This work shows that Mph1, Mms2, and the Shu complex function in distinct pathways in replication-associated recombinational repair and that the Smc5/6 complex and Esc2 prevent the accumulation of toxic recombination intermediates generated in these pathways.
Abstract
Replication-associated recombinational repair is important for genome duplication and cell survival under DNA damage conditions. Several nonclassical recombination factors have been implicated in this process, but their functional relationships are not clear. Here, we show that three of these factors, Mph1, Mms2, and the Shu complex, can act independently to promote the formation of recombination intermediates during impaired replication. However, their functions become detrimental when cells lack the Smc5/6 complex or Esc2. We show that mph1Δ, mms2Δ, and shu1Δ suppress the sensitivity to the replication-blocking agent methylmethane sulfonate (MMS) in smc6 mutants, with double deletions conferring stronger suppression. These deletion mutations also rescue the MMS sensitivity of esc2Δ cells. In addition, two-dimensional gel analysis demonstrates that mph1Δ, mms2Δ, and shu1Δ each reduce the level of recombination intermediates in an smc6 mutant when cells replicate in the presence of MMS, and that double deletions lead to a greater reduction. Our work thus suggests that Mph1, Mms2, and the Shu complex can function in distinct pathways in replication-associated recombinational repair and that the Smc5/6 complex and Esc2 prevent the accumulation of toxic recombination intermediates generated in these processes.
INTRODUCTION
Recombinational repair provides an important means to facilitate replication when DNA lesions or other obstacles are present on the template. Several modes of replication-associated recombinational repair have been proposed. These include gap filling that repairs single-stranded DNA regions left behind by the replication machinery, template switching that entails the use of newly synthesized sister strands as templates to overcome lesions on parental strands, and replication fork regression in which the newly synthesized DNA strand anneal to each other leading to DNA synthesis and/or strand invasion (Branzei and Foiani, 2007; Lambert et al., 2007; Li and Heyer, 2008; Budzowska and Kanaar, 2009; Chang and Cimprich, 2009). All modes of recombinational repair probably require core recombination proteins such as the recombinase Rad51 and mediator proteins that are essential for homology search and strand invasion (Krogh and Symington, 2004; San Filippo et al., 2008). Moreover, it is increasingly clear that additional proteins are used to couple stalled or interrupted replication with recombination. Unlike core recombination proteins, some of these factors are not crucial for other recombination processes, such as the repair of double-strand breaks in a setting that is not coupled with replication (Lambert et al., 2007; Branzei and Foiani, 2007; Li and Heyer, 2008; Budzowska and Kanaar, 2009). Analyzing the genetic relationships among these factors will help to delineate the recombination pathways operating during replication.
One such group of proteins prevents the accumulation of recombination intermediates when cells replicate in the presence of the DNA-damaging agent methylmethane sulfonate (MMS) (Branzei et al., 2006; Mankouri et al., 2007, 2009; Sollier et al., 2009). Cells lacking these proteins display high levels of Rad51-dependent X-shaped DNA molecules (X-mols) and reduced viability when grown in media containing MMS (Branzei et al., 2006; Mankouri et al., 2007, 2009; Sollier et al., 2009). Two members of this group are the evolutionarily conserved Smc5/6 complex and Esc2. The Smc5/6 complex is composed of Smc5, Smc6, and six other subunits (Hazbun et al., 2003; Zhao and Blobel, 2005; Sergeant et al., 2005; Pebernard et al., 2006; Taylor et al., 2008). One of its subunits, Mms21, possesses SUMO ligase activity that promotes the covalent linkage of SUMO to substrates (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005). In addition, the Smc5/6 complex may have the ability to tether DNA as similar SMC complexes have been shown to condense or cohese DNA strands (Murray and Carr, 2008; De Piccoli et al., 2009). The Esc2 protein contains SUMO-like domains and has no known enzymatic activities (Novatchkova et al., 2005; Raffa et al., 2006; Sollier et al., 2009). The accumulation of X-mols in mutants of the Smc5/6 complex and Esc2 suggest that these two factors can facilitate the resolution and/or limit the formation of potentially toxic recombination structures.
Another group of proteins, in contrast, promotes the formation of recombination intermediates during impaired replication. One such protein is the DNA helicase Mph1, which has been implicated in replication fork regression (Schurer et al., 2004; Prakash et al., 2005; St Onge et al., 2007; Sun et al., 2008). Another in this category is the Shu complex, which is composed of Shu1, Shu2, Psy3, and Csm2, and has been proposed to work as a type of Rad51 paralogue acting at an early recombination step (Huang et al., 2003; Shor et al., 2005; Martin et al., 2006; Mankouri et al., 2007). In addition, proteins in the error-free branch of the postreplicative repair pathway, such as Mms2, a subunit of a ubiquitin conjugating enzyme, also play a role in the formation of recombination intermediates (Branzei et al., 2008). It has been shown that the removal of Mph1 decreases the amount of X-mols in smc6 mutants and esc2Δ cells, whereas the removal of Shu1 exerts a similar effect in esc2Δ cells (Chen et al., 2009; Mankouri et al., 2009). However, it is not clear whether the Shu complex contributes to the accumulation of X-mols in mutants of the Smc5/6 complex and whether Mph1, the Shu complex and Mms2 act independently or in the same pathway. Another unresolved question is whether accumulation of X-mols underlies MMS sensitivity, because mph1Δ has been shown to suppress the MMS sensitivity of smc6 but not esc2Δ mutants (Chen et al., 2009; Mankouri et al., 2009).
