Figure 8.
Atg9 trafficking is impaired the sec7ts mutant. (A) WT (FRY162), atg1ts (FRY170 transformed with the plasmid carrying the carrying the ATG1ts allele), sec7ts (FRY326) and sec7tsatg1ts (AVY065 transformed with the plasmid carrying the carrying the ATG1ts allele) cells expressing Atg9-GFP under the control of its authentic promoters were grown at 24°C to an early log phase before being transferred at 37°C for 1 h. Cells were imaged before and after the temperature shift. (B) The experiment described in A was repeated in presence of rapamycin to study Atg9 trafficking upon autophagy activation.
Phagophore Expansion Requires Sec7
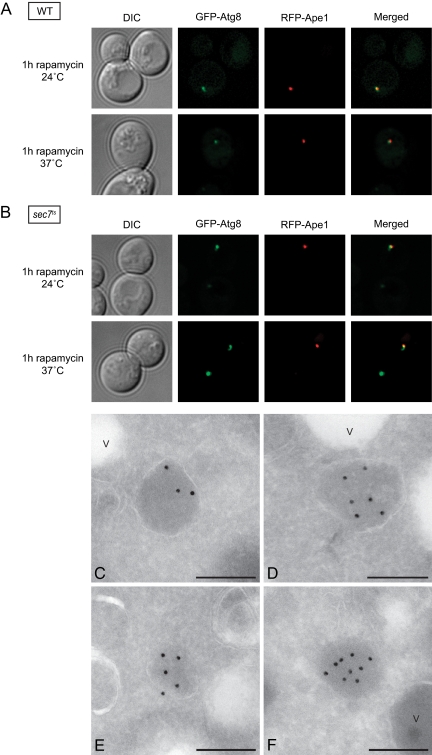
Our data show that the autophagy machinery is correctly assembled (Figures 5–7) but that autophagosomes are not formed (Figure 2A) in the absence of Sec7 function(s). This finding points to a defect in the formation, expansion, or both of the phagophore. The mechanism underlying these events is unknown and specific protein markers are not available. Consequently, the only experimental approach to follow phagophore formation and expansion is the ultrastructural analysis of the PAS by EM (Mari, Griffith, Rieter, Krishnappa, Klionsky, and Reggiori, unpublished data). To identify the PAS, we used the presence of the large electron-dense oligomers mostly formed by the precursor vacuolar protease Ape1 (prApe1) (Baba et al., 1997; Mari, Griffith, Rieter, Krishnappa, Klionsky, and Reggiori, unpublished data). After synthesis, prApe1 forms a large cytoplasmic oligomer that is transported into the vacuole by the cytoplasm to vacuole targeting (Cvt) pathway, a selective type of autophagy that operates in presence of nutrients (Baba et al., 1997; Shintani et al., 2002). Under starvation conditions, the prApe1 oligomer is delivered inside the vacuole by autophagosomes (Baba et al., 1997). The prApe1 oligomer colocalizes with the PAS to be sequestered into a double-membrane vesicle (Shintani et al., 2002). Before using it as a bona fide PAS marker in our ultrastructural analyses, we first verified by fluorescence microscopy whether the prApe1 oligomer is associated with the PAS in the sec7ts mutant at 37°C under starvation conditions. We generated WT and sec7ts strains coexpressing red fluorescent protein (RFP)-Ape1 and GFP-Atg8, and we imaged them after inducing autophagy by rapamycin addition for 1 h at either 24 or 37°C. In all conditions, the RFP-Ape1 was present in a single bright punctate structure that represents the prApe1 oligomer (Shintani et al., 2002). In WT cells, the RFP-Ape1 signal occasionally colocalized with that of GFP-Atg8 both under growing and starvation conditions (Figure 9A). This observation is in agreement with the fact that both the Cvt pathway and autophagy are rapid processes and the prApe1 oligomer only transiently localizes to PAS before being delivered into the vacuole. Importantly, the colocalization between RFP-Ape1 and GFP-Atg8 was much more prominent in sec7ts cells at restrictive temperature (Figure 9B). This phenotype is typical for numerous atg mutants where the prApe1 oligomer accumulates at the PAS because it fails to be sequestered into a double-membrane vesicle (Shintani et al., 2002). We concluded that the prApe1 oligomer is a suitable protein marker for the identification of the PAS in the sec7 mutant.
Figure 9.
