Migrating cells do not proliferate and vice versa, but the mechanism involved remains unknown. Ghosh et al. reveal how this cellular decision is made by showing that a Gαi–GIV molecular complex interacts with EGF receptor and programs growth factor signaling, triggering migration when assembled and favoring mitosis when assembly is prevented.
Abstract
Cells respond to growth factors by either migrating or proliferating, but not both at the same time, a phenomenon termed migration-proliferation dichotomy. The underlying mechanism of this phenomenon has remained unknown. We demonstrate here that Gαi protein and GIV, its nonreceptor guanine nucleotide exchange factor (GEF), program EGF receptor (EGFR) signaling and orchestrate this dichotomy. GIV directly interacts with EGFR, and when its GEF function is intact, a Gαi–GIV–EGFR signaling complex assembles, EGFR autophosphorylation is enhanced, and the receptor's association with the plasma membrane (PM) is prolonged. Accordingly, PM-based motogenic signals (PI3-kinase-Akt and PLCγ1) are amplified, and cell migration is triggered. In cells expressing a GEF-deficient mutant, the Gαi–GIV-EGFR signaling complex is not assembled, EGFR autophosphorylation is reduced, the receptor's association with endosomes is prolonged, mitogenic signals (ERK 1/2, Src, and STAT5) are amplified, and cell proliferation is triggered. In rapidly growing, poorly motile breast and colon cancer cells and in noninvasive colorectal carcinomas in situ in which EGFR signaling favors mitosis over motility, a GEF-deficient splice variant of GIV was identified. In slow growing, highly motile cancer cells and late invasive carcinomas, GIV is highly expressed and has an intact GEF motif. Thus, inclusion or exclusion of GIV's GEF motif, which activates Gαi, modulates EGFR signaling, generates migration-proliferation dichotomy, and most likely influences cancer progression.
INTRODUCTION
Cells either migrate or proliferate, but not both at the same time, a phenomenon termed migration-proliferation dichotomy (Giese et al., 1996; Fedotov and Iomin, 2007). This term was initially coined in the context of invading cancer cells, but similar observations were made during development of retinal and tumor blood vessels (Ausprunk and Folkman, 1977; Gerhardt et al., 2003) and epithelial wound healing (Gaylarde and Sarkany, 1975; Bonneton et al., 1999; Chung et al., 1999). What determines the cellular choice between migration and proliferation has remained unknown.
Previous work has established that growth factor receptors such as EGF (Chen et al., 1994a,b), VEGF (Gerhardt et al., 2003), and PDGF (De Donatis et al., 2008) receptors, can trigger both motility and mitosis and that the type and concentration of the activating ligand and the abundance and distribution of receptor influence whether cells migrate or divide. In migrating cells, a distinct set of signals (phospholipase C [PLC]γ1 and PI3-kinase [PI3K]) are amplified and coupled to actin remodeling within pseudopods at the leading edge. In proliferating cells, another set of signals (MAP-kinase [MAPK]/ERK1/2, and c-Src/STAT5b) are amplified, which leads to activation of nuclear transcription factors that drive DNA synthesis during mitosis (Haugh, 2002; Kloth et al., 2003). Ligand stimulation initiates both sets of signals, which are rapidly modulated in the immediate postreceptor phase such that migration and mitosis are executed in a mutually exclusive manner (Chen et al., 1994b; De Donatis et al., 2008). Little is known about how cells responding to growth factors make this decision.
We recently discovered a novel non-receptor GEF for Gαi, GIV, a.k.a. Girdin (Garcia-Marcos, et al., 2009). Activation of Gαi by GIV is required for growth factors (EGF and insulin) to trigger cell migration during epithelial wound healing, macrophage chemotaxis, and tumor cell migration (Ghosh et al., 2008). Others implicated GIV/Girdin in development of retinal and tumor blood vessels (Kitamura et al., 2008) and in cancer invasion and metastasis (Jiang et al., 2008) based on its promigratory effects on cells responding to VEGF and insulin-like growth factor (IGF). We have shown that the GEF motif within GIV's C terminus, which specifically binds and activates Gαi subunits (Garcia-Marcos et al., 2009, 2010) is the critical component of GIV that is required for cell migration. Activation of Gαi releases “free” Gβγ subunits, thereby enhancing Akt signaling via the Gβγ–PI3K pathway. Because Gαi and GIV constitute a key regulatory complex within the growth factor signaling network during cell migration (Ghosh et al., 2008), we investigated whether GIV's GEF function and ability to activate Gαi can influence cells to preferentially migrate or proliferate by regulating the EGF receptor (EGFR), the prototype member of the growth factor receptor tyrosine kinase family.
MATERIALS AND METHODS
Additional experimental protocols and methods are provided in the Supplemental Data.
Reagents and Antibodies
Unless otherwise indicated, all reagents were of analytical grade and obtained from Sigma-Aldrich (St. Louis, MO). Cell culture media were purchased from Invitrogen (Carlsbad, CA). Activated EGFR kinase and phospho-tyrosine monoclonal antibody (mAb) were purchased from Cell Signaling Technology (Danvers, MA). Silencer Negative Control #1 small interfering RNA (siRNA) and siRNA Gαi3 were purchased from Ambion (Austin, TX) and Santa Cruz Biotechnology (Santa Cruz, CA), respectively. Streptavidin-horseradish peroxidase (HRP), biotinylated EGF, and mouse submaxillary EGF were purchased from Invitrogen, and Rhodamine Red X-anti-HRP was from Jackson ImmunoResearch Laboratories (West Grove, PA). Antibodies against GIV that were used in this work include rabbit serum and affinity-purified anti-GIV coiled-coil immunoglobulin (Ig)G (GIV-ccAb) raised against the coiled-coil domain of GIV (Le-Niculescu et al., 2005; Ghosh et al., 2008), and affinity-purified anti-Girdin C terminus (GIV-CTAb) raised against the last 19 aa of GIV's C terminus (IBL America, Minneapolis, MN). To visualize total EGFR by immunofluorescence, mAb #225 raised against the ectodomain (gift from Gordon Gill, University of California San Diego [UCSD], La Jolla, CA; Gill et al., 1984) or polyclonal antibody (pAb) anti-EGFR against the C-terminus of EGFR (Cell Signaling Technology). Polyclonal phosphosite-specific EGFR antibodies (pY992, pY1045, and pY1068; Cell Signaling Technology), mAb pY845 (Millipore, Billerica, MA), and total EGFR (tEGFR) antibodies (Cell Signaling Technology and Santa Cruz Biotechnology) were used for immunoblotting. Rabbit polyclonal antibodies (pAb) against Gαi3 (Calbiochem, San Diego, CA) for immunofluorescence, Gαi3 (M-14, Santa Cruz Biotechnology) for immunoblotting, STAT5b and Grb2 (Santa Cruz Biotechnology), phospho-Y527 Src, PLCγ1, phospho-Y783 PLCγ1, phospho-S473 Akt, and phospho-ERK1/2 (Cell Signaling Technology), were obtained commercially. Mouse monoclonal antibodies (mAb) against phospho-Y845 EGFR and phospho-Y694/Y699-STAT5b (Millipore), Akt and EEA1 (BD Biosciences, San Jose, CA), c-Src (Santa Cruz Biotechnology), ERK1/2 (Cell Signaling Technology), and tubulin (Sigma-Aldrich) were obtained from commercial sources. Anti-mouse and anti-rabbit Alexa-594– and Alexa-488–coupled goat secondary antibodies for immunofluorescence (IF) were purchased from Invitrogen. Goat anti-rabbit and goat anti-mouse Alexa Fluor 680 or IRDye 800 F(ab`)2 for immunoblotting were from LI-COR Biosciences (Lincoln, NE). Control mouse and rabbit IgGs for immunoprecipitations were purchased from Bio-Rad Laboratories (Hercules, CA) and Sigma-Aldrich, respectively.
Plasmid Constructs, Mutagenesis, and Protein Expression
FLAG-EGFR was a generous gift from Dr. Howard A. Rockman (Duke University Medical Center, Durham, NC) (Noma et al., 2007). Cloning of Gαi3 and GIV into pGEX-4T-1 or pET28b were described previously (Garcia-Marcos et al., 2009). To transiently express C-terminal FLAG-tagged Gαi3 in Cos7 cells, Gαi3 was cloned into p3XFLAG-CMVTM-14 expression vector using BamHI and HindIII restriction enzymes. Expression and purification of His-GIV-CT (1623–1870) were done as described previously (Garcia-Marcos et al., 2009). GIV and Gαi3 mutants were generated using specific primers (sequences available upon request) following the manufacturer's instructions (QuikChange II; Stratagene, La Jolla, CA). The truncated GIVΔCT construct was generated by creating a stop codon using a similar protocol to that for creating GIV and Gαi3 mutants. RNA interference (RNAi)-resistant GIV was generated by silent mutations as described previously (Enomoto et al., 2005). To obtain glutathione transferase (GST)-tagged cytoplasmic tail of EGFR (GST-EGFR-T), EGFR cDNA (encoding aa 1046–1210) was amplified by polymerase chain reaction (PCR) and cloned into pGEX4T3 using EcoRI and XhoI sites. All constructs were checked by DNA sequencing.
Plasmids encoding GST-Gαi3, GST-EGFR, His-Gαi3, or His-GIV-CT fusion constructs were used to express these proteins in Escherichia coli exactly as described previously (Ghosh et al., 2008; Garcia-Marcos et al., 2009, 2010).