To gain a better understanding of the replication-associated recombinational repair system, we examined genetic interactions among the aforementioned proteins that modulate recombination intermediates. Our results suggest that Mph1, the Shu complex, and Mms2 can act independently to promote the formation of recombination structures, and that the Smc5/6 complex and Esc2 prevent toxicity from unresolved recombination intermediates generated by Mph1-, Shu complex-, and Mms2-dependent processes.
MATERIALS AND METHODS
Yeast Strains and Genetic Manipulations
Yeast strains are listed in Table 1. Strains in this study are derivatives of W1588-4C, a RAD5 derivative of W303 (Thomas and Rothstein, 1989). Standard yeast protocols were used for strain construction, growth, and medium preparation. The construction of smc6-56 and smc6-P4 strains has been described previously (Chen et al., 2009). To detect wild-type RAD5 and rad5-535, polymerase chain reaction (PCR) reactions using primer pair RAD5-L (5′-gcagcaggaccatgtaaacg-3′) and RAD5-R (5′-aaactcgttactccactgcg-3′) were performed followed by MnlI digestion. Wild-type RAD5 PCR products gave rise to two fragments (155 and 182 base pairs), whereas rad5-535 PCR products gave rise to three fragments (155, 120, and 62 base pairs). Genotyping of different alleles bearing the same selection marker was carried out by PCR as described below. For shu1Δ::HIS3, the primer pair Shu1 UF (5′-GTATGCGTGT GTTATAC GTGAG-3′) and Shu1 DR (5′-GATGCCTCTTTTTGGTTTCG-3′) was used. shu1Δ::HIS3 gave rise to a 1.6-kb PCR fragment, whereas wild-type SHU1 gave rise to a 0.9-kb fragment. For smc6-56 and smc6-P4, the primer pair Smc6-3163ntF (5′-CAGGTTAACAGGA AGATTGG-3′) and Myc-TADH R (5′-TAGAAGTGGCGCGAATTCAC-3′) was used. Both alleles gave rise to a 750-base pair PCR fragment. For mms2Δ::KAN, the primer pair KAN CterF (5′-CCTATGGAACTGCC TCGGTG-3′) and Mms2 DR (5′-CAAACGCAGAAGCAACTAAAT-3′) was used. mms2Δ::KAN gave rise to a 630-base pair PCR fragment. For esc2Δ::KAN, the primer pair KAN CterF and Esc2 DR (5′-GGTAGAAGAGGG TCAGCAC-3′) was used. esc2Δ::KAN gave rise to a 620-base pair PCR fragment.
Table 1.
Strains used in this studya
| Name | Genotype | Source |
|---|---|---|
| W1588-4C | MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| W2889-19B | MATa shu1Δ::HIS3 | Shor et al. (2005) |
| X2745-3A | MATa mph1Δ::KAN | This study |
| X2745-2A | MATa mph1Δ::KAN shu1Δ::HIS3 | This study |
| T592 | MATa mms2Δ::KAN | This study |
| X2739-4A | MATa mph1Δ::URA3 mms2Δ::KAN | This study |
| X2726-15C | MATa shu1Δ::HIS3 mms2Δ::KAN | This study |
| X2123-2A | MATa smc6-56-13myc::HIS3 | Chen et al. (2009) |
| T382-P4 | MATa smc6-P4–13myc::HIS3 | Chen et al. (2009) |
| X2123-3C | MATa smc6-56-13myc::HIS3 mph1Δ::KAN | Chen et al. (2009) |
| X1787-4A | MATa smc6-P4-13myc::HIS3 mph1Δ::KAN | Chen et al. (2009) |
| X2575-5A | MATa smc6-56-13myc::KAN shu1Δ::HIS3 | This study |
| X2605-12C | MATa smc6-P4-13myc::KAN shu1Δ::HIS3 | This study |
| X2607-14B | MATa smc6-56-13myc::KAN mph1Δ::URA3 shu1Δ::HIS3 | This study |
| X2605-19C | MATa smc6-P4-13myc::KAN mph1Δ::URA3 shu1Δ::HIS3 | This study |
| X2740-4C | MATa smc6-56-13myc::HIS3 mms2Δ::KAN | This study |
| X2739-5A | MATa smc6-P4-13myc::HIS3 mms2Δ::KAN | This study |
| X2819-1A | MATa smc6-56-13myc::HIS3 mms2Δ::KAN shu1Δ::HIS3 | This study |
| X2818-6A | MATa smc6-P4-13myc::HIS3 mms2Δ::KAN shu1Δ::HIS3 | This study |
| X2740-6C | MATa smc6-56-13myc::HIS3 mms2Δ::KAN mph1Δ::URA3 | This study |
| X2739-2B | MATa smc6-P4-13myc::HIS3 mms2Δ::KAN mph1Δ::URA3 | This study |
| X2622-1-20B | MATα rad5-535 LEU2 TRP1 URA3 | This study |
| X2622-1-6C | MATα rad5-535 mph1Δ::KAN LEU2 TRP1 URA3 | This study |
| X2942-2-2D | MATa rad5-535 shu1Δ::HIS3 | This study |
| X2752-4-2A | MATα rad5-535 mms2Δ::KAN LEU2 TRP1 URA3 | This study |
| W6330-3D | MATα esc2Δ::KAN ADE2 TRP1 lys2Δ leu2-ΔBsteII | R. Rothstein |
| X2853-1-3C | MATa esc2Δ::KAN mph1Δ::URA3 lys2Δ | This study |
| X2751-12B | MATa esc2Δ::KAN shu1Δ::HIS3 TRP1 | This study |
| X2750-8B | MATα esc2Δ::KAN mms2Δ::KAN TRP1 | This study |
| T749 | MATa/α smc6Δ::KAN/+ shu1Δ::HIS3/+ mph1Δ::URA3/+ | This study |
| T750 | MATa/α mms21Δ::KAN/+ shu1Δ::HIS3/+ mph1Δ::URA3/+ | This study |
| T768 | MATa/α smc6Δ::KAN/+ mms2Δ::HIS3/+ mph1Δ::URA3/+ | This study |
| T769 | MATa/α mms21Δ::KAN/+ mms2Δ::HIS3/+ mph1Δ::URA3/+ | This study |
| X3056-2A | MATa rad5-535 | This study |
| X3054-2-6A | MATa mph1Δ::KAN rad5-535 | This study |
