The RAB-5 and RAB-7 GTPases regulate endosome to lysosome trafficking. Here, we show that Caenorhabditis elegans TBC-2 functions as a RAB-5 GAP. TBC-2 colocalizes with RAB-7 on late endosomes, and requires RAB-7 for membrane localization where TBC-2 could function to antagonize RAB-5 activity during early to late endosome maturation.
Abstract
During endosome maturation the early endosomal Rab5 GTPase is replaced with the late endosomal Rab7 GTPase. It has been proposed that active Rab5 can recruit and activate Rab7, which in turn could inactivate and remove Rab5. However, many of the Rab5 and Rab7 regulators that mediate endosome maturation are not known. Here, we identify Caenorhabditis elegans TBC-2, a conserved putative Rab GTPase-activating protein (GAP), as a regulator of endosome to lysosome trafficking in several tissues. We show that tbc-2 mutant animals accumulate enormous RAB-7–positive late endosomes in the intestine containing refractile material. RAB-5, RAB-7, and components of the homotypic fusion and vacuole protein sorting (HOPS) complex, a RAB-7 effector/putative guanine nucleotide exchange factor (GEF), are required for the tbc-2(−) intestinal phenotype. Expression of activated RAB-5 Q78L in the intestine phenocopies the tbc-2(−) large late endosome phenotype in a RAB-7 and HOPS complex-dependent manner. TBC-2 requires the catalytic arginine-finger for function in vivo and displays the strongest GAP activity on RAB-5 in vitro. However, TBC-2 colocalizes primarily with RAB-7 on late endosomes and requires RAB-7 for membrane localization. Our data suggest that TBC-2 functions on late endosomes to inactivate RAB-5 during endosome maturation.
INTRODUCTION
Rab GTPases regulate intracellular trafficking by controlling the transport of vesicles between membrane compartments along the exocytic and endocytic pathways (Stenmark and Olkkonen, 2001; Zerial and McBride, 2001; Stenmark, 2009). Acting as molecular switches, Rabs alternate between an “active” guanosine triphosphate (GTP)-bound state and an “inactive” guanosine diphosphate (GDP)-bound state. In the active GTP-bound state, Rabs associate with distinct membrane domains, where they can interact with specific effector proteins to regulate vesicle budding, tethering, and fusion as well as vesicle transport along the cytoskeleton, whereas in the inactive GDP-bound state Rabs are sequestered in the cytoplasm. This is regulated by Rab guanine nucleotide exchange factors (GEFs) and Rab GTPase-activating proteins (GAPs), which promote the active GTP-bound and inactive GDP-bound states, respectively. By regulating the GTP-bound state of Rab GTPases, Rab GEFs and Rab GAPs play an important role in regulating membrane localization of Rab GTPases and in coordinating interactions between Rab GTPases and their effectors.
Active Rab5 and Rab7 localize to early and late endosomes, respectively, where they regulate trafficking of cargo from the plasma membrane to the lysosome (Stenmark, 2009). Rab5 and Rab7 are activated by distinct Rab GEFs. Several Vps9 domain proteins display GEF activity for Rab5 (Carney et al., 2006), whereas Vps39p displays GEF activity for the yeast Rab7, Ypt7p (Wurmser et al., 2000). Interestingly, GEFs for both Rab5 and Rab7 can exist in a complex with effector proteins. The Rab5 GEF Rabex-5 binds to the Rab5 effector Rabaptin-5 (Horiuchi et al., 1997), and Vps39p interacts with the Rab7 effector Vps41p and the class C vacuolar protein sorting core complex to comprise the homotypic fusion and protein sorting (HOPS) complex (Wurmser et al., 2000). Coupling of the GEF with the effector can serve to bring the effector complex within proximity of the activated Rab as well as recruit and activate more Rab proteins as part of a positive feedback loop. Activated Rab5 can also recruit the HOPS complex (the putative Rab7 GEF) as a mechanism whereby Rab5 can recruit and activate Rab7 to drive endosome maturation (Rink et al., 2005). A similar mechanism for sequential Rab activation occurs in the yeast secretory pathway where the active Rab GTPases Ypt31p and Ypt32p can stimulate activation of the Sec4p Rab, by recruiting Sec2p, the Sec4p GEF, to secretory vesicles (Ortiz et al., 2002). This positive feed forward mechanism can be coupled with a negative feedback mechanism whereby the downstream Rab could recruit the GAP for the upstream Rab (Rivera-Molina and Novick, 2009). During Rab conversion, it has been proposed that recruitment of a Rab5 GAP by activated Rab7 would facilitate the inactivation and removal of Rab5 (Rink et al., 2005).
RN-Tre and Rab GAP-5 can stimulate GTP hydrolysis by Rab5 in vitro and disrupt endocytosis when overexpressed in cell culture (Lanzetti et al., 2000; Haas et al., 2005). Furthermore, RNA interference (RNAi) of Rab GAP-5 induces a large early endosome phenotype as seen with expression of activated Rab5 (Haas et al., 2005). Rab GAP-5 has higher Rab5 GAP activity than RN-Tre, and RN-Tre displays stronger GAP activity against Rab41, suggesting that Rab41 might be the true target (Haas et al., 2005). However, RN-Tre interacts with several proteins in the epidermal growth factor receptor signaling pathway and might regulate Rab5 in a signal-specific manner that is not mimicked in the in vitro GAP assays (Matoskova et al., 1996; Lanzetti et al., 2000). Because Rab5 interacts with several different effectors to regulate different activities on early endosomes, regulation by multiple GEFs and GAPs might provide functional specificity for Rab5.
Although most Rab GEFs do not share a common recognizable catalytic domain, most known Rab GAPs have a Tre-2/Bub2/Cdc16 (TBC) catalytic domain (Bernards, 2003). There are at least 60 Rab GTPases and ∼50 TBC domain proteins in humans, most of which have not been characterized (Pereira-Leal and Seabra, 2001; Bernards, 2003). Identifying the Rab GTPase substrates of these Rab GAPs will be essential to understand how vesicular trafficking is regulated. Although in vitro binding and GAP assays have been useful in identifying cognate Rab GTPase/Rab GAP pairs, there is some promiscuity in substrate recognition in vitro (Albert and Gallwitz, 1999; Itoh et al., 2006). Other factors such as subcellular localization, posttranslational modifications, and possibly cofactors might contribute to GAP activity and substrate specificity; therefore, in vivo functional assays are needed in conjunction with the in vitro assays.
The nematode Caenorhabditis elegans has proven to be a powerful in vivo model for studying vesicle trafficking by using genetic and cell biological approaches (Fares and Grant, 2002; Poteryaev and Spang, 2008). There are 21 genes in C. elegans predicted to encode TBC domain Rab GAP proteins, of which 19 have clear mammalian orthologues (www.wormbase.org). Here, we describe the characterization of C. elegans TBC-2, a conserved TBC domain protein, by using genetics and in vivo trafficking assays in C. elegans in combination with in vitro GAP assays. We show that TBC-2 regulates endosome to lysosome trafficking and is required to maintain the correct size and localization of endosomes. TBC-2 localizes to endosomes with RAB-7, requires RAB-7 for membrane localization, but it displays the strongest GAP activity toward RAB-5. Our data are consistent with TBC-2 functioning on late endosomes to inactivate RAB-5 during Rab conversion on maturing endosomes.