Cell Culture, Transfection, and Lysis
Unless mentioned otherwise, all cell lines used in this work were cultured according to American Type Culture Collection guidelines. The 21T breast cell lines (16N, NT, and MT2) were obtained from Arthur Pardee (Dana-Farber Cancer Institute and Harvard Medical School, Boston, MA) and maintained as described previously (Band et al., 1990; Qiao et al., 2007). Ls-174T and Ls-LiM6 were obtained from Robert Bresalier (The University of Texas MD Anderson Cancer Center, Houston, TX) and grown as described previously (Bresalier et al., 1987). Ls-174T is a poorly metastatic colon cancer cell line harvested from a Duke's clinical stage B tumor. Ls-LiM6 is a highly metastatic subclone of Ls-174T cells that was selected by serial passage of Ls-174T through a murine cecum-to-liver metastasis model. Transfection was carried out using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) or Oligofectamine (Invitrogen) following the manufacturers' protocols. Transfections to transiently overexpress proteins using FLAG-EGFR, GIV-NT, or Gαi3 plasmids or protein silencing using siRNA Gαi3 were carried out exactly as described previously (Garcia-Marcos et al., 2009). HeLa cell lines stably expressing GIV-wt (HeLa-GIV-wt) or GIV-F1685A mutant (HeLa-GIV-FA) were generated as described previously (Garcia-Marcos et al., 2009). A HeLa cell line stably expressing GIV 1-1354 (GIVΔCT) was generated using the same protocol. All these cell lines were maintained in the presence of G418 (500 μg/ml, Cellgro, Mediatech, Herndon, VA). Clones were chosen for each construct that had relatively low expression levels of GIV (∼3 times higher than endogenous levels). For each construct, two separate clones were investigated, and similar results were obtained.
Lysates used as a source for EGFR or GIV in pull-down or immunoprecipitation assays were prepared by resuspending cells in lysis buffer (20 mM HEPES, pH 7.2, 5 mM Mg-acetate, 125 mM K-acetate, 0.4% Triton X-100, and 1 mM dithiothreitol [DTT], supplemented with 100 μM sodium orthovanadate, phosphatase [Sigma-Aldrich], and protease inhibitor cocktails [Roche Diagnostics]), after which they were passed through a 28-gauge needle at 4°C and cleared (10,000 × g for 5 min) before use in subsequent experiments.
Live Cell Imaging
HeLa cells were grown to confluence in DMEM with 10% serum. Experiments on cells expressing Gαi3-yellow fluorescent protein (YFP) were performed as described previously (Ghosh et al., 2008). For study of GIV-wt and GIV-FA HeLa cells serum was reduced to 0.2%, and after 6–12 h cells were scratch wounded with a 20-μl pipette tip and 0.1 nM EGF was added. Cells were immediately transferred to an incubated stage (37°C in 5% CO2) on an X81 inverted microscope (Olympus, Melville, NY) with ASI x-y-z moving stage (UCSD Neuroscience Microscopy Facility) to capture images of many fields simultaneously. Cells were imaged at 20× magnification every 10 min for up to 12 h. The extent of cell migration or division at the wound edge was assessed by counting the number of cells in each field that displayed polarized motility or successful cell division divided by the total number of cells within ∼2.5 cm (2–3 cells) of the wound edge.
Immunofluorescence
Cells were fixed at room temperature with 3% paraformaldehyde for 30 min, permeabilized (0.2% Triton X-100) for 45 min, and incubated for 1 h each with primary and then secondary antibodies as described previously (Ghosh et al., 2008). Antibody dilutions were as follows: affinity-purified GIV (ccAb), 1:30; Gαi3 (Calbiochem), 1:100; EEA1 (BD Biosciences), 1:250; EGFR (Cell Signaling Technology), 1:100; EGFR #225 mAb, 1:100; secondary goat anti-rabbit (594) and goat anti-mouse (488) Alexa-conjugated antibodies, 1:500; and 4,6-diamidino-2-phenylindole (DAPI; Invitrogen), 1:2000. For surface labeling of EGFR (see Figure 3A), HRP-conjugated EGF was generated by incubating 5 μg Strep-HRP with 0.3 μg biotinylated-EGF in 300 μl PBS overnight at 4C as described previously (Razi and Futter, 2006). Ligand-bound receptor was then visualized using Rhodamine-Red X anti-HRP (1:100; Jackson ImmunoResearch Laboratories). Images were acquired with an Axioimager M1 microscope using a 100× aperture (Plan Neofluar, 1.30 numerical aperture [NA]; Carl Zeiss), Orca-ER camera (Hamamatsu, Bridgewater, NJ), and OpenLab software (Improvision, Lexington, MA). All individual images were processed using ImageJ software (National Institutes of Health, Bethesda, MD) and assembled for presentation using Photoshop and Illustrator software (Adobe Systems, Mountain View, CA).
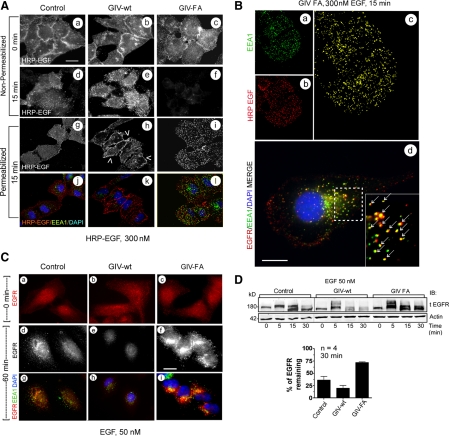
Figure 3.
GIV's GEF function prolongs EGFR localization at the PM but enhances degradation upon internalization. (A) At 15 min after ligand (HRP-EGF) stimulation, ligand-bound EGFR localizes at the PM (e, arrowheads in h) in GIV-wt cells and within intracellular compartments (i and l) in GIV-FA cells. Starved control, GIV-wt, and GIV-FA cells were labeled and stimulated with 300 nM HRP-EGF (equivalent to 50 nM EGF) at 4°C (0 min), washed with PBS, and warmed to 37°C for 15 min. They were then fixed and costained with or without prior permeabilization for HRP, EEA1, and nucleus/DAPI, and visualized by confocal microscopy. Staining for HRP without permeabilization allows selective visualization of ligand-bound receptor at the PM (a–f), whereas permeabilization allows visualization of both the PM and intracellular pools of receptor (g–l). Bar, 10 μm. (B) In GIV-FA cells, EGFR maximally colocalizes with EEA1-positive endosomes at 15 min. GIV-FA cells were stimulated with HRP-EGF for 15 min as in Figure 3A, visualized by confocal microscopy, and analyzed for colocalization of EEA1 (green; a) and HRP (red; b) using Volocity software. The yellow pixels (c) showed significant overlap (Pearson's correlation = 0.45) between HRP-EGF and EEA1. Identical results were obtained when the C terminus of EGFR was stained instead of the ligand (arrows in d). Bar, 10 μm. (C) At 60 min after ligand stimulation, EGFR is virtually undetectable in GIV-wt cells (e and h) but significant staining is seen in GIV-FA cells (f and i) compared with controls (d and g). Cells were stimulated with 50 nM EGF for 60 min and costained for EEA1 (green), tEGFR (red; anti-EGFR cytoplasmic tail), and the nucleus/DAPI (blue). Bar, 10 μm. (D) EGFR degradation is delayed in GIV-FA cells. Serum-starved control, GIV-wt, and GIV-FA HeLa cells were stimulated for 30 min with 50 nM EGF as in Figure 1C and analyzed for total EGFR (tEGFR, anti-EGFR cytoplasmic tail) and actin by immunoblotting (IB; top). Band-shifts and doublets are consistently detected that correlate with phosphorylation of EGFR. Bottom, the amount of receptor (180 kDa, full length) present at 30 min was quantified by Odyssey infrared imaging, normalized to actin, and expressed as percent remaining compared with 0 min. Results are shown as mean ± SEM.
For colocalization of EGFR with GIV and EGFR with Gαi3 (see Figure 4, B and C), HeLa cells were starved overnight in 0.2% fetal bovine serum (FBS) DMEM, stimulated with EGF (Invitrogen) for 0 or 5 min, and then processed for IF as described above. Cells were analyzed by confocal microscopy using 60× PlanApo N objective lenses (1.42 NA, oil immersion) on an inverted FluoView 1000 confocal microscope (Olympus). The microscope was equipped with DAPI (405 nm excitation, 430–470 emission), Alexa-488 (488 excitation, 505/525 emission), Alexa-594 (543 excitation, 560/620 emission), and Alexa-647 (635 excitation, 655–755 emission) filter sets, a manual stage, and a Photometrics CH350 charge-coupled device camera (Hamamatsu). All individual images were analyzed for colocalization by generating scatter-plots using Volocity software (Improvision). Images were processed using ImageJ software and assembled for presentation using Photoshop and Illustrator software (Adobe Systems).
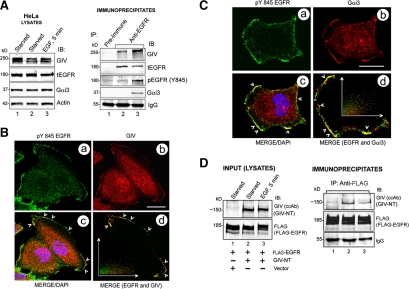
Figure 4.