| X3056-3B | MATa shu1Δ::HIS3 rad5-535 | This study |
| X3056-2C | MATa esc2Δ::KAN rad5-535 | This study |
| X3054-2-6C | MATα mph1Δ::KAN esc2Δ::KAN rad5-535 | This study |
| X3056-3C | MATa shu1Δ::HIS3 esc2Δ::KAN rad5-535 | This study |
| X3057-2-14A | MATa mms2Δ rad5-535 TRP | This study |
| X3057-1-1C | MATα mms2Δ esc2Δ rad5-535 TRP | This study |
| X2662-4A | MATα mph1Δ::URA3 | This study |
| X2662-4C | MATα csm2Δ::KAN | This study |
| X2662-4D | MATa mph1Δ::URA3 csm2Δ::KAN | This study |
| X3053-2D | MATa pol30K164R | This study |
| X3053-8B | MATa pol30-K164R smc6-P4::HIS3 | This study |
| X3005-9B | MATα pol30-K164R esc2Δ::KAN | This study |
a Strains in this study are derivatives of W1588-4C, a RAD5 derivative of W303 (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1 rad5-535; Thomas and Rothstein, 1989). When applicable, a single representative of each genotype is listed.
To test whether shu1Δ or mms2Δ suppresses the lethality caused by smc6Δ and mms21Δ, diploid cells heterozygous for smc6Δ or mms21Δ and for mph1Δ were transformed with a PCR fragment containing the HIS3 marker flanked by the upstream and downstream sequences surrounding the SHU1 or MMS2 ORF, respectively. The transformants were verified for the disruption of the SHU1 or MMS2 gene by PCR (see above) and sporulated. The confirmed diploids were dissected, and plates were incubated at 30°C for 7 d before being photographed. Tetrad analyses were performed as described in the text.
Spot assays for detecting DNA damage sensitivity were carried out as described previously (Chen et al., 2009). In brief, early to mid-log phase cells grown in rich medium (YPD) were spotted in 10-fold serial dilutions (104–10 cells) on plates containing YPD with or without MMS and were grown at 30°C. Plates were photographed 2–4 d after spotting. At least two different isolates for each genotype were tested. Each figure panel shows strains spotted on the same plate, with occasional rearrangement of lanes for presentation purposes.
Two-dimensional (2D) Gel Analysis of Recombination Intermediates
Experiments were performed as described previously (Branzei et al., 2006). In brief, cells were synchronized in G2 by adding nocodazole at a final concentration of 10 mg/ml together with DMSO at 1% (vol/vol) for ∼2.5 h. Cells were then released from nocodazole arrest into YPD medium containing MMS at a final concentration of 0.033% (vol/vol) at 30°C. At the indicated times after release, cells were collected for fluorescence-activated cell sorting (FACS) analysis and purification of DNA intermediates. The DNA samples were digested with HindIII and EcoRV and separated by 2D gel electrophoresis followed by Southern blotting with a probe against ARS305. Quantification of the X-mol signals was performed using the ImageQuant software (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Regions corresponding to monomer DNA, X-mols, and background were selected. Signal intensity (S) and area (A) for each region were calculated by the software. Background (BK) value was derived as the ratio between signal intensity and area for the background region. Values of X-mols (Vx) and monomer DNA (Vm) were calculated by the following equations: Vx = Sx – (BK × Ax) and Vm = Sm − (BK × Am), respectively. The relative amount of X-mols was derived as Vx/(Vx + Vm) and was normalized against the highest values in the plot of Figure 3, C and E. Two different strains were examined for each genotype; each set of experiments was performed twice with qualitatively identical results. Results from representative experiments are shown.
Figure 3.
shu1Δ, mms2Δ, and mph1Δ each reduces the levels of X-mols in smc6-P4 cells with double deletions conferring greater reduction. Cells were arrested using nocodazole and then released into YPD medium containing 0.033% MMS. The replication and recombination intermediates at the ARS305 region 60, 120, 180 and 240 min after release were analyzed by 2D gel electrophoresis followed by Southern blotting (see Materials and Methods). (A) Cartoons indicating the position of the probe and the replication structures. (B and D) Results of 2D gel analyses. X-shaped DNA structures are indicated by arrowheads in smc6-P4. FACS analyses are presented to the right of the gel image; and displayed from bottom to top are the profiles for asynchronous cultures, G2 arrested cells, released cultures at 60, 120, 180, and 240 min. Quantification of the 2D gel results in B and D are plotted in C and E, respectively.