MATERIALS AND METHODS
C. elegans Alleles and General Methods
General methods for the handling and culturing C. elegans were as described previously (Brenner, 1974). C. elegans var Bristol strain N2 is the wild-type parent for all strains used in this work. Escherichia coli strain HB101 was used as a food source. Specific genes and alleles are described on Wormbase (www.wormbase.org) and were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN) unless otherwise noted here. LGI: arIs37(Pmyo-3::ssGFP). LGII: mIn1, tbc-2(sv41) this study, tbc-2(tm2241) was kindly provided by Shohei Mitani (National Bioresource Project, Tokyo Women's Medical University School of Medicine, Japan), unc-4(e120), rab-7(ok511), pwIs72[Pvha-6::GFP::rab-5 + unc-119] (linkage based on difficulty creating a tbc-2(tm2241) pwIs72 strain). LGIII: unc-119(ed3). LGIV: dpy-20(e1282). LGV: rde-1(ne219). LGX: glo-1(zu391), bIs1[vit-2::GFP + rol-6(su1006)], pwIs69[Pvha-6::GFP::rab-11 + unc-119] (X-linkage based on 100% of the progeny of heterozygous males having pwIs69). Linkage unknown: Ex[GFP::lgg-1] (Melendez et al., 2003) was kindly provided by Alicia Meléndez (Queens College, Flushing, NY), pwIs50[lmp-1::GFP + Cb-unc-119], pwIs170 [Pvha-6::GFP::rab-7 + Cb-unc-119], pwIs206 [Pvha-6::GFP::rab-10 + Cb-unc-119], pwIs429(Pvha-6::mCherry::rab-7) was a kind gift from Barth Grant (Rutgers University, NJ), rrEx236[Pend-3::rde-1 + Pelt-2::rde-1 + Pinx-6::GFP] was a kind gift from Michael Hebeisen and Richard Roy (McGill University, Montreal, QC, Canada), and from this study: vhIs1[Pvha-6::mCherry::tbc-2 + Cb-unc-119], vhIs6[Pvha-6::mCherry::tbc-2 R689K + Cb-unc-119], vhIs12[Pvha-6::GFP::tbc-2 + Cb-unc-119], vhIs24[Pvha-6::GFP::rab-5 Q78L + Cb-unc-119], vhIs34[Pvha-6:: GFP::rab-7 Q68L + Cb-unc-119].
Molecular Cloning, Isolation of the Deletion Allele, and Generation of Transgenic Lines
A full-length tbc-2 cDNA was generated from a partial cDNA yk11e7 (Yuji Kohara, National Institute of Genetics, Mishima, Japan), and by reverse transcription-polymerase chain reaction (PCR), which matches the predicted coding sequence (www.wormbase.org). tbc-2 is trans-spliced to SL2 consistent with it being a downstream gene in an operon. The sv41 deletion allele was isolated by using PCR to screen a deletion library of mutagenized N2 worms as described in Kao et al. (2004). Pooled genomic DNA samples representing a total of 400,000 haploid genomes were used as templates for nested PCR reactions. The first round of PCR was performed with primers with the sequences 5′-GAG GGA ACG GAT TCA GTT CG-3′ and 5′-ATG TCT ACC CAG CTC GCC AC-3′. The second round was performed with primers with the sequences 5′-TCG GCA ACC GCT CCG ATC AG-3′ and 5′-CTC TCC GTT CGC CAC CTT CC-3′. The sv41 and tm2241 deletion alleles are homozygous viable, healthy and display similar trafficking phenotypes in several tissues (Supplemental Figure 1, A–L). tbc-2(RNAi) displays similar, albeit weaker phenotypes (Supplemental Figure 1, M–P), and the phenotypes of the deletion alleles do not get more severe when placed in trans to the chromosomal deficiency maDf4 (Supplemental Figure 1, Q–T), suggesting they are potential null or strong loss-of-function alleles. tbc-2 R689K was generated by site directed-mutagenesis. tbc-2 and tbc-2 R689K cDNAs were subcloned into a Gateway destination vector containing the vha-6 intestinal specific promoter, an N-terminal mCherry or green fluorescent protein (GFP) tag and the Cb-unc-119(+) gene (a gift from Barth Grant, Rutgers University, The State University of New Jersey, Piscataway, NJ) using Gateway technology (Invitrogen, Carlsbad, CA). Vectors containing Pvha-6::GFP::rab-5 Q79L + Cb-unc-119(+) and Pvha-6::GFP::rab-7 Q67L + Cb-unc-119(+) were gifts from Barth Grant. Integrated transgenic lines were generated by microparticle bombardment into unc-119(ed3) animals as described previously (Praitis et al., 2001).
RNA Interference
RNAi feeding was performed essentially as described in Kamath et al. (2001), using clones from the Ahringer RNAi library (Geneservice, Cambridge, United Kingdom). The following genes (clones) were tested for suppression of the tbc-2(−) intestinal phenotype: rab-1(V-1L07), rab-2/unc-108(I-1J21), rab-3(II-4J07), rab-5(I-4J01), rab-6.1(III-4H12), rab-6.2(X-7B14), rab-7(II-8G13), rab-8(I-1F11), rab-10(I-3G23), rab-11.1(I-1A15), rab-11.2(I-5H12), rab-14(X-7K17), rab-21(II-6I02), rab-28(IV-4B06), rab-37(X-2H03), rab-39(II-6L15), tag-312(X-2G09), 4R79.2(IV-8I20), F11A5.3(V-10F14), C52B11.5(X-1I10), vps-39(V-9C02), and vps-41(X-3C03). pwIs50[lmp-1::GFP] or tbc-2(tm2241); pwIs50[lmp-1::GFP] larval stage (L)4 animals placed on plates containing bacteria expressing double-stranded RNA (dsRNA) corresponding to each RNAi clone. L4 progeny born 48–72 h after plating were scored for suppression. The strong embryonic lethal phenotype of rab-5(RNAi) prevented us from analyzing the requirements for rab-5 in the intestine; therefore, we performed tissue-specific RNAi in the intestine using the RNAi-defective strain: rde-1(ne219) rescued specifically in the intestine with the extrachrosomal array rrEx236[Pend-3::rde-1 + Pelt-2::rde-1 + Pinx-6::GFP] (Richard Roy, personal communication). rde-1(ne219); pwIs50; rrEx236 and tbc-2(tm2241) unc-4(e120); rde-1(ne219); pwIs50; rrEx236 were tested for suppression for rab-5(RNAi) as described above.
Microscopy and Image Analysis
For phenotype analysis by differential interference contrast (DIC) and epifluorescence, live worms were mounted on 2% agarose pads with 10 mM levamisol as described in Wormbook (www.wormbook.com). Animals were analyzed on an Axio Zeiss A1 Imager compound microscope (Carl Zeiss, Oberkochen, Germany), and images were captured using an Axio Cam MRm camera and AxioVision software (Carl Zeiss). Statistical analysis and graphing of coelomocyte size was done on Prism 5 (GraphPad Software, San Diego, CA).
Confocal analysis was performed using a Zeiss LSM-510 Meta laser scanning microscope with 63× oil immersion lens in a single or multitrack mode by using a single or dual excitation (488 nm for GFP and/or 543 nm for mCherry), and GFP band-pass filter settings were reduced to avoid interference from the nonspecific autofluorescence in the intestine (Clokey and Jacobson, 1986; Hermann et al., 2005). Images were captured using LSM Image software (Carl Zeiss).
For Electron Microscopy, fixation was essentially done as described previously (Michaux et al., 2000). L4/young adult animals were fixed in 2% glutaraldehyde, 1% paraformaldehyde, 0.1 M cacodylate, pH 7.4, at 4°C overnight. Animals were washed with 0.1 M cacodylate buffer and fixed in 2% OsO4, dehydrated and embedded in Epon resin at 58°C for 2 d. Transverse sections through the intestine were placed onto a coated grid for observation. Pictures were taken using a FEI Technai 12 (FEI, Hillsboro, OR) equipped with a Bioscan charge-coupled device camera (model 792; Gatan, Pleasanton, CA).
To visualize acidic compartments, L4/young adults were placed onto plates containing 2 μM LysoTracker Red mixed with HB101 bacteria (Invitrogen). Animals were grown at 20°C in the dark until the progeny reached the L4/young adult stage (Hermann et al., 2005).