Gαi3, GIV, and EGFR form a ligand-regulated complex. (A) Endogenous Gαi3 and GIV coimmunoprecipitate with EGFR. Serum-starved HeLa cells were stimulated with 50 nM EGF for 5 min before lysis. Immunoprecipitation was carried out on equal aliquots of cell lysates (left) using anti-EGFR (#225, Lanes 2 and 3) and pre-immune (Lane 1) IgGs, and the bound immune complexes were analyzed for tEGFR, Gαi3, and GIV by immunoblotting (IB; right). Receptor activation was confirmed by immunoblotting for pEGFR (Y845). (B) Endogenous GIV and EGFR partially colocalize at the cell periphery upon ligand stimulation. Starved HeLa cells were stimulated with EGF for 5 min, fixed, stained for pY845 EGFR (a; green), GIV (b; red), and the nucleus/DAPI (blue) and analyzed by confocal microscopy. Merged image (c) shows patches at the PM where GIV and EGFR colocalize (yellow, arrowheads; Pearson's correlation = 0.50; overlap coefficient = 0.50). Inset in d is a scatter-plot of red and green pixels at the PM, generated using Volocity software. Bar, 10 μm. (C) Endogenous Gαi3 and EGFR partially colocalize at the cell periphery upon ligand stimulation. Starved HeLa cells were treated as described in B and stained for activated (pY845) EGFR (a; green), Gαi3 (b; red), and the nucleus/DAPI (blue), and analyzed by confocal microscopy. Merged image (c) shows patches at the PM where Gαi3 and EGFR colocalize (arrowheads, yellow; Pearson's correlation = 0.69; overlap coefficient = 0.69). A scatter plot (d) of red and green pixels at the PM was generated as described in B. Bar, 10 μm. (D) GIV's N terminus constitutively interacts with EGFR. Cos7 cells cotransfected with FLAG-EGFR and a truncated GIV construct (GIV-NT) lacking the C terminus (∼520 aa) were serum starved or treated with EGF, and immunoprecipitation was carried out on cell lysates using anti-FLAG IgG. Left, Cos7 lysates (Input) show expression of GIV-NT and FLAG-EGFR by immunoblotting (IB). RIGHT: Immunoprecipitates analyzed for EGFR and GIV-NT by IB. GIV-NT interacts with EGFR both before (lane 2) and after (lane 3) EGF stimulation. (E) GIV's C terminus (GIV-CT) interacts only with activated EGFR. FLAG-EGFR was immunopurified from starved (SS) or stimulated (EGF) Cos7 cells as described in D followed by incubation of the bead-bound receptor with purified His-GIV-CT (aa 1623–1870) overnight. Bound proteins were analyzed for FLAG-EGFR, His-GIV-CT, and Gαi3, and actin by IB. Gαi3-3XFLAG (lane 6), which is known to interact with GIV-CT (Garcia-Marcos et al., 2009), was used as a positive control. His-GIV-CT binds to the activated EGFR (lane 5) and Gαi3 (lane 6), but not to the inactive receptor (lane 4). (F and G) The C terminus of GIV directly interacts with the phosphorylated cytoplasmic tail of EGFR (EGFR-T). Equal aliquots (15 μg) of GST or GST-EGFR-T (aa 1064–1210) were phosphorylated in vitro using 5 ng of recombinant EGFR kinase and used in pull-down assays with purified His-GIV-CT (aa 1660–1870) (F) or His-Gαi3 (G). Bound proteins were visualized by IB for His, and phosphorylation of EGFR was confirmed by immunoblotting for pTyr. Phosphorylated but not unphosphorylated GST-EGFR-T directly binds His-GIV-CT (F; right lane) but not His-Gαi3 (G; right lane). (H) GIV's GEF motif is required for ligand-stimulated recruitment of Gαi3 and actin to EGFR. Lysates prepared from EGF-stimulated GIV-wt and GIV-FA cells (as described in Figure 1C) were incubated with anti-EGFR (#225) IgG (as described in Figure 2, D–G), and immune complexes were analyzed for tEGFR, phosphorylated (pY845) EGFR, Gαi3, and actin by IB. Gαi3 and actin coimmunoprecipitated with EGFR in GIV-wt (top) but not in GIV-FA cells (bottom) at 5 and 15 min. (I) Working model summarizing findings in A–H. In the starved state, GIV interacts with inactive EGFR (left) through its N terminus, whereas activation of EGFR (right) triggers a regulated interaction between the receptor tail and GIV's C terminus, thereby coupling the receptor tail to G protein signaling at the PM. NT and CT domains of GIV and the G protein are illustrated (bottom).
Immunoprecipitation, Binding Assays, and In Vitro Phosphorylation
Cell lysates (∼1–2 mg of protein) were incubated 4 h at 4°C with either 2 μg of anti-EGFR (#225) mAb (endogenous EGFR), anti-FLAG mAb (Sigma-Aldrich) (FLAG-EGFR), and their respective preimmune control IgGs. Protein G-agarose beads (Invitrogen) were added and incubated at 4°C for an additional 60 min. Beads were washed then either resuspended and boiled in SDS sample buffer, or beads were incubated with purified His-GIV-CT (aa 1623–1870) overnight, washed, and then resuspended and boiled in SDS sample buffer. For immunoprecipitations involving ligand-activated EGFR, buffers used during all steps of the process were supplemented with 100 μM sodium orthovanadate.
For experiments investigating direct interaction between GIV and EGFR, recombinant EGFR kinase (Cell Signaling Technology) was used to phosphorylate GST-tagged EGFR C terminus (EGFR-T, aa 1064–1210) in vitro according to the manufacturer's protocol. In brief, equal aliquots of GST and GST-EGFR-T were incubated with 5 ng purified kinase in the presence of 200 μM ATP (Sigma-Aldrich) at room temperature for 60 min before their use in binding assays.
The in vitro binding assays using GST-fusion proteins were carried out as described previously (Ghosh et al., 2008). In brief, purified GST-fusion proteins (15–20 μg) or GST alone (30 μg) were immobilized on glutathione-Sepharose beads and resuspended in binding buffer supplemented with nucleotides (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 0.4% [vol/vol], NP-40, 10 mM MgCl2, 5 mM EDTA, 2 mM DTT, and protease inhibitor cocktail supplemented with either 30 μM GDP or 30 μM guanosine diphosphate [GDP], 30 μM AlCl3, and 10 mM NaF) (Ghosh et al., 2008). Thereafter, [35S]Met (GE Healthcare) -labeled GIV prepared using the TnT Quick Coupled Transcription/Translation System (Promega, Madison, WI) was added to the binding buffer, and binding was carried out overnight at 4°C with constant tumbling. The following day, the beads were washed (4.3 mM Na2HPO4, 1.4 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, 0.1% [vol/vol] Tween 20, 10 mM MgCl2, 5 mM EDTA, and 2 mM DTT), and boiled in sample buffer for SDS-polyacrylamide gel electrophoresis. For Gαi3, the wash buffer was supplemented with GDP or GDP, AlCl3 and NaF during binding.
Reverse Transcription (RT)-PCR
Total RNA was isolated from each cell line using the RNeasy Mini Kit (QIAGEN, Valencia, CA). First-strand cDNA was generated by RT-PCR using the Superscript First Strand Synthesis System for RT-PCR (Invitrogen). Primers were designed using Invitrogen's Oligo Perfect Designer and evaluated with NetPrimer from Premierbiosoft. Positioning of primers was dependent on the target domains of GIV we intended to amplify (primer sequences are available upon request); glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Allele Biotechnology and Pharmaceuticals, San Diego, CA) mRNA amplified from the same samples served as an internal control. Thirty-five cycles of PCR were carried out across all experiments using JumpStart Taq DNA Polymerase (Sigma-Aldrich) to determine the presence of regions of interest. The amplified PCR products were visualized, photographed (Chemi Doc XRS; Bio-Rad Laboratories), and quantified using Quantity One SW software (Bio-Rad Laboratories). For sequence analysis (DNA Sequencing Facility, UCSD Moore's Cancer Center), PCR products were purified using QIAquik (QIAGEN). To rule out contamination due to genomic DNA, RT-PCR-minus reactions were run as a one step reaction using total RNA for PCR, eliminating the step of cDNA synthesis using reverse transcriptase. To rule out gene fusion events at the C terminus of GIV, rapid amplification of cDNA ends (3`RACE kit; Invitrogen) reactions were carried out on some of the cell lines (HT29, Ls174T, and MCF7). To determine the strength of 19th intron as a splice site, the donor and acceptor splice site strengths were scored by the Splice Site Prediction program by Neural Networks (http://www.fruitfly.org/seq_tools/splice.html), which takes other known splicing sequences into account.
Statistical Analysis
Each experiment presented in the figures is representative of at least three independent experiments. Statistical significance (p value) between various conditions was assessed with the Student's t test. All graphical data presented was prepared using GraphPad software (GraphPad Software, San Diego, CA).
RESULTS
GIV's GEF Function and Gαi Activation Lead to Decreased Proliferation and Increased Migration
To investigate how GIV's GEF function affects cell migration and proliferation, we used live cell imaging to compare the behavior of HeLa cells stably expressing either siRNA-resistant wild-type GIV (GIV-wt cells) or a GEF-deficient GIV F1685A mutant (GIV-FA cells) incapable of interacting with or activating the G protein (Garcia-Marcos et al., 2009). Endogenous GIV was depleted in both cell lines using previously validated siRNA oligonucleotides (Enomoto et al., 2005; Ghosh et al., 2008). Cells were stimulated with EGF in combination with scratch wounding, and both migration and proliferation at the wound edge were simultaneously monitored by live cell imaging. We found that GIV-wt cells typically migrated into the wound, whereas GIV-FA cells showed reduced migration but ∼50% more mitotic events at the wound edge (Supplemental Movies 1 and 2; Figure 1A). Both GIV-wt and GIV-FA cells maintained their distinct behavior with (Figure 1A) or without (Supplemental Figure S1C) depletion of endogenous GIV, indicating that the GEF-deficient GIV F1685A mutant behaves as a dominant-negative protein. The mitotic index as determined by levels of phospho-histone H3, an indicator of mitosis (Hans and Dimitrov, 2001), was consistently high in GIV-FA cells, intermediate in control (untransfected) cells, and low in GIV-wt cells after scratch wounding (Supplemental Figure S2, A and B). Analysis of growth curves at steady state confirmed that expression of GIV-wt suppressed, whereas expression of GIV-F1685A enhanced cell proliferation (data not shown). Thus, in the absence of GIV's GEF function, cells display decreased motility but increased proliferation. Overall these results demonstrate that upon growth factor stimulation, cells with an intact GEF motif preferentially migrate, whereas cells with a disrupted GEF motif preferentially proliferate.
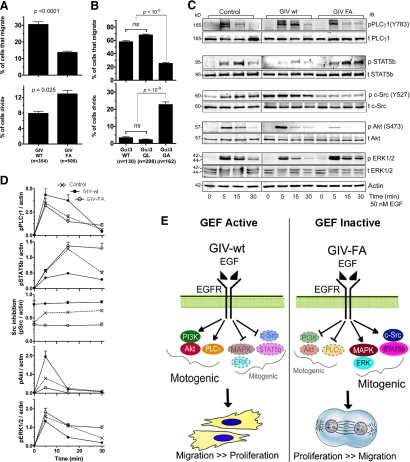
Figure 1.