RESULTS
Mph1, the Shu Complex and Mms2 Have Nonoverlapping Functions
Previous studies have shown that mutants of Mph1 and the Shu complex display moderate MMS sensitivity and have epistatic relationships with rad51Δ (Schurer et al., 2004; Shor et al., 2005; Mankouri et al., 2007; Chen et al., 2009). To further understand the recombination processes in which Mph1 and the Shu complex participate, we analyzed the genetic interaction between mph1Δ and deletion of SHU1, which encodes a subunit of the Shu complex. As shown in Figure 1A, mph1Δ shu1Δ double mutants were more sensitive to MMS than either single mutant. A similar interaction was found between mph1Δ and the deletion of the gene encoding another Shu complex subunit, Csm2 (Figure 1B). These results indicate that Mph1 and the Shu complex have nonoverlapping functions under conditions of replicative stress.
Figure 1.
Mph1, the Shu complex, and Mms2 have nonoverlapping functions in DNA repair. Wild-type (WT) and mutant cell cultures were diluted and spotted onto YPD plates with or without the indicated concentration of MMS. mph1Δ enhances the MMS sensitivity of shu1Δ (A), csm2Δ (B), and mms2Δ (C) cells. shu1Δ enhances the MMS sensitivity of mms2Δ cells (D).
Next, we examined how Mph1 and the Shu complex are related to Mms2, which also contributes to the formation of recombination intermediates when cells replicate in the presence of MMS (Branzei et al., 2008). As shown in Figure 1, C and D, both mph1Δ and shu1Δ enhanced the MMS sensitivity of mms2Δ cells, indicating that Mms2 has functions distinct from those of Mph1 and Shu1. The synthetic interactions described above suggest that Mph1, the Shu complex, and Mms2 can function in separate pathways, although they do not exclude the possibility that these proteins may work with each other under certain circumstances.
Both shu1Δ and mms2Δ Rescue the MMS Sensitivity of Two smc6 Mutants
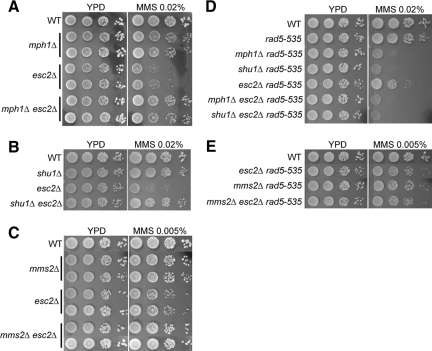
The Smc5/6 complex has been shown to prevent unregulated or incomplete recombination reactions involving Mph1 (Chen et al., 2009). We asked whether it also affects recombinational processes involving Shu1 and Mms2. To address this, genetic interactions between two previously characterized smc6 alleles, smc6-P4 and smc6-56, and shu1Δ and mms2Δ were examined. smc6-P4 and smc6-56 cells display severe MMS sensitivity, which is suppressed by mph1Δ or its helicase mutations (Onoda et al., 2004; Chen et al., 2009; Sollier et al., 2009). We found that the MMS sensitivity of both smc6 mutants was also suppressed by either shu1Δ (Figure 2, A and B) or mms2Δ (Figure 2, C and D), indicating that like Mph1, the actions of the Shu complex and Mms2 also become toxic when the Smc5/6 complex is defective.
Figure 2.
Suppression of MMS sensitivity in smc6 mutants by mph1Δ, shu1Δ, and mms2Δ. (A and B) mph1Δ shu1Δ confers greater suppression of MMS sensitivity of smc6-P4 (A) and smc6-56 (B) cells than either mph1Δ or shu1Δ alone. (C and D) mph1Δ mms2Δ confers greater suppression of MMS sensitivity of smc6-P4 (C) and smc6-56 (D) cells than either mph1Δ or mms2Δ alone. (E and F) shu1Δ mms2Δ confers greater suppression of the MMS sensitivity of smc6-P4 (E) and smc6-56 (F) cells than either shu1Δ or mms2Δ alone.
The strengths of suppression resulting from MPH1, SHU1, or MMS2 deletion in smc6-P4 or smc6-56 cells were different. mph1Δ conferred better suppression than shu1Δ (Figure 2, A and B) and mms2Δ (Figure 2, C and D), and suppression by mms2Δ was greater than that by shu1Δ (Figure 2, E and F). These results are consistent with the idea that Mph1, the Shu complex and Mms2 can function in different processes, and that the Mph1-dependent process causes the most toxicity when the Smc5/6 complex is defective.
Combinatorial Deletions of MPH1, SHU1, and MMS2 Confer Greater Suppression of MMS Sensitivity of smc6 Mutants than Any Single Deletion
To further test the idea that Mph1, the Shu complex and Mms2 have independent functions, we examined whether their combinatorial deletions can result in additive effects in smc6 mutants. As shown in Figure 2, A and B, the mph1Δ shu1Δ double mutant alleviated the MMS sensitivity of smc6-P4 and smc6-56 cells to a greater degree than either single mutant. Similarly, mph1Δ mms2Δ (Figure 2, C and D) and shu1Δ mms2Δ (Figure 2, E and F) resulted in better suppression in both smc6-P4 and smc6-56 cells than the corresponding single deletions. These results strongly support the notion that Mph1, the Shu complex and Mms2 can act independently and that the Smc5/6 complex is genetically linked with the processes involving these three proteins.