Expression, Purification, and High-Throughput GAP Assay
GST fusions of full-length mammalian Rab GTPases were expressed and purified as described previously (Pan et al., 2006). The C. elegans full-length Rab GTPases and the TBC-2 construct (residues 410–908) were subcloned into pGEX-6P-1 (GE Healthcare, Little Chanfont, Buckinghamshire, England) and 6xHis SUMO-pET fusion vectors, respectively. Expression plasmids were transformed into BL21(DE3) Codon plus RIL cells (Stratagene, La Jolla, CA). Cells were grown at 22°C in 2× YT media containing 100 mg/l ampicillin (GST fusion) or 50 mg/l kanamycin (SUMO fusion) to an optical density at 600 nm of 0.4 and induced with 0.05 mM isopropyl β-d-thiogalactoside for 16 h. Cells were harvested and lysed in 50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, 14.2 mM 2-mercaptoethenol, 0.1 mM phenylmethylsulfonyl fluoride, 0.2 mg/ml lysozyme, and 0.01 mg/ml DNase. After supplementing with 0.5% Triton X 100, lysates were clarified via centrifugation at 15,500 rpm, at 4°C for 45 min. Lysates for GST Rab GTPases were incubated with glutathione-Sepharose beads, and then washed with 50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, and 14.2 mM 2-mercaptoethenol. Bound proteins were eluted with 10 mM reduced glutathione and further purified over a Superdex 200 gel filtration column. The lysate for the SUMO-TBC-2 fusion was passed through a Ni2+ Sepharose column (GE Healthcare), washed with 50 mM Tris-Cl, pH 8.0, 500 mM NaCl, 10 mM 2-mercaptoethenol, 15 mM imidazole and bound protein was eluted with 300 mM imidazole in 50 mM Tris-Cl, pH 8.0, 100 mM NaCl, 10 mM 2-mercaptoethenol. The 6xHis Sumo fusion tag was removed by digestion with 6xHis-sumoase, passed through a Ni2+ Sepharose column, and further purified over HiTrap Q HP ion exchange and Superdex 200 gel filtration columns (GE Healthcare).
GAP assays were performed in microplate format using GTP-loaded Rabs, a TBC-2 construct (residues 410–908) spanning the THR-TBC domain, and a fluorescent phosphate sensor consisting of N-[2-(1-maleimidyl) ethyl]-7-(diethylamino) coumarin-3-carboxamide=−labeled E. coli phosphate binding protein (Brune et al., 1994), which senses the Pi produced after GTP hydrolysis. Rab GTPases were loaded with GTP by incubating 0.5–1 mg of protein with a 25-fold molar excess of GTP in a buffer consisting of 20 mM Tris, pH 8.0, 150 mM NaCl, and 5 mM EDTA for 1 h at room temperature. Unbound nucleotide was removed using 10 ml d-salt column (Pierce Protein Research Products, Thermo Fisher Scientific, Rockford, IL), pre-equilibrated with 20 mM Tris, pH 8.0, and 150 mM NaCl. GAP reactions were initiated by mixing a solution of GTP loaded Rabs and the phosphate sensor with varying concentrations of TBC-2 (410–908) in 20 mM Tris-Cl, pH 8.0, 150 mM NaCl, and 10 mM MgCl2 in 96 well half area microplates (Corning Life Sciences, Lowell, MA). The final reaction mixture (100 ml) contained 2 mM Rab GTPase, 2.5 mM phosphate sensor, and 0–2 mM TBC-2 (410–908). Real-time single turnover kinetics of intrinsic and GAP accelerated GTP hydrolysis were monitored at excitation and emission wavelengths of 425 and 457 nm, respectively, using a Safire microplate spectrometer (Tecan, Männedorf, Switzerland). Data were analyzed by fitting globally to a pseudofirst-order Michaelis–Menten model function I(t) = (I∞ − I0)(1 − exp(−kobst)) + I0 where kobs = kintr + (kcat/Km)[GAP]. The catalytic efficiency (kcat/Km) and intrinsic rate constant for GTP hydrolysis (kintr) were treated as global parameters, whereas the initial (I0) and final (I∞) emission intensities were treated as local parameters.
RESULTS
C. elegans TBC-2 Is Homologous to Human TBC1D2 and TBC1D2B
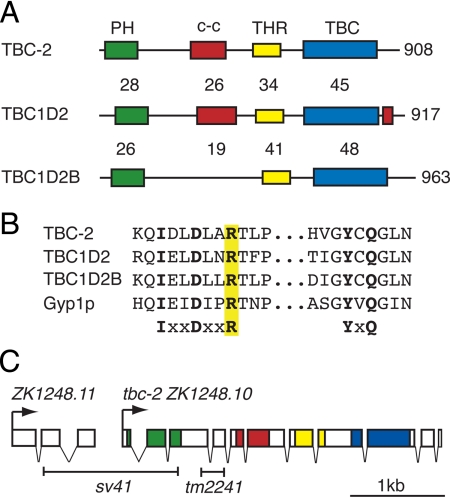
Among the 21 predicted C. elegans TBC domain proteins (www.wormbase.org), TBC-2 has a unique domain structure, consisting of an amino-terminal pleckstrin homology (PH) domain, a central coiled-coil domain, and a carboxy-terminal TBC domain (Figure 1A). TBC-2 is most similar to human TBC1D2 (also so known as PARIS-1 or Armus) and TBC1D2B (Zhou et al., 2002; Frasa et al., 2010). These proteins share a conserved domain structure with the highest degree of homology within the TBC domain. Outside of the known domains they share an additional region of homology amino-terminal to the TBC domain that we call the TBC1D2 homology region (THR). The arginine- and glutamate-finger residues required for catalytic activity are conserved suggesting that they might function as a Rab GAP (Figure 1B) (Albert et al., 1999; Pan et al., 2006).
Figure 1.
TBC-2: protein homology, gene structure, and deletion alleles. (A) C. elegans TBC-2 (NP_495156) is homologous to the human TBC1D2 (Q9BYX2) and TBC1D2B (Q9UPU7) proteins. They share a conserved PH domain (green), coiled-coil domain (c-c, red) and TBC domain (blue). The homology between TBC-2 and its mammalian homologues to mainly limited to the three domains with the exception of a region we designated the THR (yellow). The percentage of amino acid identity between TBC-2 and the two human homologues is shown above each domain. (B) Alignment of the TBC catalytic motifs IXXDXXR and YXQ containing the conserved arginine (R; highlighted yellow) and glutamine (Q) residues conserved in C. elegans TBC-2, human TBC1D2, and TBC1D2B and yeast Gyp1p (NP_014713). (C) Genomic structure of tbc-2 ZK1248.10 and the upstream gene ZK1248.11. The regions encoding the different domains in TBC-2 are highlighted in corresponding colors. Shown below are the regions deleted by the deletion alleles sv41 and tm2241. The sv41 allele removes most of ZK1248.11 in addition to the 5′ region of tbc-2. The tm2241 allele removes intron three resulting in a frame shift introducing a premature stop codon.
tbc-2(−) Phenotypes in the Intestine, Coelomocytes, and Oocytes
To determine the biological function of tbc-2, we analyzed two homozygous viable deletion alleles, tm2241 and sv41, that are potential null or strong loss-of-function alleles (Figure 1C; see Materials and Methods). Consistent with a potential role in vesicular trafficking, we find that tbc-2 mutants accumulate large vesicular structures in the intestine (Figure 2, A–F, and Supplemental Figure 1). These intestinal vesicles differ from those of previously reported trafficking mutants, such as rab-10, rme-1, ppk-3, and tat-1, in that tbc-2 mutant vesicles usually accumulate “refractile material” resembling that normally found in the cytoplasm of the intestinal cells (Sulston and Horvitz, 1977; Chen et al., 2006; Nicot et al., 2006; Ruaud et al., 2009).