A Gαi–GIV complex imparts migration-proliferation dichotomy by modulating motogenic and mitogenic signals initiated by EGFR. (A) GIV-wt cells preferentially migrate, whereas GIV-FA cells preferentially proliferate. Confluent monolayers of HeLa cells stably expressing either siRNA-resistant wild-type GIV (GIV-wt) or GEF-deficient GIV (GIV-FA) (see Supplemental Figure S1) were depleted of endogenous GIV, starved for 6–12 h in 0.2% FBS, scratch wounded, and then treated with 0.1 nM EGF. The edge of the wound was monitored by live cell imaging for 12 h (see Supplemental Movies 1 and 2). The total number of cells along the edge of the wound that demonstrated polarized migration into the wound or completed cell division in response to EGF were counted and expressed as the percent of cells analyzed. Bar graphs display the percent of GIV-wt versus GIV-FA cells that migrate (above) or divide (below) during the first 12 h. Results are shown as mean ± SEM (n = cells from 14 to 15 randomly chosen fields from one representative experiment). (B) Active Gαi3 (Q204L) promotes migration, whereas inactive Gαi3 (G203A) promotes mitosis. HeLa cells transfected with Gαi3wt-YFP, Gαi3(QL)-YFP, or Gi3(GA)-YFP were grown to confluence and stimulated by scratch wounding in the presence of 10% serum to initiate EGFR signaling (Tetreault et al., 2007). Cells were then imaged for 8 h as they migrated into the wound (see Supplemental Movies 3 and 4; as in A). Bar graphs display the percent of cells at the edge of the wound that migrate or divide. Results are shown as mean ± SEM (n = cells from 25 to 30 randomly chosen fields from 2 to 3 independent experiments). (C and D) GIV-wt cells enhance PLCγ1 and Akt and suppress c-Src, ERK1/2 and STAT5b signals in response to EGF, whereas GIV-FA cells show the opposite signaling profile. (C) Serum-starved GIV-wt, GIV-FA, and control (untransfected) HeLa cells were stimulated with 50 nM EGF, and whole cell lysates were analyzed for total (t)- and phospho (p)-PLCγ1, c-Src, Akt, ERK1/2, and STAT5b and actin by immunoblotting (IB). (D) Kinetics of EGF-initiated signaling pathways in HeLa cell lines determined by phospho-protein:actin ratios at each time point after EGF stimulation and expressed as fold increase in activation normalized to t = 0 min. Although all three cell lines achieved similar enhancement of phosphorylation of PLCγ1 at 5 min, this enhancement was sustained in GIV-wt but rapidly reduced in GIV-FA cells by 15 min. For STAT5b, phosphorylation was enhanced and sustained up to 30 min GIV-FA cells but not in GIV-wt cells. Inhibitory phosphorylation of Src at Y527 was increased in GIV-wt and suppressed in GIV-FA, but the ligand-dependent variations seen in control cells were lost in GIV-wt and GIV-FA cells. Phosphorylation of Akt was enhanced (peak at 5 min) in GIV-wt and suppressed in GIV-FA cells. Finally, the peak activity of ERK1/2 at 5 min was suppressed in GIV-wt but enhanced in GIV-FA; the latter also displayed sustained ERK activity until 30 min. Results are shown as mean ± SEM (n = 4); p < 0.05 for comparison between GIV-wt and GIV-FA with regard to ERK, Src, PLCγ1, and STAT5b at 15 min and for Akt at 5 min. (E) Summary of the effects of GIV's GEF function on EGF-initiated signaling pathways and the phenotypic outcome. In the presence of a Gi–GIV complex (GEF Active), motogenic cascades are preferentially enhanced and migration is triggered. In the absence of the complex (GEF Inactive), mitogenic cascades are preferentially enhanced and mitosis is triggered.
Because GIV-wt and GIV-FA cells differ by a single point mutation that specifically disrupts GIV's GEF activity and prevents activation of Gαi3 (Garcia-Marcos et al., 2009), we asked whether G protein activity alone is sufficient to create this phenotypic dichotomy. We stimulated HeLa cells expressing YFP-tagged Gαi3-wt or the constitutively inactive Gαi3-G203A (GA) or active (GTPase-deficient) Gαi3-Q204L (QL) mutant (Hermouet et al., 1991) by scratch wounding, and then we monitored cell motility and division simultaneously by live cell imaging. We found that cells expressing Gαi3-wt-YFP and Gαi3-QL-YFP frequently displayed polarized migration into the wound but rarely (∼3%) underwent mitosis, whereas in cells expressing the inactive Gαi3 mutant, cell migration was reduced by ∼50% and mitosis was increased sixfold (Supplemental Movies 3 and 4; and Figure 1B). These findings demonstrate that migration is triggered and proliferation is suppressed when GIV's GEF function is intact or and Gαi3 is constitutively active. By contrast, when GIV's GEF function is selectively disrupted and Gαi3 is inactive, proliferation is triggered and migration is suppressed. Together, these results suggest that Gαi3 activity as determined by the GEF domain of GIV is a critical determinant of cellular preference for migration versus proliferation.
GIV's GEF Function Regulates EGFR Signaling
Next we assessed the effects of disrupting GIV's GEF function on EGF signaling. GIV-wt, GIV-FA, and control HeLa cells were stimulated with EGF and analyzed by immunoblotting for activation (phosphorylation) of four major EGFR-stimulated signals—Akt, PLCγ1, ERK1/2 MAPK, and c-Src/STAT5b (Haugh, 2002; Kloth et al., 2003). GIV-wt cells showed enhanced Akt and PLCγ1 phosphorylation but attenuated STAT5b and ERK1/2 phosphorylation, whereas GIV-FA cells showed the opposite, i.e., enhanced ERK1/2 and STAT5b but attenuated Akt and PLCγ1 signaling (Figure 1, C and D). c-Src activity, as determined by dephosphorylation of pY527 on Src, which coincides with entry into mitosis (Bagrodia et al., 1991), was decreased in GIV-wt and increased in GIV-FA cells compared with controls (Figure 1, C and D). These distinct signaling profiles were also seen in GIV-wt and GIV-FA cells depleted of endogenous GIV (Supplemental Figure S3A), indicating that the differences in signaling were a direct consequence of the presence or absence of a functional GEF motif. Contrasting profiles of Akt and ERK1/2 activation were also evident when GIV-wt and GIV-FA cells were stimulated by scratch wounding (data not shown), indicating that selective signal amplification occurs irrespective of the mode of receptor activation. These results demonstrate that the presence of the GEF motif in GIV-wt cells leads to amplification of Akt and PLCγ1 signals that are predominantly motogenic (i.e., required for motility) and concomitantly suppress c-Src/STAT5b and ERK1/2 signals that are predominantly mitogenic, whereas mutation of the GEF domain in GIV-FA cells leads to a mirror image response (Figure 1E). We conclude that GIV's C-terminal GEF motif, which activates Gαi (Garcia-Marcos et al., 2009), is the critical component of the decision to migrate or proliferate.
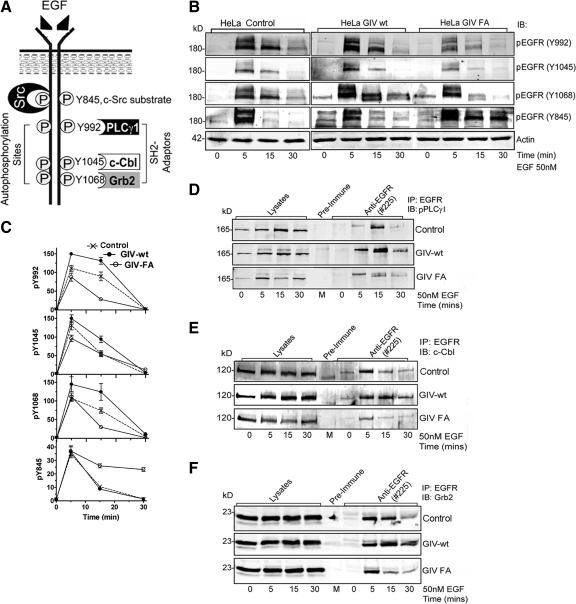
GIV's GEF Function Regulates EGFR Phosphorylation and Recruitment of Scr Homology (SH)2-Adaptors
EGFR signaling to downstream effectors is mediated through ligand-induced tyrosine autophosphorylation of the EGFR tail and SH2 adaptor recruitment to the resultant phosphotyrosines (Schlessinger, 2002). We next investigated whether GIV's GEF activity regulates these two early events in EGFR signaling. Serum starved GIV-wt, GIV-FA, and control HeLa cells were stimulated with EGF, and EGFR autophosphorylation was assessed at Y992, Y1045, and Y1068, (docking sites of the SH2 adaptors, PLCγ1, cCbl, and Grb2) and at Y845 (the substrate for c-Src kinase that triggers mitosis; Tice et al., 1999), using site-specific phospho-Tyr antibodies (Figure 2A). Promigratory GIV-wt cells displayed enhanced EGFR autophosphorylation at Y992, Y1045, and Y1068, whereas Src-dependent phosphorylation at Y845 was similar to controls (Figure 2, B and C; and Supplemental Figure S3B). By contrast, proproliferative GIV-FA cells showed attenuated EGFR autophosphorylation at Y992, Y1045, and Y1068 but sustained phosphorylation at Y845 (Figure 2, B and C; and Supplemental Figure S3B). These results demonstrate that GIV's GEF function is required to enhance EGFR autophosphorylation and that in the absence of a GEF motif the receptor is preferentially phosphorylated by c-Src.
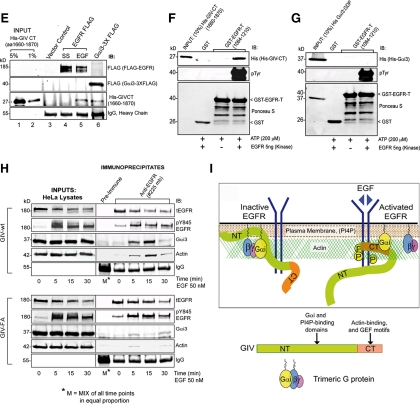
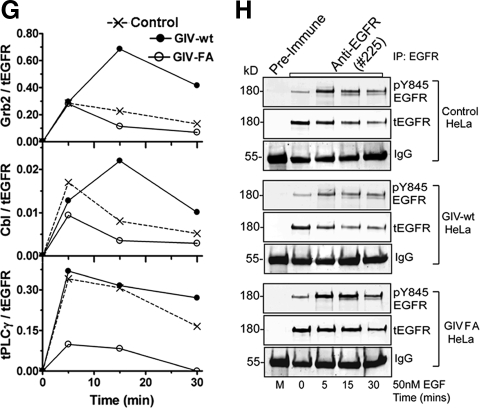
Figure 2.