Sec7 is essential for the phagophore expansion. Wild-type (strain AVY047; A) and sec7ts (strain AVY048; B) strains expressing RFP-Ape1 and carrying the pCuGFPATG8416 plasmid were grown at 24°C to an early log phase. Rapamycin was then added to the cultures, which were kept either at 24°C or transferred to 37°C for 1 h. Cells were finally visualized by fluorescence microscopy. The prApe1 oligomer is associated to the PAS in the sec7ts mutant at restrictive temperatures. (C–F) The sec7ts (AFM69-1A) strain was grown at 24°C to an early log phase before being transferred to 37°C for 1 h in the SD-N medium. Cells were processed for IEM as described in Materials and Methods and the PAS identified by labeling the cryosections with an anti-Ape1 antibody followed by protein A–15-nm gold particle conjugates. V, vacuole. Bar, 200 nm. DIC, differential interference contrast.
Next, the sec7ts strain was grown at permissive temperature before being transferred to 37°C for 1 h into starvation medium. Cells were then processed for IEM as described in Materials and Methods, and ultrathin cryosections were labeled with antibodies recognizing Ape1. As shown previously by conventional EM, inspection of the cryosections revealed two populations of sec7ts cells with different morphologies and also in this case, we focused on those without a degenerated aspect. In agreement with previous IEM studies, the prApe1 oligomer appeared as an electron-dense circular structure with a diameter of ∼150–200 nm (Figure 9, C–F; (Griffith et al., 2008; Mari, Griffith, Rieter, Krishnappa, Klionsky, and Reggiori, unpublished data). Astonishingly, a membrane was frequently seen surrounding the preApe1 oligomer (Figure 9, C and D). In some cases, the enclosure seemed to be incomplete (Figure 9E). These observations and the fact that the prApe1 oligomer is not associated with a lipid bilayer (Figure 8F) generate a view in which this proteinaceous complex is partially enwrapped into a cisterna. Based on this projection that is reminiscent of the structure of the mammalian phagophore (Mizushima et al., 2001; Sou et al., 2008) and our fluorescence microscopy data showing the presence of all Atg proteins at this site (Figures 5–7), we deduced that the observed membranous rearrangements are phagophores or partially expanded phagophores. Importantly, these structures had a smaller diameter than that of autophagosomes, 150–250 versus 400–900 nm (Baba et al., 1997), indicating a defect in the supply of lipid bilayers required to form a full-size autophagosome. Consequently, we concluded that Sec7 and by extension the Golgi complex are essential for the expansion of the phagophore.
DISCUSSION
Activation of Arf GTPases by Sec7 and the Gea Proteins at the Golgi Is Essential for Autophagy
To gain insight into the role of the Golgi complex in autophagy, we extended a previous study showing that Sec7 is necessary for autophagy (Reggiori et al., 2004b) to other factors mediating crucial tasks of this organelle. In particular, we wanted to reveal the exact role of Sec7 in autophagy. More precisely, it had remained unclear whether Sec7 Golgi functions play a role in autophagy or whether this GEF is also operating at the PAS. Furthermore, it is still unknown which part of the Sec7 protein was implicated in autophagy and that could be relevant to understand its precise molecular contribution. Thus, we looked into the involvement of Sec7 in autophagy and in particular into the role of its downstream effectors.
Using the specific GEF inhibitor BFA, we demonstrated that the GEF activity of Sec7 is essential for autophagy (Figure 3). Three yeast proteins in the secretory pathway have a Sec7 domain that is inhibited by BFA: Sec7, Gea1, and Gea2 (Peyroche et al., 2001). Unexpectedly, we have found that the two redundant Gea proteins are also essential for autophagy (Figure 3). Although Sec7 and the Gea proteins are localized at different Golgi compartments (Peyroche et al., 2001; Chantalat et al., 2003), they have the Arf GTPases as common effector proteins. Our data show that the arf1ts arf2Δ mutant has an autophagy defect (Figures 3 and 4A and 4B), indicating that the activation of Golgi Arf proteins by the GEFs Sec7, Gea1, and Gea2 plays a key role in autophagy. Sec7 does not colocalize with the PAS protein marker Atg8 under growing or starvation conditions (Figure 5), suggesting that the activation of Arf1 and Arf2 occurs at the Golgi complex. This supports the notion that one or more functions of the Golgi are essential for autophagy. The relevance of the membrane flow out of the Golgi in autophagy is also elicited by the fact that secretory mutants such as sec2ts, sec4ts, and ypt31tsypt32Δ display a strong impairment in this catabolic pathway (Geng et al., 2010). Not all the secretory mutants, however, seem to have a block in autophagy. For example, this process proceeds normally in pik1ts cells (data not shown), which fail to generate the phosphatidylintositol-4-phosphate necessary for the conventional transport through the Golgi (Audhya et al., 2000), suggesting that a different membrane exit mechanism out of the Golgi is crucial for autophagy.