Figure 2.
tbc-2(−) phenotypes in the intestine, coelomocytes, and oocytes. (A and B) DIC images of wild-type (A) and tbc-2(tm2241) (B) animals. The intestine is composed of 20 polarized epithelial cells with an apical membrane facing the intestinal lumen (arrow) and basolateral membranes facing the pseudocoelomic space and neighboring organs (boundaries marked with bracket). Examples of the large vesicles observed in tbc-2(−) animals are marked with arrowheads. (C–F) Electron micrographs of transverse sections of wild-type (C) and tbc-2(tm2241) (D–F) animals. (C and D) A white line demarcates the basolateral boundaries of the intestine (int) from other tissues such as the gonad (go), and the intestinal lumen is marked with an arrow. (E and F) Higher magnification images of the large tbc-2(tm2241) vesicles and the amorphous material present inside the vesicles. (G and H) Epifluorescence images of coelomocytes containing internalized ssGFP in arIs37[Pmyo-3::GFP]; dpy-20(e1282) (G) and tbc-2(tm2241); arIs37[Pmyo-3::GFP] (H) animals. (I) Scatter dot plot of the largest ssGFP-positive vesicle in each of 60 coelomocytes from arIs37[Pmyo-3::GFP]; dpy-20(e1282) (wild-type) and tbc-2(tm2241); arIs37[Pmyo-3::GFP] animals. The difference between the two groups is statistically significant in an unpaired t test (p < 0.0001). Error bars represent the mean with a 95% confidence interval (p < 0.05). (J–M) DIC (J and L) and epifluorescence (K and M) images of oocytes with internalized YP170::GFP in bIs1[vit-2::GFP] (J and K) and tbc-2(tm2241); bIs1[vit-2::GFP] (L and M) animals. Bar, 10 μm (A, B, G, H, J, K, L, and M), 5 μm (C and D), 0.5 μm (E) and 0.2 μm (F).
To determine whether tbc-2 mutants are defective in either endocytic or exocytic trafficking, we tested tbc-2 mutants in trafficking assays in coelomocytes and oocytes (Fares and Grant, 2002). Six coelomocytes reside in the pseudocoelom that exhibit a high rate of endocytosis of fluid-phase molecules and resemble macrophages or scavenger cells. The coelomocytes readily take up a signal secreted GFP (ssGFP), that is expressed in body wall muscles and secreted into the pseudocoelom (Fares and Greenwald, 2001a). In the maturing oocytes, a yolk protein::GFP fusion, YP170::GFP, is secreted from the intestine and internalized by the oocytes (Grant and Hirsh, 1999).
We find that tbc-2 mutants do not show defects in the secretion or the uptake of ssGFP or YP170::GFP, but both GFP proteins are found in larger vesicles. In coelomocytes, we find that the ssGFP accumulates in larger than average vesicles, which are sometimes interconnected by tubules (Figure 2, G–I; data not shown). The large ssGFP-positive vesicle phenotype is similar to that reported for the late endocytic regulator cup-5 (Fares and Greenwald, 2001b; Hersh et al., 2002; Treusch et al., 2004). In oocytes, the YP170::GFP accumulates in larger vesicles that seem to be more concentrated in the perinuclear region, whereas in wild-type animals, the YP170::GFP seems to be more evenly distributed throughout the cytoplasm (Figure 2, J–M; and Supplemental Figure 1). tbc-2(−) oocytes can be easily distinguished from wild-type under DIC optics based on the coarse granular appearance of the vesicles that are often YP170::GFP positive. These phenotypes suggest that TBC-2 regulates trafficking of endocytic vesicles after internalization and corroborate the recent finding that TBC-2 mediates phagocytic trafficking of apoptotic cell corpses (Li et al., 2009).
The tbc-2(−) Intestinal Vesicles Are Large Endosomes
To determine whether the tbc-2(−) intestinal vesicles are endocytic in nature, we compared the localization of markers for early endosomes (GFP::RAB-5), late endosomes/lysosomes (GFP::RAB-7 and mCherry::RAB-7, LMP-1::GFP and LysoTracker Red) and recycling endosomes (GFP:: RAB-10 and GFP::RAB-11) in the intestine of both wild-type and tbc-2(−) animals (Treusch et al., 2004; Hermann et al., 2005; Chen et al., 2006). Although some of the tbc-2(tm2241) intestinal vesicles were positive for GFP::RAB-5 (Figure 3, A and B), the majority and the largest vesicles were positive for GFP::RAB-7 (Figure 3, C and D) and LMP-1::GFP (Figure 3, E and F). LMP-1::GFP and mCherry::RAB-7 can localize on the same vesicles (Figure 3, G and H). Similarly, GFP::RAB-7 is found on the large vesicles of tbc-2(sv41) and tbc-2(RNAi) animals (Supplemental Figure 1). LysoTracker Red staining indicates that acidic lysosomal compartments are present in tbc-2(tm2241) intestinal cells and that the large late endosomes are not acidic compartments, but are sometimes found adjacent to lysosomes (Figure 3, I and J). No significant accumulation of either recycling Rab GTPases, GFP:: RAB-10 (Figure 3, K and L) or GFP::RAB-11 (Figure 3, M and N), were seen on the tbc-2(tm2241) vesicles. Therefore, the large intestinal vesicles in tbc-2(−) animals are primarily late endosomes.
Figure 3.
tbc-2(−) intestinal vesicles are large endosomes. (A–P) Confocal images of wild-type (A, C, E, G, I, K, M, and O) and tbc-2(tm2241) (B, D, F, H, J, L, N, and P) animals carrying the following integrated transgenes: pwIs72 [Pvha-6::GFP::rab-5] (A and B), pwIs170 [Pvha-6::GFP::rab-7] (C and D), pwIs50 [lmp-1::GFP] (E and F), pwIs50 [lmp-1::GFP] and pwIs429[Pvha-6::mCherry::rab-7] (G and H), pwIs50 [lmp-1::GFP] fed LysoTracker Red (red) (I and J), pwIs206 [Pvha-6::GFP::rab-10] (K and L), pwIs69 [Pvha-6::GFP::rab-11] (M and N), and an extrachromosomal array carrying GFP::lgg-1 (Melendez et al., 2003) (O and P). Arrowheads denote examples of GFP::RAB-5–positive vesicles (B), vesicles with internalized GFP::RAB-7 (D), LMP-1::GFP (F), or GFP::LGG-1 (P), and LysoTracker Red-positive vesicles adjacent to LMP-1::GFP vesicles (J). Bar, 5 μm.
The accumulation of refractile material in the tbc-2(−) intestinal vesicles suggests that autophagy might contribute to the tbc-2(−) phenotype (Figure 2, B, D–F). Mammalian LC3 (Atg8p in yeast) is a specific marker for autophagosomes. It is covalently linked to phosphatidylethanolamine during autophagosome formation and remains associated even after autophagosome fusion with the lysosome (Kabeya et al., 2000). To determine whether the tbc-2(−) intestinal vesicles are autophagosomes, we looked at the localization of the C. elegans LC3 homologue (LGG-1) fused to GFP (Melendez et al., 2003). We find that GFP::LGG-1 is not present on the tbc-2(tm2241) intestinal vesicles, but that it is present inside the vesicles, suggesting that fusion of autophagosomes with the late endosome has occurred (Figure 3, O and P). Furthermore, by electron microscopy we observe that the tbc-2(tm2241) intestinal vesicles are not surrounded by a double membrane characteristic of an autophagosome (Figure 2, C–F). In addition, the refractile material does not seem to be surrounded by internal membranes as would be expected for multivesicular bodies. The presence of the GFP marker proteins, in particular LMP-1::GFP and GFP::LGG-1, inside the vesicles suggests that the refractile material is in part nondegraded protein (Figure 3).
The Arginine-Finger Is Required for TBC-2 Function in the Intestine
The catalytic domain of TBC-2 contains the conserved amino acids shown to be important for GAP function (Figure 1B). Mutation of the arginine-finger to a lysine has been shown to abolish GAP function (Albert et al., 1999). To determine whether TBC-2 might function as a Rab GAP, we tested whether the catalytic Arginine was required for TBC-2 function in the intestine. We find that expression of a wild-type mCherry::TBC-2 fusion in the intestine can completely rescue the tbc-2(tm2241) phenotype (Figure 4, A, G, M, and S), whereas a predicted catalytically inactive mutant, mCherry:: TBC-2 R689K, expressed at comparable levels failed to rescue (Figure 4, D, J, P, and V). In addition, we find that transgenic lines with high expression of a GFP::TBC-2 R689K in a wild-type background can phenocopy a tbc-2(−) intestinal phenotype, suggesting that arginine-to-lysine mutants can act in a dominant-negative manner (data not shown). Therefore, the catalytic arginine-finger is required for TBC-2 function, suggesting it might act as a Rab GAP.