GIV's GEF function enhances autophosphorylation of EGFR and SH2 adaptor recruitment. (A) Schematic representation of EGFR showing tyrosine autophosphorylation sites and Src phosphorylation site in the cytoplasmic tail and the corresponding SH2 adaptors (PLCγ1, c-Cbl, and Grb2) that are specifically recruited to these sites. (B and C) GIV-wt and GIV-FA cells have distinct profiles of EGFR phosphorylation. (B) Serum-starved GIV-wt, GIV-FA, and control HeLa cells were stimulated with EGF as in described in Figure 1C, and whole cell lysates were analyzed for phospho-EGFR by using p-Tyr site-specific antibodies to Y845, Y992, Y1045, and Y1068 by immunoblotting (IB). GIV-wt cells showed increased EGFR autophosphorylation at Y992 at both 5 and 15 min and at Y1045 and Y1068 at 15 min, whereas GIV-FA cells showed sustained phosphorylation at Y845 at 15 and 30 min. (C) Receptor activation was quantified as described in Figure 1D and is expressed as fold increase in activation normalized to t = 0 min. Results are shown as mean ± SEM (n = 3). (D–H) Recruitment of SH2 adaptors to ligand-activated EGFR is enhanced in GIV-wt but inhibited in GIV-FA cells. Lysates of EGF-stimulated control, GIV-wt, and GIV-FA cells (prepared as in described in Figure 1C) were incubated with anti-EGFR (#225) IgG. Immunoprecipitated complexes were analyzed for adaptor recruitment by immunoblotting (IB) for pPLCγ1 (D), c-Cbl (E), and Grb2 (F). (G) Ratio of SH2 adaptor:tEGFR (y-axis) plotted over time (x-axis). The pPLCγ1:tEGFR and c-Cbl:t EGFR ratios are high in GIV-wt but low in GIV-FA cells at all time points (D and E) after ligand stimulation, whereas the Grb2:tEGFR ratio peaks to a similar extent in all cells at 5 min but is sustained at high levels in GIV-wt cells at 15 and 30 min (F). Results are representative of three separate experiments. (H) Successful immunoprecipitation of ligand-activated receptor in the above assays was confirmed using pY845 EGFR and anti-EGFR cytoplasmic tail (tEGFR).
SH2-adaptor recruitment to the above autophosphorylation sites on the EGFR tail (Figure 2A) was evaluated in GIV-wt versus GIV-FA cells by immunoprecipitating the receptor and analyzing the receptor-bound complexes for Grb2, c-Cbl, and phospho-PLCγ1 by immunoblotting. On ligand stimulation, recruitment of phospho-PLCγ1, c-Cbl, and Grb2 to ligand-activated EGFR was increased in GIV-wt and decreased in GIV-FA cells compared with controls (Figure 2, D–H). These results demonstrate that the GIV's GEF activity affects two of the earliest events in EGF signaling—receptor phosphorylation and SH2 adaptor recruitment, indicating that the Gαi–GIV complex may function at the level of the receptor tail as well as the immediate postreceptor phase.
GIV's GEF Function Is a Key Determinant of EGFR Localization and Degradation
The distribution of ligand-activated EGFR between the plasma membrane (PM) and endosomes is known to affect EGFR signaling (Haugh, 2002) in that motogenic PI3K and PLCγ signals are initiated by ligand-activated receptor largely or exclusively at the PM, whereas mitogenic MAPK-ERK1/2 and c-Src signals can be propagated from endosomes (Murphy et al., 2009). To find out whether the different signaling profiles we observed in GIV-wt and GIV-FA cells are accompanied by differences in distribution of EGFR we compared the localization of ligand-bound EGFR in GIV-wt and GIV-FA cells. Cell surface EGFR was labeled in chilled (4°C), serum-starved cells using HRP-tagged EGF (Razi and Futter, 2006), after which cells were warmed to 37°C for 15 min and then fixed and analyzed for the distribution of the ligand (HRP-EGF)-bound receptor by immunofluorescence. To selectively visualize ligand-bound receptor at the PM, HRP staining was carried out without detergent permeabilization, whereas to visualize both the PM and internalized pools permeabilization was performed before staining. At 0 min, GIV-wt, GIV-FA, and control cells showed a similar pattern and intensity of HRP-EGF labeling on the cell surface, indicating that these cells have similar amounts of EGFR available at the surface for ligand binding (Figure 3A, a–c). However, at 15 min after ligand stimulation a larger pool of ligand-bound EGFR remained as punctate staining on the PM in GIV-wt cells (Figure 3Ae) compared with controls (Figure 3Ad), whereas EGFR was virtually undetectable at the PM in GIV-FA cells (Figure 3Af), and most of the receptor was found in internal compartments (Figure 3A, i and l). To establish the nature of these compartments, we costained HRP-EGF (Figure 3B, a–c) or EGFR [using anti-EGFR mAb; Burke et al., 2001)] (Figure 3Bd) with markers of early (EEA1) or late (CI-MPR; data not shown) endosomes. We found that both HRP-EGF and the receptor colocalized with EEA1, indicating that in GIV-FA cells the ligand-bound receptor was in early endosomes. These data indicate that although there were similar amounts of receptor on the surface at steady state (Figure 3A, a–c), upon ligand stimulation the duration and extent of receptor association with the PM is enhanced in GIV-wt and decreased in GIV-FA cells. In the latter, the receptor is rapidly (within 15 min) compartmentalized within EEA-1–positive early endosomes. Similar results were obtained with either high (50 nM) or low (0.1 nM) concentrations of EGF or when cells were stimulated by scratch wounding (data not shown). At later time points after ligand stimulation (60 min), a significant amount of the receptor was still detectable in the juxtanuclear region in GIV-FA but not in GIV-wt cells (Figure 3C). When total EGFR was quantified by immunoblotting at 30 min after ligand stimulation, ∼20% remained in GIV-wt, ∼40% in controls, and ∼74% in GIV-FA cells (Figure 3D and Supplemental Figure S3B). Thus, GIV-wt promotes and GIV-FA slows EGFR degradation. Together, these results indicate that GIV's GEF activity enhances the receptor pool at the PM but promotes EGFR degradation upon internalization. When the GEF function is disrupted, the cell surface pool of receptor is reduced, the receptor stays longer in endosomes, and degradation of the receptor is delayed. Thus, GIV's GEF motif regulates both spatial and temporal aspects of EGFR signaling.
Gαi, GIV, and EGFR Form a Ligand-regulated Signaling Complex
Based on our findings that disruption of the Gi-GIV interaction affects EGFR signaling and localization, we investigated whether GIV and/or Gαi interacts with EGFR. When we immunoprecipitated EGFR from HeLa (Figure 4A) and Cos7 (Supplemental Figure S4A) cells and immunoblotted for Gαi3 and GIV, both Gαi3 and GIV coimmunoprecipitated with endogenous EGFR. GIV interacted with the receptor under both starved (lane 2) and EGF-stimulated (lane 3) conditions, and ligand stimulation increased GIV–EGFR complexes. By contrast Gαi3 interacted only after EGF stimulation, suggesting that ligand stimulation is required for recruitment of Gαi3. These results demonstrate the existence of a Gαi3-GIV-EGFR complex in vivo. Moreover, by confocal microscopy ligand-activated EGFR partially colocalized with GIV (Figure 4B) and Gαi3 (Figure 4C and Supplemental S4B) at the PM in HeLa cells (analyzed using scatter plots), suggesting that the Gαi-GIV-EGFR signaling complexes form at the PM.
Next, we asked whether the N terminus (GIV-NT) or the C terminus (CT) of GIV participates in formation of the Gαi3–GIV–EGFR complex. We found that GIV-NT (aa 1–1354) coimmunoprecipitated with EGFR under both starved conditions and after EGF stimulation (Figure 4D, lanes 2 and 3). By contrast, GIV-CT (aa 1623–1870), which contains the crucial actin binding (Enomoto et al., 2005) and GEF (Garcia-Marcos et al., 2009) domains, bound only after EGF stimulation (Figure 4E, lane 5). Thus, the interaction between GIV-NT and EGFR is constitutive, whereas the interaction between GIV-CT and EGFR occurs exclusively upon receptor activation. These results support the following model (Figure 4I): In the starved state, GIV couples to EGFR via its NT, and upon ligand stimulation the GIV-EGFR interaction is reinforced by additional recruitment of GIV's CT.
To investigate whether GIV-CT or Gαi3 directly binds to activated EGFR, we used in vitro-phosphorylated, GST-tagged cytoplasmic tail of EGFR (EGFR-T) in pull-down assays with purified His-GIV-CT or His-Gαi3. We found that the phosphorylated EGFR-T directly bound GIV-CT (Figure 4F, last lane) but not Gαi3 (Figure 4G, last lane), suggesting that the Gαi3-EGFR interaction seen in vivo (Figure 4A) is indirect.
GIV's GEF Motif Couples G Protein Activity to EGFR Signaling
To investigate whether the GEF motif in the C terminus of GIV participates in the formation of the Gαi3–GIV–EGFR complex, we immunoprecipitated EGFR from GIV-wt and GIV-FA cells and analyzed the receptor-bound proteins for Gαi3 by immunoblotting. In GIV-wt cells, EGFR transiently and maximally associated with Gαi3 at 15 min after ligand stimulation, whereas in GIV-FA cells Gαi3 recruitment was virtually abolished (Figure 4H). Thus, EGFR–Gαi3 complexes form only when GIV's GEF motif is intact, indicating that EGFR and the G protein are probably linked via GIV (Figure 4I).