Arf GTPases are important for vesicle formation (Nie et al., 2003; Gillingham and Munro, 2007) and it is plausible that they have a similar function in autophagy. Golgi-associated Arf GTPases are involved in the generation of both COPI- and clathrin-coated vesicles (Nie et al., 2003; Gillingham and Munro, 2007). Our analysis of the ret2ts, ret3ts, chc1Δ, and gea1Δ gea2Δ mutants, however, revealed that neither COPI nor clathrin-coated vesicle formation is required for autophagy in yeast (Figures 3 and 4 and Supplemental Figure 4). Our data are consistent with the recent report that in mammalian cells the coatomer is not necessary for the formation of autophagosmes but rather for its maturation by fusion with early endosomes (Razi et al., 2009), a step that seems not to occur in yeast (Reggiori et al., 2004b). There are other less well-characterized types of carriers exiting the Golgi and their formation could also require the Arf GTPases (De Matteis and Luini, 2008). Future studies are necessary to identify the Arf1 and Arf2 downstream effector(s) and the carriers essential for autophagy.
The Golgi Plays a Crucial Role in the Phagophore Expansion
The biogenesis of an autophagosome can be divided into four distinct steps, e.g., induction and assembly of the PAS, phagophore formation and expansion, autophagosome completion, and autophagosome fusion with the vacuole. Our fluorescence microscopy and IEM data show that the formation of both the PAS and the phagophore are not affected by the sec7-4 mutation (Figures 5, 6, and 9). Complete autophagosomes, however, are not formed in sec7ts and arf1ts arf2Δ cells (Figures 2, 4, and 9), revealing that exit from the Golgi is necessary for the expansion of the phagophore into an autophagosome.
How could a block in the exit from the Golgi complex interfere with the phagophore expansion? The formation of an autophagosome relies on the delivery of a large quantity of lipid bilayers to the PAS. Some of these lipid bilayers are initially supplied by the Atg9-containing structures (Mari et al., unpublished data), but the origin of the rest of them is still unknown. The correct assembly of the Atg proteins at the PAS in sec7 cells (Figures 5 and 6) shows that at least one of the Atg9-positive compartments is able to reach the perivacuolar location that will become the PAS. A possible hypothesis explaining the phagophore expansion block observed in the sec7 strain (Figure 9) could nevertheless be that the transport of additional Atg9-containing membranes to the PAS is essential for the completion of an autophagosome. Another likely hypothesis is that Golgi-derived vesicles carry the membranes necessary for the biogenesis of an autophagosome. This is supported by two recent works; (Geng et al., 2010), show that two other post-Golgi proteins beside Sec7, namely, the GEF Sec2 and its GTPase Sec4, are required for autophagy. Furthermore, another recent investigation has revealed that trans-Golgi–derived vesicles can fuse with the phagophore to form autophagosomes in an Atg5/Atg7-independent type of autophagy (Nishida et al., 2009). The fact that the growing extremities of the phagophores and a section of the complete autophagosome can be decorated with lectins that recognize glycans exclusively present in post-Golgi membranes supports the idea that Golgi-membranes are necessary for autophagosome formation (Yamamoto et al., 1990). It cannot be excluded a priori, however, that both Golgi-derived and Atg9-containing membranes are necessary for the phagophore expansion.
Alternatively, a dysfunctional Golgi could interfere with the homeostasis of another organelle that is directly involved in providing the lipid bilayers to the forming double-membrane vesicles. Atg8 becomes lipidated after autophagy induction and once the protein is conjugated to phosphatidylethanolamine it localizes to the PAS (Geng and Klionsky, 2008). However, Atg8 is also present on small vesicles in the cytoplasm (Kirisako et al., 1999). The origin of these vesicles is still unclear, but they could be derived from Golgi membranes. Interestingly, Atg8 has been detected on membranes of putative Golgi origin in sec7 cells kept at 37°C for 3h in presence of nutrients (Reggiori et al., 2004b). In addition, in mammalian cells the Atg8 orthologue GATE-16 localizes to the Golgi (Sagiv et al., 2000; Reggiori et al., 2004b).
In conclusion, our work has revealed a role for the Golgi complex in autophagy, in particular in the events leading to the expansion of the phagophore into an autophagosome. Future studies are needed to identify which factors essential for autophagy depend on Golgi functions. This information is crucial to understand the molecular mechanism of this vital pathway for the eukaryotic cell.