Figure 4.
TBC-2 rescue and localization. Confocal images of tbc-2(tm2241) animals expressing either vhIs1[Pvha-6::mCherry::tbc-2] (A–C, G–I, M–O, and S–U), or vhIs6[Pvha-6::mCherry::tbc-2 R689K] (D–F, J–L, P–R, and V–X) in combination with pwIs72[Pvha-6::GFP::rab-5] (A–F), pwIs170[Pvha-6:: GFP::rab-7] (G–L), pwIs50[lmp-1::GFP] (M-R), pwIs69[Pvha-6::GFP::rab-11] (S–X). Bar, 5 μm.
rab-5 and rab-7 Are Required for the tbc-2(−) Phenotypes
If TBC-2 functions as a Rab GAP, then the tbc-2(−) phenotypes might be due to too much Rab GTPase activity. To identify potential targets of TBC-2 activity, we assayed 21 Rab GTPases by RNAi (or with available mutants) for suppression of the tbc-2(tm2241) intestinal phenotype (see Materials and Methods). We observed that rab-5(RNAi) and rab-7(RNAi), or the rab-7(ok511) deletion mutant, strongly suppressed the large vesicle phenotype in the intestine of tbc-2(tm2241) animals (Figure 5, A–D; data not shown). No suppression was observed with rab-11.1(RNAi), a glo-1(zu391) mutant, or RNAi of any of the remaining 17 Rab GTPases (Figure 5, E and F; data not shown). Thus, rab-5 and rab-7 are specifically required for the tbc-2(−) intestinal phenotype.
Figure 5.
rab-5, rab-7, and the HOPS complex are required for tbc-2(−) intestinal phenotype. (A–J) DIC images (A, C, E, G, and I) and epifluorescence images (B, D, F, H, and J) of the intestines of tbc-2(tm2241) animals expressing LMP-1::GFP in different mutant and RNAi backgrounds. (A and B) tbc-2(tm2241) unc-4(e120); rde-1(ne219); pwIs50 [lmp-1::GFP]; rrEx236[Pend-3::rde-1 + Pelt-2::rde-1 + Pinx-6::GFP] animals fed dsRNA corresponding to rab-5. This strain was used for intestinal specific RNAi (see Materials and Methods). (C and D) tbc-2(tm2241) rab-7(ok511); pwIs50 [lmp-1::GFP] animals. (E–J) tbc-2(tm2241); pwIs50 [lmp-1::GFP] fed dsRNA corresponding to rab-11.1 (E and F), vps-39 (G and H), or vps-41 (I and J). Confocal images of pwIs170[Pvha-6::GFP::rab-7] (K and L), and tbc-2(tm2241); pwIs170[Pvha-6::GFP::rab-7] (M and N) animals fed dsRNA targeting vps-39. (L and N) are higher magnification images from K and M, respectively. Bar, 10 μm (B and K) and 2 μm (L).
In oocytes, rab-5(RNAi) blocks YP170::GFP internalization, whereas rab-7(RNAi) results in internalized YP170::GFP accumulating in larger vesicles adjacent to the plasma membrane (Grant and Hirsh, 1999). The distribution of YP170::GFP to the cell periphery in a rab-7(RNAi) background is opposite the distribution seen in tbc-2(tm2241) oocytes (Figure 6, A–D). We find that YP170::GFP is localized to the cell periphery in both rab-7(RNAi) and tbc-2(tm2241) rab-7(RNAi) animals (Figure 6, C–F), indicating that RAB-7 activity is required for distribution of YP170::GFP in tbc-2(−) oocytes.
Figure 6.
rab-7 is required for the YP170::GFP distribution in tbc-2(−) oocytes. DIC (A, C, and E) and epifluorescence (B, D, and F) images of oocytes of tbc-2(tm2241) (A and B), rab-7(RNAi) (C and D) and tbc-2(tm2241) rab-7(RNAi) (E and F) animals expressing YP170::GFP. Bar, 10 μm (D).
Constitutively Active RAB-5 Q78L Phenocopies the tbc-2(−) Intestinal Phenotype
If the tbc-2(−) phenotypes are due to too much Rab GTPase activity, then expression of the constitutively active form of the target Rab GTPase might induce the same phenotype. In mammalian cells, GTP hydrolysis-deficient mutants Rab5 Q79L and Rab7 Q67L are locked in the GTP-bound conformation resulting in constitutive activity (Stenmark et al., 1994; Meresse et al., 1995; Vitelli et al., 1997). We expressed the analogous C. elegans mutants, GFP::RAB-5 Q78L and GFP::RAB-7 Q68L, in the intestine of wild-type animals to determine whether they can mimic the tbc-2(−) intestinal phenotype. We found that animals expressing GFP::RAB-5 Q78L, but not GFP::RAB-7 Q68L, displayed the large intestinal vesicle similar to the tbc-2 deletion mutants (Figure 7, A–D). By confocal microscopy, we find that GFP::RAB-5 Q78L is present on smaller vesicles along with mCherry::RAB-7; however, the larger vesicles are almost exclusive for mCherry::RAB-7 (Figure 7, E–G). This phenotype is caused by expression of the gfp::rab-5 Q68L transgene, because it is suppressed by RNAi targeting gfp (Figure 7H). Like the tbc-2(−) intestinal phenotype, the GFP::RAB-5 Q78L intestinal vesicles could be suppressed by rab-7(RNAi) (Figure 7I). None of the GFP::RAB-7 Q68L transgenic lines induced a significant phenotype (Figure 7, C and D). This could be explained by the fact that all of the GFP::RAB-7 Q68L lines generated had lower expression levels than the GFP::RAB-5 Q78L lines, or the activated RAB-7 alone might not be sufficient to mimic the tbc-2(−) phenotype. Our data show that activation of RAB-5 is sufficient to recruit RAB-7 and generate large late endosomes in C. elegans and suggest that TBC-2 might function as a RAB-5 GAP. The colocalization of GFP::RAB-5 Q78L and mCherry::RAB-7 on the enlarged vesicles is consistent with the model whereby activated Rab5 recruits Rab7.
Figure 7.
Expression of RAB-5 Q78L mimics the tbc-2(−) intestinal phenotype in a RAB-7 and HOPS complex dependent manner. (A–D) DIC images (A and C) and epifluorescence images (B and D) of the intestine of vhIs24 [Pvha-6::GFP::rab-5 Q78L] (A and B) and vhIs34[Pvha-6::GFP::rab-7 Q68L] (C and D) transgenic animals. (E–G) Confocal images of the intestine of a strain expressing both vhIs24 [Pvha-6::GFP::rab-5 Q78L] and pwIs429[Pvha-6:: mCherry::RAB-7]. Arrowheads denote smaller vesicles positive for both GFP::RAB-5 Q78L and mCherry::RAB-7, and asterisks denotes larger mCherry::RAB-7–positive vesicles almost devoid of GFP::RAB-5 Q78L. (H–K) DIC images of the intestine of vhIs24 [Pvha-6:: GFP::rab-5 Q78L] transgenic animals fed with dsRNA targeting GFP (H), rab-7 (I), vps-39 (J), or vps-41 (K). Bar, 10 μm (D and K) and 5 μm (G).
The HOPS Complex Is Required for the tbc-2(−) and RAB-5 Q78L Phenotypes
The highly conserved class C vacuolar protein sorting/HOPS complex contains six subunits (Vps39, Vps41, Vps11, Vps18, Vps16, and Vps33) that function as both a GEF and effector for Ypt7p, the yeast orthologue of Rab7 (Wurmser et al., 2000). Furthermore, the mammalian HOPS complex has been found to interact with GTP-bound Rab5 where it could function to recruit and activate Rab7 during endosome maturation (Rink et al., 2005). To determine whether the HOPS complex is required for the large endosome phenotype, we performed RNAi on the two specific subunits, the putative RAB-7 GEF vps-39 and the RAB-7 effector vps-41. We find that RNAi of vps-39 or vps-41 suppressed the tbc-2(tm2241) (Figure 5, G–N) and GFP::RAB-5 Q78L (Figure 7, J and K) large endosome phenotypes in the intestine. Thus, the large endosome phenotype of tbc-2(−) and GFP::RAB-5 Q78L animals share the same requirements for RAB-7 and the HOPS complex. Of note, we found that vps-39(RNAi) potently suppressed the tbc-2(tm2241) intestinal phenotype but did not affect the membrane localization of GFP::RAB-7 (Figure 5, K–N), suggesting that VPS-39 is not required as a GEF.