GIV's GEF Activity Is Dysregulated in Cancer Cells
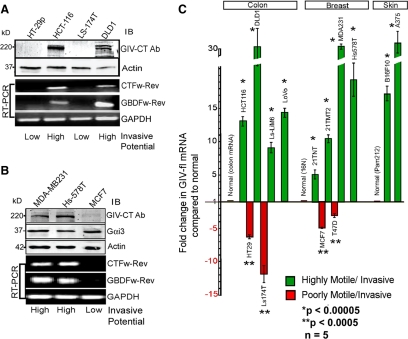
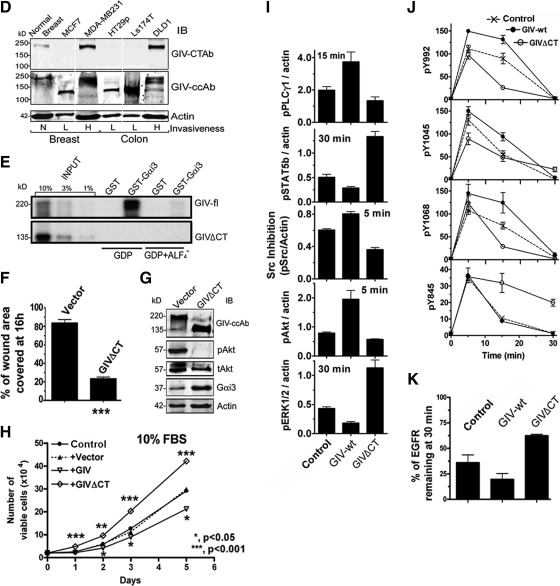
EGFR signaling has been implicated in the generation of migration-proliferation dichotomy during cancer invasion (Athale et al., 2005). Previously, we demonstrated that GIV's GEF function is required for the prometastatic properties of cancer cells (Garcia-Marcos et al., 2009) and that full-length GIV (GIV-fl) is expressed in some but not all cancer cells (Ghosh et al., 2008). Here, we investigated whether the presence or absence of GIV's GEF function differentiates slow growing, highly motile colon cancer cells from rapidly growing, poorly motile cells. Using an antibody against the C-terminal 19 aa of GIV, we detected GIV-fl (∼250 kDa) in slowly growing highly motile/highly invasive (DLD1 and HCT-116) cells but not in rapidly growing poorly motile colon cancer cells (Lelievre et al., 1998; Brattain et al., 1999) with low invasiveness (HT29 and LS174T) (Figure 5A). When we analyzed these cells for expression of GIV-fl mRNA by RT-PCR using two C-terminal GIV domain-specific primers (CT and G binding domain [GBD]) (Supplemental Figure S5A), we found that wherever GIV-fl protein was undetectable the transcript was also virtually abolished (Figure 5A). Similar results were obtained on three breast cancer cell lines with different growth and invasive potential (Howlin et al., 2008): GIV-fl (protein and mRNA) was expressed only in slow-growing, highly motile/invasive cells and undetectable in fast-growing, poorly motile cells with low invasiveness (Figure 5B), whereas levels of Gαi3 were equal. In contrast, a GIV-fl transcript was always detected in normal colon and breast epithelia (Supplemental Figure S5, B and C). Further comparison of the abundance of PCR products (GBD through C terminus) in an extended set of cells revealed that across three different carcinomas GIV-fl was consistently up-regulated ∼3- to 30-fold in highly invasive cells and down-regulated ∼5- to 12-fold in poorly invasive cells (Figure 5C). We confirmed that the up-regulated GIV message in highly invasive cells is wild-type in sequence and that the translated protein selectively binds Gαi3 in the presence of GDP, but not GDP · AlF4− (data not shown), indicating that GIV retains its properties as a nonreceptor GEF (Garcia-Marcos et al., 2009). These results indicate that altered expression of GIV, but not Gαi3, is the key feature that determines whether a functional complex can be assembled in vivo: GIV can interact with Gαi3 in slow-growing, highly motile/invasive cells that express GIV-fl, but not in fast-growing, poorly invasive cells that suppress GIV-fl.
Figure 5.
Full-length GIV is expressed in highly invasive, but not in poorly invasive, cancer cells. (A and B) Full-length GIV (GIV-fl) is detectable only in highly invasive variants of breast and colon carcinoma cells. Lysates of colon (A) and breast (B) cancer cell lines with low or high invasiveness were analyzed for expression of GIV-fl (using GIV-CTAb), Gαi3, and actin by immunoblotting (IB; top) and for GIV and GAPDH mRNA by RT-PCR (bottom). (C) GIV transcript is up-regulated in highly invasive and down-regulated in poorly invasive cancer cells. The relative abundance of amplified cDNA products spanning the C terminus (see Supplemental Figure S5A) in RT-PCR assays (35 cycles) on several colon, breast, and skin cancer cells is expressed as fold change compared with their respective normal controls. GIV was up-regulated ∼3- to 30-fold in highly motile cells (green bars) and down-regulated ∼5- to 12-fold in poorly invasive cells (red bars).
In Fast-growing, Poorly Invasive Cancer Cells, GIV Is Replaced by GIVΔCT, a Truncated Variant Lacking the C-Terminal GEF Motif
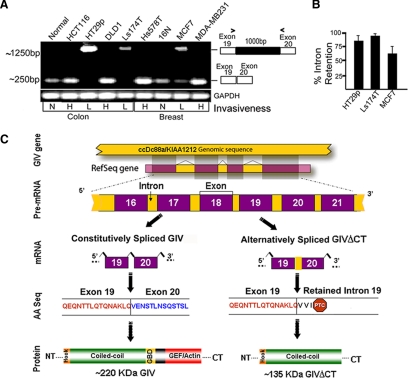
Because metastasis-related genes often undergo alternative splicing leading to aberrant expression (Weber, 2008; Srebrow and Kornblihtt, 2006), we asked whether dysregulation of GIV-fl expression occurs due to alternative splicing. Using RT-PCR and a series of oligonucleotide primers, we investigated the N terminus of GIV and detected transcripts of predicted size (∼250 base pairs) and sequence in highly invasive cancer cells and their respective normal controls (Figure 6A and Supplemental Figure S6B). In poorly invasive colon (Ls174T and HT29) and breast (MCF7) cancer cells, this product was either accompanied by or virtually replaced by an additional, larger (∼1250-base pair) PCR product (Figure 6A and Supplemental Figure S6A). This larger PCR product appeared only when primer pairs flanked the 19th intron. Thus, we suspected that the amplified RNA transcript is the product of an alternative splicing event due to failure of processing and resultant retention of the 19th intron. We confirmed this by sequence analysis and named the aberrant transcript GIV-intron retention 19 (GIV-IR19). In poorly invasive cells, the abundance of GIV-IR19 was inversely proportional to that of GIV-fl—the higher the percentage of intron retention (IR; Figure 6B), the less GIV-fl mRNA (Figure 5C) or protein (Figure 5, A and B). These results indicate that in poorly invasive cells, IR effectively disrupted translation resulting in down-regulation of GIV-fl expression, a phenomenon known to occur in other instances during oncogenesis (Dreyfuss et al., 2002).
Figure 6.
Poorly invasive cancer cells express an alternatively spliced C-terminal truncated GIV protein (GIVΔCT) that fails to bind Gαi3 and enhances mitosis in response to EGF. (A) Retention of intron 19 in GIV mRNA occurs exclusively in poorly invasive cancer cells. RT-PCR was carried out on mRNA isolated from normal (N) and colon and breast cancer cells with high (H) or low (L) invasiveness by using exon 19 forward and exon 20 reverse primers (primer sequences available upon request). Although normal (N) cells and highly (H) metastatic cells yielded the expected ∼250-bp PCR product, poorly (L) invasive cells yielded a larger ∼1250-bp product. (B) Percent of intron retention varies among poorly invasive cancer cells. The percent of IR (calculated as [GIV mRNA retaining intron 19]/[GIV mRNA retaining intron 19 + GIV mRNA with intron 19 processed] × 100) was variable across cell lines: ∼95, ∼85, and ∼50–60% in Ls-174T, HT29, and MCF7 cells, respectively. (C) Proposed scheme for the generation of GIV-IR19 isoform with an in-frame stop codon by alternative splicing of GIV pre-mRNA. Shown are the constitutive and alternative pre-mRNA splicing events, the corresponding GIV mRNA isoforms (GIV and GIV-IR19) generated, the translated amino acid (AA) sequences, and the predicted 220-and 135-kDa protein products (GIV and GIVΔCT). PTC, premature stop codon. (D) A C-terminal truncated protein (GIVΔCT) is expressed exclusively in poorly invasive cells with low metastatic potential. Lysates of normal breast (N) or cancer cells with low (L) or high (H) metastatic potential were immunoblotted (IB) for GIV using GIV-CTAb (against the C-terminal 18 aa of GIV) and GIV-ccAb (against the coiled-coil domain of GIV) and actin. An ∼135-kDa truncated protein is expressed in cells with low (L) invasive potential, whereas GIV-fl is expressed in cells with high (H) invasive potential. (E) GIV-fl, but not GIVΔCT binds to GDP-bound Gαi3. In vitro-translated, [35S]Met-labeled full-length GIV (aa1-1870; top) and GIVΔCT (aa1-1354; bottom) were incubated with ∼15 μg of GST-Gαi3 or GST immobilized on glutathione-agarose beads in the presence of GDP or GDP · AlF4−. Bound GIV was quantified by autoradiography. GIV-fl bound Gαi3 preferentially in the presence of GDP as shown previously (Garcia-Marcos et al., 2009), whereas GEF-deficient GIVΔCT showed no binding. (F–H) GIVΔCT cells proliferate but do not migrate or enhance Akt in scratch wound assays. (F) Confluent monolayers of HeLa cells stably expressing GIVΔCT or vector controls were induced to migrate by scratch wounding, and the area of wound covered at 16 h was quantified as described in Figure 1A. Results are expressed as mean ± SEM, n = 3. (G) Lysates prepared from cells in F were analyzed for GIV, pAkt, tAkt, Gαi3, and actin by IB. (H) Growth curves for cells lines stably expressing GIV-wt (▿), GIVΔCT (♢), vector (▴) or control HeLa (●) in media supplemented with 10% serum. (I) GIVΔCT inhibits motogenic signals but enhances mitogenic signals in response to EGF. GIVΔCT, GIV-wt, and control HeLa cells were stimulated with 50 nM EGF for the indicated times, and analyzed as described in Figure 1, C and D. pAkt, pPLCγ1, c-Src, pERK1/2, and STAT5b were quantified by Odyssey infrared imaging, normalized to actin, and expressed as fold change compared with untransfected controls. Results are shown as mean ± SEM, n = 3. (J and K) GIVΔCT inhibits EGFR autophosphorylation and delays receptor down-regulation in response to EGF. (J) Extent of receptor phosphorylation at Y992, Y1045, Y1068, and Y845 was measured as described in Figure 2C and expressed as fold increase in activation at 30 versus 0 min. Results are shown as mean ± SEM, n = 4. (K) GIVΔCT, GIV-wt, and control HeLa cells were stimulated with 50 nM EGF as described above and analyzed as described in Figure 3D and expressed as %EGFR remaining at 30 min versus 0 min.