Supplementary Material
ACKNOWLEDGMENTS
We thank Ben Glick, Andreas Conzelmann, Daniel Klionsky, Catherine Jackson, Wilhelm Just, Maria Mazón, Yoshinori Ohsumi, Ignacio Sandoval, and Takahiro Shintani for reagents; Judith Klumperman, Daniel Klionsky, and Iryna Monastyrska for critical reading of the manuscript; and Marc van Peski and René Scriwanek for assistance with the preparation of the figures. F. R. is supported by the Netherlands Organization for Health Research and Development (ZonMW-VIDI-917.76.329) and by the Utrecht University (High Potential grant).
Abbreviations used:
- BFA
brefeldin A
- EM
electron microscopy
- GEF
guanine nucleotide exchange factor
- IEM
immunoelectron microscopy
- PAS
phagophore assembly site.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-04-0345) on May 5, 2010.
REFERENCES
- Anders N., Jurgens G. Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell Mol. Life Sci. 2008;65:3433–3445. doi: 10.1007/s00018-008-8227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A., Foti M., Emr S. D. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol. Biol. Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Osumi M., Scott S. V., Klionsky D. J., Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J. Cell Biol. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J., Wieland F., Moelleken J. COPI-mediated transport. J. Membr. Biol. 2006;211:65–79. doi: 10.1007/s00232-006-0859-7. [DOI] [PubMed] [Google Scholar]
- Bugnicourt A., Mari M., Reggiori F., Haguenauer-Tsapis R., Galan J. M. Irs4p and Tax4p: two redundant EH domain proteins involved in autophagy. Traffic. 2008;9:755–769. doi: 10.1111/j.1600-0854.2008.00715.x. [DOI] [PubMed] [Google Scholar]
- Casanova J. E. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- Cecconi F., Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantalat S., Courbeyrette R., Senic-Matuglia F., Jackson C. L., Goud B., Peyroche A. A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol. Biol. Cell. 2003;14:2357–2371. doi: 10.1091/mbc.E02-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell. 2005;16:3438–3453. doi: 10.1091/mbc.E04-10-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P., Demolliere C., Hennecke S., Duden R., Letourneur F. Delta- and zeta-COP, two coatomer subunits homologous to clathrin-associated proteins, are involved in ER retrieval. EMBO J. 1996;15:1792–1798. [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Rieder S. E., Emr S. D. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis M. A., Luini A. Exiting the Golgi complex. Nat. Rev. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Deitz S. B., Wu C., Silve S., Howell K. E., Melancon P., Kahn R. A., Franzusoff A. Human ARF4 expression rescues sec7 mutant yeast cells. Mol. Cell Biol. 1996;16:3275–3284. doi: 10.1128/mcb.16.7.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989;8:2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor E. C., Chen C. Y., Emr S. D., Graham T. R. ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell. 1998;9:653–670. doi: 10.1091/mbc.9.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Klionsky D. J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. Protein modifications: beyond the usual suspects review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nair U., Yasamura-Yorimitsu K., Klionsky D. J. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2010;21:2257–2269. doi: 10.1091/mbc.E09-11-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt B., Kordas T. J., Thompson C. M., Patel P., Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J. Biol. Chem. 1998;273:15818–15829. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
- Gillingham A. K., Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Graham T. R., Scott P. A., Emr S. D. Brefeldin A reversibly blocks early but not late protein transport steps in the yeast secretory pathway. EMBO J. 1993;12:869–877. doi: 10.1002/j.1460-2075.1993.tb05727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J., Mari M., De Maziere A., Reggiori F. A cryosectioning procedure for the ultrastructural analysis and the immunogold labelling of yeast Saccharomyces cerevisiae. Traffic. 2008;9:1060–1072. doi: 10.1111/j.1600-0854.2008.00753.x. [DOI] [PubMed] [Google Scholar]
- He C., Song H., Yorimitsu T., Monastyrska I., Yen W. L., Legakis J. E., Klionsky D. J. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. L., Casanova J. E. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Jones S., Jedd G., Kahn R. A., Franzusoff A., Bartolini F., Segev N. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics. 1999;152:1543–1556. doi: 10.1093/genetics/152.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang W. P., Stromhaug P. E., Klionsky D. J. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002;277:763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Reggiori F., Peter M. Selective types of autophagy in yeast. Biochim. Biophys. Acta. 2009;1793:1404–1412. doi: 10.1016/j.bbamcr.