TBC-2 Can Catalyze GTP Hydrolysis on RAB-5 and RAB-7 In Vitro
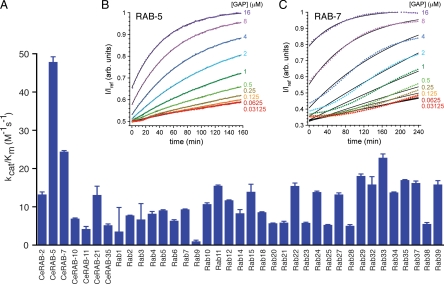
To determine whether TBC-2 can catalyze GTP hydrolysis on RAB-5, RAB-7 or other Rab GTPase(s), we performed an unbiased high-throughput optical GTPase assay that measures gamma phosphate release in real time. We tested whether the TBC domain (residues 410–908) of TBC-2 could stimulate the GTPase activity of 30 mammalian Rab GTPases and seven C. elegans Rab GTPases implicated in endocytic trafficking. By measuring initial velocity we found that TBC-2 displayed the highest catalytic efficiency with C. elegans RAB-5 and lower activity with C. elegans RAB-7 (Figure 8A). In a global fit to a Michaelis–Menten model function, the catalytic efficiency of TBC-2 for RAB-5 (33 ± 4.8 M−1 s−1) is approximately twofold higher than for RAB-7 (15 ± 0.46 M−1 s−1) (Figure 8, B and C). Lowering the Mg2+ concentration slowed the reaction but did not change the selectivity toward RAB-5 versus RAB-7 (Supplemental Figure 2). The activity of TBC-2 for RAB-5 is considerably weaker than that seen for previously characterized TBC domain proteins; however, the in vitro data are in agreement with our in vivo findings and those of Li et al. (2009) suggesting that TBC-2 is a RAB-5 GAP.
Figure 8.
TBC-2 catalyzes GTP hydrolysis by RAB-5 in vitro. (A) Catalytic efficiency (kcat/Km) plot based on the initial velocity measurements of TBC-2 on the C. elegans (Ce) and mammalian Rab GTPases. (B and C) A global fitting analysis to a Michaelis–Menten model function (in the limit where [TBC-2] < Km) for RAB-5 (B) and RAB-7 (C) at different concentrations of TBC-2. Solid lines represent the fitted model functions.
RAB-7 Is Important for TBC-2 Localization on Late Endosomes/Lysosomes
Little is known about the subcellular localization of TBC domain proteins or how they are recruited to their cognate Rab GTPases. TBC-2 has a PH domain, a putative lipid-binding domain, suggesting that TBC-2 might localize to specific membrane compartments. To determine the subcellular localization of TBC-2, we analyzed the localization of the rescuing functional mCherry::TBC-2 fusion protein as well as mCherry::TBC-2 R689K in the intestine with the GFP-tagged endocytic markers. We find that mCherry::TBC-2 localizes to puncta and primarily colocalizes with RAB-7 and LMP-1 in the intestine, with some RAB-5 puncta, but not with RAB-11, suggesting that it localizes primarily with RAB-7 on late endosomes (Figure 4, A–C, G–I, M–O, and S–U). Catalytically inactive Rab GAPs display stronger binding with their cognate Rab GTPases (Will and Gallwitz, 2001; Haas et al., 2005); therefore, we analyzed the localization of mCherry::TBC-2 R689K. We find that the catalytically inactive TBC-2 R689K still shows a strong colocalization with RAB-7 and LMP-1 on the large late endosomes (Figure 4, J–L and P–R). Consistent with RAB-5 being a catalytic target of TBC-2, we see colocalization of mCherry::TBC-2 R689K with RAB-5 (Figure 4, D–F), although to a lesser extent than RAB-7. No colocalization was detected with GFP::RAB-11 and mCherry::TBC-2 R689K (Figure 4, V–X). Thus, TBC-2 colocalizes predominantly with RAB-7 on late endosomes and to a lesser extent with RAB-5.
The colocalization of TBC-2 and RAB-7 on late endosomes suggested the possibility that RAB-7 could recruit TBC-2 to endosomes. To determine whether RAB-7 is required for TBC-2 late endosome localization, we analyzed GFP::TBC-2 localization in rab-7(RNAi)–treated animals (Figure 9). Although GFP::TBC-2 was present on endosome membranes of control animals (Figure 9, A and B), GFP::TBC-2 membrane localization was greatly reduced in rab-7(RNAi) animals (Figure 9, C and D), indicating that RAB-7 is required for GFP::TBC-2 membrane localization.
Figure 9.
RAB-7 is required for TBC-2 localization to membranes. Confocal images of tbc-2(tm2241); vhIs12 [Pvha-6::GFP::tbc-2] animals fed an empty vector (A and B) or dsRNA targeting rab-7 (C and D). B and D are higher magnification images from A and C, respectively. Experiments were performed in the tbc-2(tm2241) background as GFP::TBC-2 membrane localization was more robust, presumably due to competition with endogenous TBC-2. Bar, 10 μm (A) and 2 μm (B).
DISCUSSION
The trafficking of cargo from the cell membrane to the lysosome requires the concerted activity of the Rab5 and Rab7 GTPases, whose activities are regulated by Rab GEFs and Rab GAPs (Stenmark, 2009). Through genetic and phenotypic analysis of C. elegans tbc-2 deletion mutants, we demonstrate a role for the conserved TBC-2 protein in regulating RAB-5/RAB-7–mediated endosomal trafficking in several tissues. Our biochemical analysis shows that TBC-2 can catalyze GTP hydrolysis by RAB-5 consistent with TBC-2 acting as a RAB-5 GAP. However, we show that TBC-2 colocalizes with RAB-7 on late endosomes and requires RAB-7 for membrane localization. Our data suggest a model whereby TBC-2 might function on late endosomes to inactivate RAB-5 during endosome maturation.
The tbc-2(−) Phenotypes Are Consistent with Excessive RAB-5 and RAB-7 Activity
In yeast and mammalian cells, Ypt7p and Rab7, can mediate homotypic fusion of vacuoles and late endosomes/lysosomes (Haas et al., 1995; Papini et al., 1997; Bucci et al., 2000). Expression of constitutively active Rab7 Q67L in mammalian cells results in enlarged late endocytic structures (Meresse et al., 1995; Vitelli et al., 1997; Bucci et al., 2000), and can enhance homotypic fusion between late endosomes in vitro (Papini et al., 1997). Our data showing the accumulation of large endosomes in coelomocytes, oocytes, and in particular the large RAB-7– and LMP-1–positive late endosomes in the intestine of tbc-2(−) animals would be consistent with too much RAB-7 activity. Similarly, the perinuclear distribution of YP170::GFP in tbc-2(−) oocytes are consistent with a role for mammalian Rab7 mediating dynein and microtubule-dependent transport of late endosomes/lysosomes to a perinuclear local (Meresse et al., 1995; Vitelli et al., 1997; Press et al., 1998; Bucci et al., 2000; Jordens et al., 2001; Johansson et al., 2007). Our findings that rab-7(RNAi) and the rab-7(ok511) mutation can suppress the tbc-2(−) large late endosome phenotype in the intestine and the distribution of YP170::GFP in the oocyte indicate the RAB-7 is required for the tbc-2(−) phenotypes and suggest that TBC-2 negatively regulates the activity of RAB-7.