GIV-IR19 contains an in-frame stop codon within the retained 19th intron (Figure 6C) and is predicted to translate into a truncated protein without the C terminus. We asked whether this aberrant splice variant is translated in fast-growing, poorly invasive cells. Although GIV-fl was undetectable using GIV-CTAb that recognizes the C terminus (Figure 5, A and B), a ∼135-kDa protein was detected with GIV-cc antibody which recognizes the N-terminal coiled-coil domain (Le-Niculescu et al., 2005; Figure 6D) indicating that this variant lacks the C-terminus (GIVΔCT). We assessed if GIVΔCT, which lacks the C-terminal GEF motif (Garcia-Marcos et al., 2009), could bind Gαi3 in a pull-down assay, and, as anticipated, GIVΔCT did not bind Gαi3 in vitro (Figure 6E); therefore, it is incapable of assembling a functional complex with Gαi3 in vivo.
GEF-deficient GIVΔCT Modulates EGFR Signaling, Inhibits Migration, and Enhances Proliferation
To investigate the consequences of replacing GIV-fl with GIVΔCT, we generated a HeLa cell line expressing GIVΔCT in which the ratio of GIV-fl:GIVΔCT was identical to that found in poorly invasive MCF7 cells (intron retention, ∼65%; Figure 6B). We found that cells expressing GIVΔCT behaved like cells expressing the GEF-deficient, dominant-negative GIV-F1685A mutant in that they migrated poorly (Figure 6F); failed to enhance Akt in scratch wound assays (Figure 6G); and grew more rapidly (Figure 6H) than GIV-wt and controls, indicating that lack of GIV's C terminus renders cells immotile and highly proliferative. On stimulation with EGF, cells expressing GIVΔCT also displayed a signaling profile identical to that observed in GIV-FA cells, in that expression of GIVΔCT suppressed motogenic signals (Akt and PLCγ1), enhanced mitogenic signals (ERK1/2 and STAT5b) (Figure 6I), suppressed EGFR-autophosphorylation (Figure 6J), and delayed receptor degradation (Figure 6K). Thus, disruption of the GEF function is the common denominator that accounts for the identical phenotypes of cells expressing GIV-FA or GIVΔCT.
Furthermore, exogenous expression of GIV-fl in poorly invasive MCF7 cells at high levels using plasmid cDNA (where introns are excluded) enhanced migration and Akt activity (data not shown), suggesting that when alternative splicing is bypassed, GIV-fl is translated and can enhance Akt signaling and trigger epithelial cell migration. GIV-fl induced migration in a dose-dependent manner and increased Akt activity (data not shown), demonstrating that expression of GIV-fl is necessary and sufficient to enhance Akt activation and trigger epithelial cell migration. These findings indicate that the presence or absence of GIV's C-terminus containing its GEF motif can enable or disable Gαi-GIV interaction and thereby trigger migration or mitosis in different cancer cells.
Cells Expressing GEF-deficient GIVΔCT Predominate Early and Those Expressing GIV-wt Are Enriched Later during Progression of Colorectal Carcinomas
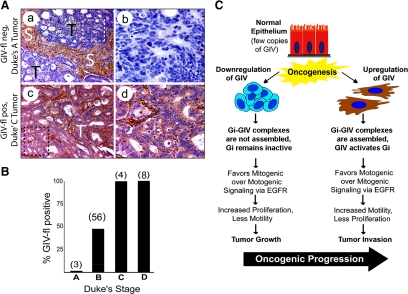
Next, we investigated whether alternative splicing of GIV's C terminus takes place in tumors in vivo during cancer invasion—a process that relies heavily on EGFR signaling (Wells et al., 2002). We carried out immunohistochemistry (IHC) for GIV on paraffin-embedded human colorectal cancers representing various stages of invasive disease (Duke's stages A through D, where A denotes locally restricted tumor without spread and D indicates the presence of distant macroscopic metastases). In normal colon, moderate amounts of GIV-fl as determined with GIV-CTAb were detected in epithelial cells lining the crypt bases (Supplemental Figure S7c). In the epithelia of noninvasive (Duke's A) tumors (Figure 7A, a and b; and B) GIV-fl was virtually undetectable. Among tumors of intermediate stage (Duke's B), ∼50% expressed GIV-fl (Figure 7B), whereas all invasive tumors of advanced stages (Duke's C and D) stained positive for GIV-fl (Figure 7B). In keeping with the previously reported high expression of GIV-fl in mesenchymal cells (Enomoto et al., 2005; Ghosh et al., 2008; Jiang et al., 2008; Kitamura et al., 2008), the stroma stained very strongly in both normal and cancer tissues (Figure 7A and Supplemental Figure S7b). Both normal and tumor tissue stained homogeneously positive for GIV's N terminus using GIV-ccAb irrespective of their clinical stage (data not shown). Thus, noninvasive tumors stained positive for GIV's N terminus, but not for GIV's C terminus, and invasive tumors stained positive for both. These results validate that the phenomenon of alternative splicing observed in cancer cell lines also occurs in tumors in situ. As a result during early, noninvasive stages of tumor growth cells in which GIV is alternatively spliced to generate GEF-deficient GIVΔCT predominate, whereas during late invasive stages the proportion of cells that express full-length GIV progressively increases. These results indicate that expression of GIV-fl is first down-regulated by alternatively splicing to generate a GEF-deficient variant during tumor growth and then up-regulated during tumor invasion (Figure 7C).
Figure 7.
In colorectal carcinomas, expression of GIV-fl is suppressed in early stages of noninvasive tumor growth and increased in late stages of tumor invasion. (A) Cells deficient in GIV-fl dominate noninvasive tumors, whereas those with increased GIV-fl are found in invasive tumors. Paraffin embedded human colon cancer samples were analyzed for GIV-fl by immunohistochemistry using GIV-CTAb. Panels on the left display representative fields from either noninvasive (Duke's A) tumors of early clinical stage (a and b) or invasive (Duke's C/D) tumors of late clinical stages (c and d). Noninvasive Duke's A tumor cells (T; a and b) stain negatively for GIV-fl whereas invasive Duke's C and D tumors (c and d) are strongly positive (d). Stroma (S) consistently stained strongly positive in all tumors irrespective of their clinical stage/invasiveness. b and d are higher magnification views of the boxed regions on the left. (B) Percentage of GIV-fl–positive tumors increases with increasing clinical stage of colorectal carcinoma. GIV expression was analyzed in tumors (as in A) of variable clinical stages by three independent observers with >95% congruence. Bar graphs comparing the proportion of tumors that were scored as positive for full-length GIV expression within each Duke's clinical stage: 0% for Dukes A, ∼48% for B, and 100% for C and D. The total number of tumors examined within each clinical stage is indicated in parentheses. (C) Working model. Expression of GIV-fl undergoes bipartite dysregulation during oncogenesis. In some cancer cells (left), GIV is down-regulated by alternative splicing such that these cells cannot assemble functional Gαi–GIV complexes and fail to activate Gαi. Consequently, EGF signaling is programmed such that mitogenic signals are favored over motogenic pathways. This pattern of signal transduction triggers mitosis and suppresses migration/invasion. By contrast, in other cancer cells (right) the GIV-fl transcript and protein are up-regulated. In these cells functional Gαi–GIV complexes are assembled, via which GIV activates Gαi, and EGF-signaling is programmed such that motogenic signals are preferentially amplified over mitogenic pathways. This pattern of signal transduction triggers migration/invasion and suppresses mitosis in cells overexpressing GIV-fl. The highly proliferative cells in which GIV is down-regulated dominate the noninvasive tumor first and highly invasive cells in which GIV-fl is up-regulated are enriched later during metastatic invasion, suggesting that GIV may influence tumor growth and invasiveness during oncogenic progression.
DISCUSSION
A Gαi–GIV Molecular Complex Establishes Migration-Proliferation Dichotomy
Here, we describe a novel role for GIV's GEF domain and G protein activation in dictating whether cells migrate or proliferate. We show that GIV-wt cells which contain a functional GEF motif and assemble a Gαi–GIV complex frequently migrate but rarely divide in response to EGF. By contrast, GIV-FA cells or GIVΔCT cells that express GEF-deficient variants of GIV, which are defective in their ability to interact with or activate Gαi (Garcia-Marcos et al., 2009), frequently divide but rarely migrate. Cells expressing active or inactive Gi3 mutants also display preferential motility or mitosis, respectively, indicating that Gαi activation status is sufficient for dictating cellular decision making. Overall, these results indicate that the presence or absence of GIV's GEF domain, which activates Gαi is the key to establishing this striking phenotypic dichotomy upon growth factor stimulation.