shu1Δ, mms2Δ, and mph1Δ Each Reduces the Levels of X-mols in smc6-P4 Cells, with Double Deletions Conferring Greater Reduction
The alleviation of MMS sensitivity in smc6 mutants by mph1Δ, mutations in its helicase domain, and rad51Δ have been attributed to their ability to reduce the levels of potentially toxic recombination intermediates (Branzei et al., 2008; Chen et al., 2009). Here, we examined whether shu1Δ and mms2Δ can also attenuate X-mol accumulation in smc6-P4 cells. Synchronized cells were released into the cell cycle and allowed to replicate in the presence of sublethal doses of MMS. DNA from smc6-P4 and smc6-P4 shu1Δ cells was extracted at intervals and analyzed by 2D agarose gel electrophoresis (2D gel) using a probe for the early firing replication origin ARS305 (Figure 3A). As shown in Figure 3, B and C, shu1Δ, like mph1Δ, reduced the levels of X-mols in smc6-P4 cells. Similarly, mms2Δ also decreased the amounts of X-mols in smc6-P4 cells (Figure 3, D and E). These results correlate well with the observed genetic suppression shown in Figure 2 and indicate that the Smc5/6 complex is required to prevent the accumulation of X-mols generated by Shu1- and Mms2-dependent processes, in addition to those generated by the Mph1-dependent process. Because Mms2 catalyzes proliferating cell nuclear antigen (PCNA; Pol30) polyubiquitination at lysine 164, promoting template switching, we examined whether mutating this residue to arginine can mimic mms2Δ in the suppression of X-mol levels. As shown in Supplemental Figure 1, pol30-K164R, like mms2Δ, reduced the X-mol levels in smc6-P4 cells. This result is in line with previous findings (Moldovan et al., 2007; Branzei et al., 2008) and suggests that Mms2-mediated PCNA polyubiquitination is partly responsible for X-mol accumulation in smc6-P4 cells.
Considering our genetic results showing that mph1Δ shu1Δ, mph1Δ mms2Δ, and shu1Δ mms2Δ yielded greater suppression of smc6 MMS sensitivity than each single deletion, we asked whether these double deletions have additive effects on X-mol levels in smc6 mutants. As shown in Figure 3, B and C, mph1Δ shu1Δ reduced the levels of X-mols in smc6-P4 cells to a greater degree than either single deletion. Similarly, when mph1Δ or shu1Δ was combined with mms2Δ, they conferred a larger reduction in X-mol levels than each single deletion at times when the majority of the cells were undergoing replication (Figure 3, D and E). These results suggest that Mph1, Shu1, and Mms2 can act independently to promote the formation of recombination structures. The similar suppression patterns observed for MMS sensitivity and X-mol levels in all cases strongly indicate that unresolved recombinational structures contribute to the MMS sensitivity of smc6 mutants.
mph1Δ, but Not shu1Δ or mms2Δ, Rescues the Lethality of smc6Δ and mms21Δ
The Smc5/6 complex is not only required for replication under DNA damage conditions but is also essential for normal growth (Murray and Carr, 2008; De Piccoli et al., 2009). Although its essential functions are not completely understood, they are linked with the Mph1-dependent recombinational process, because mph1Δ rescues the lethality caused by deleting the genes encoding Smc5/6 complex subunits (Chen et al., 2009). Considering the observed suppression effects of shu1Δ and mms2Δ in smc6 mutants, we asked whether those deletions can also rescue the inviability of cells lacking the Smc5/6 complex. To do this, a copy of SHU1 or MMS2 was deleted in diploid cells that are heterozygous for mph1Δ and for either smc6Δ or deletion of the gene encoding the Mms21 subunit of the Smc5/6 complex. As summarized in Figure 4, E and F, and shown as examples in Figure 4, A–D, neither shu1Δ nor mms2Δ conferred viability to smc6Δ or mms21Δ cells. Control mph1Δ smc6Δ and mph1Δ mms21Δ cells were viable (Figure 4, A–F) and grew with a doubling time of 6.3 and 5.5 h, respectively (Figure 4G). Furthermore, deletion of either SHU1 or MMS2 did not improve the growth of smc6Δ mph1Δ or mms21Δ mph1Δ cells (Figure 4, A–F). These results suggest that the role of the Smc5/6 complex in the Mph1- but not Shu1- or Mms2-dependent processes is essential for cell viability, further illustrating the difference in the functions of these three proteins.
Figure 4.
mph1Δ, but not shu1Δ or mms2Δ, rescues the lethality of smc6Δ and mms21Δ. (A–D) Representative tetrads from diploid strains with the indicated genotypes are shown. Spore clones with relevant genotypes are labeled with indicated symbols. Genotypes for spore clones containing smc6Δ and mms21Δ were deduced from sibling spore clones. (E–F) Summary of tetrad analysis for the diploids depicted in A–D. The numbers of viable spores versus total spores for each genotype are given (viable/total) for diploids shown in A and C (E) and for the diploids shown in B and D (F). (G) The doubling time of strains with indicated genotype.