In mammalian cells, constitutively active Rab5 Q79L can induce large early endosomes (Stenmark et al., 1994; Lawe et al., 2002), and more recent data demonstrate that Rab5 Q79L can recruit Rab7 and other late endosome markers to these large endosomes (Rink et al., 2005; Wegener et al., 2010). In C. elegans, we show that the tbc-2(−) large late endosome phenotype is suppressed by rab-5(RNAi) and mimicked by expression of constitutively active RAB-5 Q78L. The fact that expression of RAB-5 Q78L induces large RAB-7–positive late endosomes that can be suppressed by rab-7(RNAi) suggests that activated RAB-5 not only recruits RAB-7 but also requires RAB-7 for large endosome formation. Therefore, the negative regulation of RAB-7 by TBC-2 might be an indirect consequence of TBC-2 antagonizing the activity of RAB-5. These results are consistent with the Rab conversion model of endosome maturation, whereby Rab5 recruits the HOPS complex which then recruits and activates Rab7 to convert the Rab5-positive early endosome into a Rab7-positive late endosome (Rink et al., 2005). We find that RNAi of the HOPS complex components, vps-39 and vps-41, strongly suppress the large late endosome phenotype of tbc-2(−) and RAB-5 Q78L expressing animals. However, we find that vps-39(RNAi) animals still retain GFP::RAB-7 on vesicles, implying that RAB-7 is still in a GTP-bound state. Therefore, VPS-39 is probably required in a different capacity, rather than as a RAB-7 GEF for the tbc-2(−) intestinal phenotype. Furthermore, it suggests that in C. elegans, VPS-39 is either not a RAB-7 GEF or acts redundantly with an unidentified GEF. Together, our phenotypic and genetic data are consistent with TBC-2 negatively regulating RAB-5 and RAB-7 during endosome maturation.
A role for C. elegans TBC-2 in the phagocytic degradation of apoptotic cell corpses was recently described (Li et al., 2009). In accordance with our data, they find that expression of activated RAB-5 Q78L could mimic the tbc-2(−) phenotype, the delayed degradation of apoptotic cell corpses. Interestingly, RAB-7 and LMP-1 fail to be recruited to the phagosome in tbc-2(−) animals and the phenotype does not require RAB-7 activity. Therefore, activation of RAB-5 might recruit and activate RAB-7 during endosome maturation, but not during phagosome maturation. This may reflect the mechanistic differences by which endosomal regulators are recruited to apoptotic cell corpse containing phagosomes (Kinchen et al., 2008).
TBC-2 Is a RAB-5 GAP
Our genetic and phenotypic data, and the requirement for the catalytic arginine 689 of TBC-2 for rescuing activity strongly support the idea that TBC-2 functions as a Rab GAP. Using an in vitro GAP assay that measures GTP hydrolysis in real-time, we show that TBC-2 specifically catalyzes GTP hydrolysis by RAB-5 and to a lesser extent RAB-7. However, the in vitro activity of this TBC-2 protein fragment was considerably weaker than that for previously described Rab GAPs. We found that the catalytic domain of TBC-2 was unstable; however, by including the THR domain and expressing it as an N-terminal SUMO fusion we were able to obtain enough soluble protein for our assays. Deletion of the N-terminal domains preceding the catalytic domains of the yeast Rab GAPs Gyp1p and Gyp7p resulted in greater in vitro catalytic activity, indicating that these domains might play a regulatory role in vivo (Albert et al., 1999). It is possible that the THR domain could have an autoinhibitory activity similar to N-terminal domains of Gyp1p and Gyp7p. Alternatively, TBC-2 could require the N-terminal PH and coiled-coil domains, posttranslational modifications, or the interaction with a cofactor for optimal activity. Because TBC-2 can catalyze GTP hydrolysis by RAB-7 at half the activity of RAB-5, we cannot rule out the possibility that TBC-2 might also directly regulate RAB-7 in vivo. Interestingly, a mammalian homologue of TBC-2, Armus (an isoform of TBC1D2), has recently been found to display GAP activity toward Rab7 in vitro (Frasa et al., 2010). Nonetheless, our in vitro results are consistent with our in vivo data and those of Li et al. (2009) indicating that TBC-2 functions as a RAB-5 GAP, but they also suggest that TBC-2 could function as a RAB-7 GAP.
TBC-2 Might Function on Late Endosomes to Regulate Endosome Maturation
Although TBC-2 localizes to phagosomes with similar timing as RAB-5, before RAB-7 recruitment (Li et al., 2009), we find that TBC-2 primarily colocalizes with RAB-7. Furthermore, we find that RAB-7 is required for TBC-2 membrane localization, suggesting different mechanisms might exist for TBC-2 recruitment to phagosomes versus endosomes.
A countercurrent GAP cascade has recently been demonstrated during Rab conversion in the secretory pathway (Rivera-Molina and Novick, 2009), and it has been proposed that Rab7 might recruit a Rab5 GAP to complete Rab conversion on maturing endosomes (Rink et al., 2005). Localization of TBC-2 with RAB-7 on late endosomes suggests that TBC-2 could function in this capacity. Although RAB-7 is required for TBC-2 membrane localization, further analysis will be required to determine whether RAB-7 actively recruits or regulates TBC-2 activity.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barth Grant, Michaël Hebeisen, and Richard Roy for freely sharing unpublished strains and reagents. We thank May Simaan and Stephane Laporte for assistance and the use of the McGill University Health Centre Research Institute Confocal Microscopy Facility and Jeannie Mui and Hojatollah Vali (McGill University Facility for Electron Microscopy Research) for assistance with electron microscopy. We thank Agneta Rönnlund, Yelena Bernstein, and Monni Begum for technical assistance and Meera Sundaram (University of Pennsylvania) for previous support. We thank Shohei Mitani, Yuji Kohara, Alicia Meléndez, and the Oklahoma Knockout consortium for strains and reagents. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by Cancerfonden (grant 08-0411) (to S. T.), the National Institutes of Health National Institute of General Medical Sciences (grant GM-56324) (to D.G.L.), and Canada Institutes of Health Research (grant MOP-86719) (to C.E.R.). C.E.R. is a Canada Research Chair in Signaling and Development. The Research Institute at the McGill University Health Centre is supported in part by Le Fonds de la Recherche en Santé du Quebec.
Abbreviations used:
- dsRNA
double-stranded RNA
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- HOPS
homotypic fusion and protein sorting
- PH
pleckstrin homology
- ssGFP
signal secreted green fluorescent protein
- TBC
Tre-2/Bub2/Cdc16
- THR
TBC1D2 homology region.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-11-0947) on May 12, 2010.