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Lay D., Grosshans B. L., Heid H., Gorgas K., Just W. W. Binding and functions of ADP-ribosylation factor on mammalian and yeast peroxisomes. J. Biol. Chem. 2005;280:34489–34499. doi: 10.1074/jbc.M503497200. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., Glick B. S. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- Ludovico P., Sousa M. J., Silva M. T., Leao C., Corte-Real M. Saccharomyces cerevisiae commits to a programmed cell death process in response to acetic acid. Microbiology. 2001;147:2409–2415. doi: 10.1099/00221287-147-9-2409. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto A., Hatano M., Kobayashi Y., Kabeya Y., Suzuki K., Tokuhisa T., Ohsumi Y., Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monastyrska I., He C., Geng J., Hoppe A. D., Li Z., Klionsky D. J. Arp2 links autophagic machinery with the actin cytoskeleton. Mol. Biol. Cell. 2008;19:1962–1975. doi: 10.1091/mbc.E07-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E., Gulbis J. M., Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human arno and analysis of the interaction with ARF GTPase. Cell. 1998;92:415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- Nie Z., Hirsch D. S., Randazzo P. A. Arf and its many interactors. Curr. Opin. Cell Biol. 2003;15:396–404. doi: 10.1016/s0955-0674(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Noda T., Kim J., Huang W. P., Baba M., Tokunaga C., Ohsumi Y., Klionsky D. J. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol. 2000;148:465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Matsuura A., Wada Y., Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Obara K., Sekito T., Niimi K., Ohsumi Y. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J. Biol. Chem. 2008;283:23972–23980. doi: 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C. L. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell. 1999;3:275–285. doi: 10.1016/s1097-2765(00)80455-4. [DOI] [PubMed] [Google Scholar]
- Peyroche A., Courbeyrette R., Rambourg A., Jackson C. L. The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 2001;114:2241–2253. doi: 10.1242/jcs.114.12.2241. [DOI] [PubMed] [Google Scholar]
- Razi M., Chan E. Y., Tooze S. A. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F. 1. Membrane origin for autophagy. Curr. Top. Dev. Biol. 2006;74:1–30. doi: 10.1016/S0070-2153(06)74001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J. Cell Sci. 2006;119:2903–2911. doi: 10.1242/jcs.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Monastyrska I., Shintani T., Klionsky D. J. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2005a;16:5843–5856. doi: 10.1091/mbc.E05-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D. J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005b;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004a;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C. W., Nair U., Shintani T., Abeliovich H., Klionsky D. J. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004b;15:2189–2204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Wang C. W., Stromhaug P. E., Shintani T., Klionsky D. J. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003;278:5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. S., Klionsky D. J., Banta L. M., Emr S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagiv Y., Legesse-Miller A., Porat A., Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell. 2002;3:825–837. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Suzuki K., Kamada Y., Noda T., Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem. 2001;276:30452–30460. doi: 10.1074/jbc.M102346200. [DOI] [PubMed] [Google Scholar]
- Smaczynska-de Rooij I., Costa R., Ayscough K. R. Yeast Arf3p modulates plasma membrane PtdIns(4,5)P2 levels to facilitate endocytosis. Traffic. 2008;9:559–573. doi: 10.1111/j.1600-0854.2008.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou Y. S., et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Takeshige K., Baba M., Tsuboi S., Noda T., Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P. C., Lee S. W., Liu Y. W., Chu C. W., Chen K. Y., Ho J. C., Lee F. J. Afi1p functions as an Arf3p polarization-specific docking factor for development of polarity. J Biol. Chem. 2008;283:16915–16927. doi: 10.1074/jbc.M802550200. [DOI] [PubMed] [Google Scholar]
- van der Vaart A., Mari M., Reggiori F. A picky eater: exploring the mechanisms of selective autophagy in human pathologies. Traffic. 2008;9:281–289. doi: 10.1111/j.1600-0854.2007.00674.x. [DOI] [PubMed] [Google Scholar]
- Vellai T. Autophagy genes and ageing. Cell Death Differ. 2009;16:94–102. doi: 10.1038/cdd.2008.126. [DOI] [PubMed] [Google Scholar]
- Vyas J. M., Van der Veen A. G., Ploegh H. L. The known unknowns of antigen processing and presentation. Nat. Rev. Immunol. 2008;8:607–618. doi: 10.1038/nri2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Masaki R., Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J. Histochem. Cytochem. 1990;38:573–580. doi: 10.1177/38.4.2319125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.