shu1Δ, mph1Δ, and mms2Δ All Alleviate the MMS Sensitivity of esc2Δ Cells
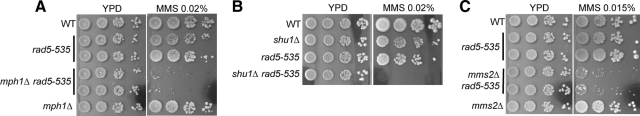
In addition to the Smc5/6 complex, Esc2 is also required to prevent the accumulation of X-mols. esc2Δ cells exhibit increased levels of these DNA structures and are moderately sensitive to MMS. It was recently found that both mph1Δ and shu1Δ decrease the levels of X-mols in esc2Δ cells when replication takes place in MMS-containing media, but neither improves esc2Δ cell survival in MMS-containing media (Mankouri et al., 2009). This lack of correlation in the suppression of two ostensibly related defects in esc2Δ cells is in contrast with our observations in smc6 mutants. Although both studies used strains in the W303 background, we noticed that the previous study used strains containing the rad5-535 mutation (Mankouri et al., 2009). Rad5 is a multifunctional protein that can work with Mms2 as a ubiquitin ligase to polyubiquitinate PCNA, and as a DNA translocase potentially promoting replication fork regression (Johnson et al., 1994; Ulrich and Jentsch, 2000; Hoege et al., 2002; Ulrich, 2003; Blastyak et al., 2007). These two functions of Rad5 are independent of each other, because mutations impairing its ATP binding and translocase activity exhibit synergistic relationship with mms2Δ (Chen et al., 2005). Rad5-535 contains a glycine-to-arginine change at amino acid 535 within the ATP-binding consensus sequence required for its translocase activity (Fan et al., 1996). This change results in mild MMS sensitivity, presumably due to a partial defect in the Rad5 translocase function (Fan et al., 1996). This observation prompted us to examine the genetic relationship of rad5-535 with mph1Δ, shu1Δ, and mms2Δ, and its potential effects on esc2Δ cells.
We first examined whether rad5-535 affects the MMS sensitivity of mph1Δ and shu1Δ cells. As shown in Figure 5, A and B, rad5-535 exacerbated the MMS sensitivity of both mph1Δ and shu1Δ mutants. It also aggravated the MMS sensitivity of mms2Δ cells, which is consistent with the previous observation that mutations affecting Rad5 translocase activity sensitize mms2Δ (Figure 5C [Chen et al., 2005]). These synergistic interactions suggest that the repair process hampered by rad5-535 is distinct from those involving Mph1, Shu1, and Mms2. Second, the genetic interactions between mph1Δ/shu1Δ/mms2Δ and esc2Δ in strains containing wild-type RAD5 were examined. As shown in Figure 6, A–C, mph1Δ, shu1Δ and mms2Δ all improved the survival of esc2Δ cells that contain wild-type RAD5 on MMS-containing media. These results show that shu1Δ and mph1Δ can in fact rescue the MMS sensitivity of esc2Δ cells, in agreement with their suppression of X-mol accumulation. However, this observed effect was lost when cells contained rad5-535 (Figure 6, D and E), suggesting that this mutation can mask the suppression of MMS sensitivity by mph1Δ, shu1Δ and mms2Δ in esc2Δ cells.
Figure 5.
rad5-535 exacerbates the MMS sensitivity of mph1Δ, shu1Δ, and mms2Δ cells. (A–C) Wild-type (WT) and mutant cell cultures were diluted and spotted onto YPD plates with or without the indicated concentration of MMS. rad5-535 enhances the MMS sensitivity of mph1Δ (A), shu1Δ (B), and mms2Δ (C) cells.
Figure 6.
mph1Δ, shu1Δ, and mms2Δ improve esc2Δ cell growth on MMS-containing media in RAD5, but not rad5-535, background. Wild-type (WT) and mutant cell cultures were diluted and spotted onto YPD plates with or without the indicated concentration of MMS. In A–C, strains are in RAD5 background, and the MMS sensitivity of esc2Δ is suppressed by mph1Δ (A), shu1Δ (B), and mms2Δ (C). In D–E, strains contain rad5-535 as indicated.
DISCUSSION
Recombinational repair is important for genome duplication under conditions of replicative stress or DNA damage. Apart from the classical recombination proteins, additional factors that function more specifically in this process have been identified recently (Lambert et al., 2007; Branzei and Foiani, 2007; Li and Heyer, 2008; Budzowska and Kanaar, 2009; Chang and Cimprich, 2009). These factors are critical for understanding the pathways coupling replication and recombinational repair. The functional relationships among a subset of these factors were analyzed in this study. Our results suggest that Mph1, the Shu complex, and Mms2, three factors involved in the formation of recombination intermediates, have nonoverlapping functions and may represent distinct pathways in replication-associated recombinational repair. This conclusion is supported by several observations. First, mph1, shu1, and mms2 deletion mutations exhibited additive genetic interactions. Moreover, their double deletion mutations conferred better suppression of the MMS sensitivity of smc6-P4 and smc6-56 cells than the corresponding single deletions. Significantly, the same suppression patterns were observed in the levels of recombination intermediates in smc6-P4 cells by using 2D gel analyses. The disparate effects of double versus single deletions in all situations argue against these proteins functioning in one pathway and support the idea that they promote recombination via different routes.