REFERENCES
- Albert S., Gallwitz D. Two new members of a family of Ypt/Rab GTPase activating proteins. Promiscuity of substrate recognition. J. Biol. Chem. 1999;274:33186–33189. doi: 10.1074/jbc.274.47.33186. [DOI] [PubMed] [Google Scholar]
- Albert S., Will E., Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim. Biophys. Acta. 2003;1603:47–82. doi: 10.1016/s0304-419x(02)00082-3. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune M., Hunter J. L., Corrie J. E., Webb M. R. Direct, real-time measurement of rapid inorganic phosphate release using a novel fluorescent probe and its application to actomyosin subfragment 1 ATPase. Biochemistry. 1994;33:8262–8271. doi: 10.1021/bi00193a013. [DOI] [PubMed] [Google Scholar]
- Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. Rab 7, a key to lysosome biogenesis. Mol. Biol. Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney D. S., Davies B. A., Horazdovsky B. F. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Chen C. C., Schweinsberg P. J., Vashist S., Mareiniss D. P., Lambie E. J., Grant B. D. RAB-10 is required for endocytic recycling in the Caenorhabditis elegans intestine. Mol. Biol. Cell. 2006;17:1286–1297. doi: 10.1091/mbc.E05-08-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clokey G. V., Jacobson L. A. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech. Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- Fares H., Grant B. Deciphering endocytosis in Caenorhabditis elegans. Traffic. 2002;3:11–19. doi: 10.1034/j.1600-0854.2002.30103.x. [DOI] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics. 2001a;159:133–145. doi: 10.1093/genetics/159.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares H., Greenwald I. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat. Genet. 2001b;28:64–68. doi: 10.1038/ng0501-64. [DOI] [PubMed] [Google Scholar]
- Frasa M. A., et al. Armus is a Rac1 effector that inactivates Rab7 and regulates E-cadherin degradation. Curr. Biol. 2010;20:198–208. doi: 10.1016/j.cub.2009.12.053. [DOI] [PubMed] [Google Scholar]
- Grant B., Hirsh D. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell. 1999;10:4311–4326. doi: 10.1091/mbc.10.12.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. K., Fuchs E., Kopajtich R., Barr F. A. A GTPase-activating protein controls Rab5 function in endocytic trafficking. Nat. Cell Biol. 2005;7:887–893. doi: 10.1038/ncb1290. [DOI] [PubMed] [Google Scholar]
- Haas A., Scheglmann D., Lazar T., Gallwitz D., Wickner W. The GTPase Ypt7p of Saccharomyces cerevisiae is required on both partner vacuoles for the homotypic fusion step of vacuole inheritance. EMBO J. 1995;14:5258–5270. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G. J., Schroeder L. K., Hieb C. A., Kershner A. M., Rabbitts B. M., Fonarev P., Grant B. D., Priess J. R. Genetic analysis of lysosomal trafficking in Caenorhabditis elegans. Mol. Biol. Cell. 2005;16:3273–3288. doi: 10.1091/mbc.E05-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh B. M., Hartwieg E., Horvitz H. R. The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc. Natl. Acad. Sci. USA. 2002;99:4355–4360. doi: 10.1073/pnas.062065399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H., et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Itoh T., Satoh M., Kanno E., Fukuda M. Screening for target Rabs of TBC (Tre-2/Bub2/Cdc16) domain-containing proteins based on their Rab-binding activity. Genes Cells. 2006;11:1023–1037. doi: 10.1111/j.1365-2443.2006.00997.x. [DOI] [PubMed] [Google Scholar]
- Johansson M., Rocha N., Zwart W., Jordens I., Janssen L., Kuijl C., Olkkonen V. M., Neefjes J. Activation of endosomal dynein motors by stepwise assembly of Rab7-RILP-p150Glued, ORP1L, and the receptor betaIII spectrin. J. Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens I., Fernandez-Borja M., Marsman M., Dusseljee S., Janssen L., Calafat J., Janssen H., Wubbolts R., Neefjes J. The Rab7 effector protein RILP controls lysosomal transport by inducing the recruitment of dynein-dynactin motors. Curr. Biol. 2001;11:1680–1685. doi: 10.1016/s0960-9822(01)00531-0. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. S., Martinez-Campos M., Zipperlen P., Fraser A. G., Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao G., Tuck S., Baillie D., Sundaram M. V. C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development. 2004;131:755–765. doi: 10.1242/dev.00987. [DOI] [PubMed] [Google Scholar]
- Kinchen J. M., Doukoumetzidis K., Almendinger J., Stergiou L., Tosello-Trampont A., Sifri C. D., Hengartner M. O., Ravichandran K. S. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetti L., Rybin V., Malabarba M. G., Christoforidis S., Scita G., Zerial M., Di Fiore P. P. The Eps8 protein coordinates EGF receptor signalling through Rac and trafficking through Rab5. Nature. 2000;408:374–377. doi: 10.1038/35042605. [DOI] [PubMed] [Google Scholar]
- Lawe D. C., Chawla A., Merithew E., Dumas J., Carrington W., Fogarty K., Lifshitz L., Tuft R., Lambright D., Corvera S. Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J. Biol. Chem. 2002;277:8611–8617. doi: 10.1074/jbc.M109239200. [DOI] [PubMed] [Google Scholar]
- Li W., Zou W., Zhao D., Yan J., Zhu Z., Lu J., Wang X. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 2009;136:2445–2455. doi: 10.1242/dev.035949. [DOI] [PubMed] [Google Scholar]
- Matoskova B., Wong W. T., Nomura N., Robbins K. C., Di Fiore P. P. RN-tre specifically binds to the SH3 domain of eps8 with high affinity and confers growth advantage to NIH3T3 upon carboxy-terminal truncation. Oncogene. 1996;12:2679–2688. [PubMed] [Google Scholar]
- Melendez A., Talloczy Z., Seaman M., Eskelinen E. L., Hall D. H., Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- Meresse S., Gorvel J. P., Chavrier P. The rab7 GTPase resides on a vesicular compartment connected to lysosomes. J. Cell Sci. 1995;108:3349–3358. doi: 10.1242/jcs.108.11.3349. [DOI] [PubMed] [Google Scholar]
- Michaux G., Gansmuller A., Hindelang C., Labouesse M. CHE-14, a protein with a sterol-sensing domain, is required for apical sorting in C. elegans ectodermal epithelial cells. Curr. Biol. 2000;10:1098–1107. doi: 10.1016/s0960-9822(00)00695-3. [DOI] [PubMed] [Google Scholar]
- Nicot A. S., Fares H., Payrastre B., Chisholm A. D., Labouesse M., Laporte J. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol. Biol. Cell. 2006;17:3062–3074. doi: 10.1091/mbc.E05-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz D., Medkova M., Walch-Solimena C., Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J. Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M., Lambright D. G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Papini E., Satin B., Bucci C., de Bernard M., Telford J. L., Manetti R., Rappuoli R., Zerial M., Montecucco C. The small GTP binding protein rab7 is essential for cellular vacuolation induced by Helicobacter pylori cytotoxin. EMBO J. 1997;16:15–24. doi: 10.1093/emboj/16.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal J. B., Seabra M. C. Evolution of the Rab family of small GTP-binding proteins. J. Mol. Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Poteryaev D., Spang A. Application of RNAi technology and fluorescent protein markers to study membrane traffic in Caenorhabditis elegans. Methods Mol. Biol. 2008;440:331–347. doi: 10.1007/978-1-59745-178-9_25. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press B., Feng Y., Hoflack B., Wandinger-Ness A. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 1998;140:1075–1089. doi: 10.1083/jcb.140.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- Rivera-Molina F. E., Novick P. J. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl. Acad. Sci. USA. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruaud A. F., Nilsson L., Richard F., Larsen M. K., Bessereau J. L., Tuck S. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 2009;10:88–100. doi: 10.1111/j.1600-0854.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stenmark H., Olkkonen V. M. The Rab GTPase family. Genome Biol. 2001;2:REVIEWS3007. doi: 10.1186/gb-2001-2-5-reviews3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H., Parton R. G., Steele-Mortimer O., Lutcke A., Gruenberg J., Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J. E., Horvitz H. R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Treusch S., Knuth S., Slaugenhaupt S. A., Goldin E., Grant B. D., Fares H. Caenorhabditis elegans functional orthologue of human protein h-mucolipin-1 is required for lysosome biogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli R., Santillo M., Lattero D., Chiariello M., Bifulco M., Bruni C. B., Bucci C. Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 1997;272:4391–4397. doi: 10.1074/jbc.272.7.4391. [DOI] [PubMed] [Google Scholar]
- Wegener C. S., Malerod L., Pedersen N. M., Prodiga C., Bakke O., Stenmark H., Brech A. Ultrastructural characterization of giant endosomes induced by GTPase-deficient Rab5. Histochem. Cell Biol. 2010;133:41–55. doi: 10.1007/s00418-009-0643-8. [DOI] [PubMed] [Google Scholar]
- Will E., Gallwitz D. Biochemical characterization of Gyp6p, a Ypt/Rab-specific GTPase-activating protein from yeast. J. Biol. Chem. 2001;276:12135–12139. doi: 10.1074/jbc.M011451200. [DOI] [PubMed] [Google Scholar]
- Wurmser A. E., Sato T. K., Emr S. D. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M., McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Toth M., Hamman M. S., Monahan S. J., Lodge P. A., Boynton A. L., Salgaller M. L. Serological cloning of PARIS-1, a new TBC domain-containing, immunogenic tumor antigen from a prostate cancer cell line. Biochem. Biophys. Res. Commun. 2002;290:830–838. doi: 10.1006/bbrc.2001.6257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.