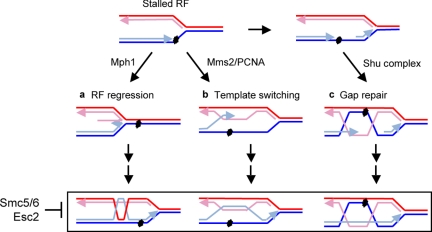
The presence of multiple recombinational repair processes underlines the importance and complexity of the task of rescuing impaired replication. It remains to be seen how Mph1, the Shu complex, and Mms2 differently promote recombination. One possibility is that they act on different structures generated during replication perturbation, such as collapsed forks versus single-stranded DNA gaps. Indeed, Mph1 orthologues have been proposed to catalyze replication fork regression, the Shu complex to facilitate single-strand gap repair, and the Mms2-mediated pathway to promote template switching (Komori et al., 2004; Shor et al., 2005; Ulrich, 2005; Martin et al., 2006; Mankouri et al., 2007; Branzei et al., 2008; Gari et al., 2008; Sun et al., 2008; Unk et al., 2010; Whitby, 2010). Based on the potential functions of these proteins, one plausible model for how they act independently in replication-associated recombinational repair is illustrated in Figure 7. In this model, Mph1 catalyzes fork regression when leading strand synthesis is impeded. Subsequently, the leading strand is extended using the lagging strand as template, and the resulting product can either be regressed back (not shown) or processed by nucleases and engaged in recombination (Figure 7, a). Independently, the Mms2-mediated pathway leads to template switching, for example, the blocked leading strand invades and uses the newly synthesized lagging strand as template to synthesize DNA over the lesion (Figure 7, b). In addition, gaps on the leading or lagging strand can be repaired by Shu complex-mediated recombination, in which this complex may facilitate joint molecule formation in the absence of end invasion (a process proposed in Cunningham et al., 1980) or contribute to recombination over damaged DNA (Figure 7, c). This model provides a good explanation for the observed genetic interactions and takes into consideration the known properties of the proteins. However, other possibilities also exist; for example, these protein factors may work on similar replication intermediates but in different ways depending on their unique interactions and activities. Further study of the functions of these proteins will provide a better understanding of recombinational repair pathways. Because we found that pol30-K164R, like mms2Δ, reduces X-mol levels in smc6-P4 cells and esc2Δ cells (Supplemental Figure 1), the contribution of Mms2 to recombination intermediates probably requires PCNA polyubiquitination. Furthermore, because the DNA translocase activity of Rad5 functions differently from Mph1, the Shu complex, and Mms2, additional complementary routes exist for the rescue of stalled replication.
Figure 7.
Candidate recombinational repair processes that require Mph1, the Shu complex, and Mms2 and the roles of the Smc5/6 complex and Esc2 in these processes. For details, see text. Note that only one of the possible modes of template switching is drawn in b.
Another conclusion supported by our observations is that the Smc5/6 complex is required to prevent the accumulation of recombination structures generated by Mph1-, Shu-, and Mms2-dependent processes (Figure 7) and that this function is crucial for cell survival under replicative stress. Although Mph1 and the Shu complex are thought to function exclusively in recombination, Mms2 has additional roles. However, we favor the interpretation that the Smc5/6 complex affects its roles in recombinational repair, because the removal of Mms2 or proteins functioning with it rescues both the accumulation of X-mols and MMS sensitivity in mutants of the Smc5/6 complex (this study; Branzei et al., 2008). Among the three proteins, the action of Mph1 seems to be most deleterious when the Smc5/6 complex is defective, because mph1Δ exhibited the strongest suppression of MMS sensitivity in smc6 mutants and is the only mutation that rescued the lethality of cells lacking the Smc5/6 complex. Considering that the Smc5/6 complex physically interacts with Mph1 (Chen et al., 2009), an attractive model is that this complex directly modulates Mph1 in fork regression or other processes. The roles of the Smc5/6 complex in the other two pathways remain to be determined, but they could involve direct modulation of protein factors and/or tethering of DNA.
Our finding that the removal of Mph1, Shu1, or Mms2 (in the RAD5 background) resulted in better growth of esc2Δ cells under DNA damage conditions indicates that Esc2, like the Smc5/6 complex, also influences the processes involving these three proteins. These results, in conjunction with the reports by Mankouri et al. (2009), suggest a good correlation between the suppression of X-mol accumulation and MMS sensitivity in esc2Δ cells, similar to the situation in smc6 cells. Therefore, the accrual of X-mols is probably a major underlying defect that accounts for the poor survival of these cells under DNA damage conditions. Although the Smc5/6 complex and Esc2 exhibit similar genetic interactions with Mph1, Shu1, and Mms2, they have at least partly different functions, as suggested by the synthetic interactions of their mutants and by the lack of physical interaction between the two factors (Sollier et al., 2009). It is also noteworthy that Sgs1, a Holliday junction dissolving enzyme that also prevents the accumulation of recombination intermediates, does not affect the Mph1-dependent process (Chen et al., 2009; Mankouri et al., 2009). The necessity of multiple regulators of the multiple replication-associated recombinational processes underlines the need to coordinate recombination steps in dealing with different types of lesions formed when replication is perturbed. Because all the proteins investigated here are conserved in humans, it is likely that similar functional circuitries also exist in higher eukaryotic cells. Molecular dissection of these processes in yeast will thus provide important clues for understanding how recombinational repair facilitates replication in human cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Rothstein (Columbia University, New York) for yeast strains and the method of genotyping rad5-535; H. Klein (New York University, New York) for the pol30-K164R strain; P. Sarangi in the Zhao laboratory for constructing the mms2Δ strain; and C. Cremona, P. Sarangi, L. Hang, W. Holloman, S. Keeney, J. Petrini, N. Lue, and P. Thorpe for comments on the manuscript. This work was supported by the Associazione Italiana per la Ricerca sul Cancro, European Research Council grant 242928 (to D. B.), and National Institutes of Health grant R01-GM080670 (to X. Z.).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0050) on May 5, 2010.
REFERENCES
- Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., Watts F. Z. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A., Pinter L., Unk I., Prakash L., Prakash S., Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Rep. 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Branzei D., Vanoli F., Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Budzowska M., Kanaar R. Mechanisms of dealing with DNA damage-induced replication problems. Cell Biochem. Biophys. 2009;53:17–31. doi: 10.1007/s12013-008-9039-y. [DOI] [PubMed] [Google Scholar]
- Chang D. J., Cimprich K. A. DNA damage tolerance: when it's OK to make mistakes. Nat. Chem. Biol. 2009;5:82–90. doi: 10.1038/nchembio.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Davies A. A., Sagan D., Ulrich H. D. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 2005;33:5878–5886. doi: 10.1093/nar/gki902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-H., Choi K., Szakal B., Arenz J., Duan X., Ye H., Branzei D., Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham R. P., DasGupta C., Shibata T., Radding C. M. Homologous pairing in genetic recombination: recA protein makes joint molecules of gapped circular DNA and closed circular DNA. Cell. 1980;20:223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- De Piccoli G., Torres-Rosell J., Aragon L. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 2009;17:251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- Fan H. Y., Cheng K. K., Klein H. L. Mutations in the RNA polymerase II transcription machinery suppress the hyperrecombination mutant hpr1 of Saccharomyces cerevisiae. Genetics. 1996;142:749–759. doi: 10.1093/genetics/142.3.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., Decaillet C., Stasiak A. Z., Stasiak A., Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Hazbun T. R., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Huang M. E., Rio A. G., Nicolas A., Kolodner R. D. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc. Natl. Acad. Sci. USA. 2003;100:11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. E., Prakash S., Prakash L. Yeast DNA repair protein Rad5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem. 1994;269:28259–28262. [PubMed] [Google Scholar]
- Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., Ishino Y. Cooperation of the N-terminal Helicase and C-terminal endonuclease activities of Archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004;279:53175–53185. doi: 10.1074/jbc.M409243200. [DOI] [PubMed] [Google Scholar]
- Krogh B. O., Symington L. S. Recombination proteins in yeast. Annu. Rev. Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Lambert S., Froget B., Carr A. M. Arrested replication fork processing: interplay between checkpoints and recombination. DNA Rep. 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Li X., Heyer W. D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D. Shu proteins promote the formation of homologous recombination intermediates that are processed by Sgs1-Rmi1-Top3. Mol. Biol. Cell. 2007;18:4062–4073. doi: 10.1091/mbc.E07-05-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri H. W., Ngo H. P., Hickson I. D. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in S. cerevisiae. Mol. Biol. Cell. 2009;20:1683–1694. doi: 10.1091/mbc.E08-08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin V., Chahwan C., Gao H., Blais V., Wohlschlegel J., Yates J. R., 3rd, McGowan C. H., Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan G. L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Murray J. M., Carr A. M. Smc5/ 6, a link between DNA repair and unidirectional replication? Nat. Rev. Mol. Cell Biol. 2008;9:177–182. doi: 10.1038/nrm2309. [DOI] [PubMed] [Google Scholar]
- Novatchkova M., Bachmair A., Eisenhaber B., Eisenhaber F. Proteins with two SUMO-like domains in chromatin-associated complexes: the RENi (Rad60-Esc2-NIP45) family. BMC Bioinform. 2005;6:22. doi: 10.1186/1471-2105-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoda F., Takeda M., Seki M., Maeda D., Tajima J., Ui A., Yagi H., Enomoto T. SMC6 is required for MMS-induced interchromosomal and sister chromatid recombinations in S. cerevisiae. DNA Rep. 2004;3:429–439. doi: 10.1016/j.dnarep.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R., 3rd, Boddy M. N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R., Krejci L., Van Komen S., Anke Schurer K., Kramer W., Sung P. S. cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- Raffa G. D., Wohlschlegel J., Yates J. R., 3rd, Boddy M. N. SUMO-binding motifs mediate the Rad60-dependent response to replicative stress and self-association. J. Biol. Chem. 2006;281:27973–27981. doi: 10.1074/jbc.M601943200. [DOI] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Schurer K. A., Rudolph C., Ulrich H. D., Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeant J., Taylor E., Palecek J., Fousteri M., Andrews E. A., Sweeney S., Shinagawa H., Watts F. Z., Lehmann A. R. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol. Cell. Biol. 2005;25:172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E., Weinstein J., Rothstein R. A genetic screen for top3 suppressors in Saccharomyces cerevisiae identifies SHU1, SHU2, PSY3 and CSM2: four genes involved in error-free DNA repair. Genetics. 2005;169:1275–1289. doi: 10.1534/genetics.104.036764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., Branzei D. The S. cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction mediated intra-S repair. Mol. Biol. Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Onge R. P., Mani R., Oh J., Proctor M., Fung E., Davis R. W., Nislow C., Roth F. P., Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W., Nandi S., Osman F., Ahn J. S., Jakovleska J., Lorenz A., Whitby M. C. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. M., Copsey A. C., Hudson J. J., Vidot S., Lehmann A. R. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol. Cell. Biol. 2008;28:1197–1206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. J., Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Ulrich H. D. Protein-protein interactions within an E2-RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J. Biol. Chem. 2003;278:7051–7058. doi: 10.1074/jbc.M212195200. [DOI] [PubMed] [Google Scholar]
- Ulrich H. D., Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich H. D. The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. Chembiochem. 2005;6:1735–1743. doi: 10.1002/cbic.200500139. [DOI] [PubMed] [Google Scholar]
- Unk I., Hajdu I., Blastyak A., Haracska L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Rep. 2010;9:257–267. doi: 10.1016/j.dnarep.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Whitby M. C. The FANCM family of DNA helicases/translocases. DNA Repair. 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.