Previous work predicted a central role for EGFR in migration-proliferation dichotomy (Athale et al., 2005) and demonstrated that the signaling pathways that lead to motility or cell proliferation diverge at the immediate postreceptor phase (Chen et al., 1994b). We have defined the point of divergence by showing that when GIV is able to bind to the EGFR tail and activate Gαi cells are biased to migrate. We further demonstrate that the phenotypic dichotomy imparted by the Gαi–GIV complex stems from its ability to differentially amplify or attenuate signaling cascades initiated by EGFR. In response to EGF, GIV-wt cells show enhanced motogenic (PI3K and PLCγ1) and concomitantly suppressed mitogenic signals (ERK, STAT5b, and c-Src), whereas GIV-FA cells show the opposite. Our finding that G protein activity plays a key role in orchestrating this migration-proliferation dichotomy is also consistent with previous work demonstrating that migration is triggered by active Gαi3 (Ghosh et al., 2008), but mitosis is enhanced by inactive Gαi (Cho and Kehrl, 2007). We conclude that both G protein and growth factor signaling operate through GIV and participate in establishing migration-proliferation dichotomy and that the presence or absence of GIV-dependent Gαi activation is crucial for it to take place. However, it currently remains unknown whether activation of Gαi by GIV's GEF motif eventually mediates its downstream effects via effectors of Gαi and/or Gβγ subunits.
From these findings, we provide evidence that Gαi and both the N and C termini of GIV participate in orchestrating phenotypic dichotomy in the cell's response to growth factors. We found that cells in which GIV was depleted differed significantly in their signaling profile from those lacking the C terminus of GIV (GIVΔCT) or those selectively lacking a functional C-terminal GEF motif (GIV-FA). In GIVΔCT and GIV-FA cells, motogenic pathways are inhibited and mitogenic pathways are enhanced, whereas in cells depleted of endogenous full-length GIV both pathways are inhibited (Supplemental Figure S3A). These differences suggest that the C-terminal GEF motif is required to enhance motogenic and suppress mitogenic pathways; and that in its absence, the N terminus of GIV is sufficient to propagate mitogenic pathways. Based on the insights our results provide on the GIV-EGFR interaction (Figure 4E), we speculate that constitutive interaction between GIV's N terminus and EGFR could contribute to such selective signaling. The nonselective suppression of signals we observe in cells without GIV may also explain why these cells were characterized previously as growth deficient and immotile (Anai et al., 2005; Enomoto et al., 2005). Similarly, we have shown that without Gαi, GIV-dependent signaling and cell migration are inhibited (Ghosh et al., 2008), and cells are rendered proapoptotic (Ohman et al., 2002). We conclude that both Gαi and GIV are required for cell migration and mitosis, however, activation of Gαi by GIV biases the cells to migrate.
GIV's GEF Function Links G Proteins to Growth Factor Receptors
Indirect stimulation of growth factor receptors by GPCR/G protein intermediates—so-called transactivation—is well established (Luttrell et al., 1999). Signaling via G proteins after direct stimulation of growth factor receptors has also been described previously (Waters et al., 2004; Dhanasekaran, 2006), but there is little mechanistic insight into how this could occur. Here, we provide evidence that GIV's GEF motif can serve as a common platform for growth factor and G protein signaling by directly linking EGFR to Gαi subunits. We show that in starved cells GIV's N terminus interacts with EGFR, and upon ligand stimulation the GIV-EGFR interaction is further strengthened by recruitment of GIV's C terminus to the autophosphorylated EGFR tail and recruitment of Gαi3 to the GIV–EGFR complex. This Gαi–GIV–EGFR ternary complex is formed exclusively in GIV-wt, but not in GEF-deficient GIV-FA or GIVΔCT cells, indicating that GIV's C-terminal GEF motif is required for Gαi3 to interact with the activated EGFR. Consequently, efficient coupling of growth factor signaling to Gαi-dependent Akt enhancement and actin remodeling occurs in GIV-wt cells but not in GIV-FA cells (Garcia-Marcos et al., 2009). Where on the receptor tail GIV binds and which phosphorylation events regulate this interaction remain unknown. Given that a variety of growth factors (VEGF, insulin, and IGF1) (Enomoto et al., 2006) trigger cell migration via GIV, it is possible that their receptors share a common mechanism.
The Gαi–GIV Molecular Complex Regulates Temporal and Spatial Aspects of EGFR Signaling
We demonstrate that GIV's GEF activity affects EGFR phosphorylation, distribution, and rate of degradation. We show that in highly motile GIV-wt cells with an intact GEF motif, EGFR autophosphorylation at Y992, Y1045, and Y1068 and the corresponding adaptor recruitment (PLCγ, cCbl, and Grb2) are enhanced, whereas in rapidly proliferating, GEF-deficient GIV-FA cells these events are suppressed. This is in keeping with a previous report that EGFR autophosphorylation is essential for motility but not for mitosis (Chen et al., 1994a).
The integrity of GIV's GEF motif is a key determinant of both receptor distribution and downstream signaling pathways. In GIV-wt cells with an intact GEF motif, ligand-activated EGFR (visualized using HRP-EGF) remains at the PM for an extended period, but once internalized, receptor degradation is accelerated. When GIV's GEF function is selectively disrupted (GIV-FA cells), EGFR is depleted at the PM, its association with endosomes is increased, and its degradation is delayed. We also show that upon EGF stimulation, motogenic PI3K and PLCγ1 signals are selectively enhanced in GIV-wt cells and motility is triggered (Figure 1C), whereas in GIV-FA cells mitogenic c-Src-STAT5b/ERK1/2 signals are propagated, and proliferation is enhanced.
These differences in receptor localization and contrasting profiles of EGFR-initiated signals we observe in GIV-wt versus GIV-FA cells are in keeping with previous reports (Rijken et al., 1991; Haugh, 2002; Howlin et al., 2008; Iyer et al., 2008) that motogenic signals are initiated and coupled to actin remodeling exclusively by receptors at the PM to preferentially trigger motility, whereas internalized receptors preferentially propagate mitogenic signals, presumably from endosomes (Murphy et al., 2009). These differences in EGFR signaling have been attributed to the levels of phosphatidylinositol-4,5-bisphosphate (PI4,5P2), a critical and common substrate of the two key motogenic enzymes PI3K and PLCγ1, which are enriched at the PM but depleted at endosomes (Haugh, 2002). We conclude that motogenic PI3K and PLCγ1 signals are enhanced in the presence of an intact GEF motif, probably due to the persistence of activated receptor at the PI4,5P2-enriched PM, and inhibited in the absence of a GEF motif, probably due to accumulation of activated receptor in the PI4,5P2-depleted endosomes. That the rates of receptor degradation are accelerated in GIV-wt and delayed in GIV-FA cells may reflect a function of GIV in sorting and trafficking of EGFR (Simpson et al., 2005), perhaps modulated by differential recruitment of c-Cbl (Figure 2, A, E, and G), the ubiquitin ligase that promotes endolysosomal degradation of EGFR (Levkowitz et al., 1998).
The distribution of EGFR in GIV-FA cells (decreased at the PM but increased within endosomes) is strikingly similar to that reported previously in cells overexpressing c-Src (Ware et al., 1997). Moreover, we found that increased Src activity in GIV-FA cells is associated with sustained receptor phosphorylation at Y845, a c-Src substrate site, as well as hyperactivation of ERK1/2. Enhanced ERK1/2 activation in spite of reduced Grb2 recruitment could be mediated through Src (Frame, 2004). That highly motile GIV-wt cells restrict ERK activity to low levels is consistent with the computational analysis of proteomic networks of the EGFR pathway (Janes and Lauffenburger, 2006), which predicts that low ERK phosphorylation favors high-speed motility. Our results suggest that increased Src activity may contribute to the spatiotemporal changes in EGFR signaling and enhanced mitosis we observe in the absence of GIV's GEF motif.
We conclude that the presence or absence of GIV's GEF function affects cellular phenotypes by modulating the activity of G proteins that in turn regulates spatial and temporal aspects of EGFR signaling. The molecular mechanisms by which GIV's GEF function helps govern EGFR distribution and regulate its fate remain to be elucidated.
The Gαi–GIV Molecular Complex Imparts Migration-Proliferation Dichotomy in Cancer Cells
We demonstrate that in human breast and colon cancer cells the assembly of the Gαi–GIV molecular complex depends on the differential expression of GIV's GEF motif. In rapidly proliferating, poorly invasive cancer cell lines, GIV is alternatively spliced to produce a dominant-negative variant without the critical C terminus (GIVΔCT). Cells expressing GIVΔCT lack the GEF motif and cannot assemble the Gαi–GIV complex. HeLa cells expressing GIVΔCT behave like GIV-FA cells in that they inhibit motogenic signals and selectively propagate mitogenic signals in response to EGF and are immotile and proliferate rapidly, features that facilitate early tumor growth (Bernards and Weinberg, 2002). In contrast, in highly invasive cancer cells expression of GIV-fl mRNA and protein is up-regulated, and these cells behave like the GIV-wt HeLa cells in that they migrate efficiently (Howlin et al., 2008) via activation of Gαi (Ghosh et al., 2008; Garcia-Marcos et al., 2009) but proliferate more slowly (Lelievre et al., 1998; Howlin et al., 2008) and require GIV for efficient tumor invasion (Jiang et al., 2008). These features are known to hinder early tumor growth but are required later for metastatic progression (Bernards and Weinberg, 2002). To the best of our knowledge, this is the first example of a protein that undergoes bipartite dysregulation during oncogenesis: In slow-growing, highly invasive cancer cells constitutive splicing allows translation of GIV-fl and maintains high levels of GIV-fl mRNA and protein, whereas in rapidly growing, poorly invasive cells alternative splicing of GIV yields a GEF-deficient GIVΔCT variant and restricts GIV-fl mRNA and protein to low levels. This pattern of GIV expression in cancer cells with two different phenotypes is in keeping with the profiles of EGFR signaling and behavior we observe in our HeLa cell lines.
We also demonstrate that expression of GIVΔCT is restricted only to poorly invasive cancer cells. On subsequent analysis of the splice site, we found it to be “weak” based on the lack of homology to consensus mammalian splice signals (Supplemental Figure S6D). However, the weakness of the splice site alone cannot explain the missplicing event observed in poorly invasive breast and colon cancer cells because the sequence is recognized and this intron is efficiently excised in healthy cells or their highly invasive variants (Figure 6A). In addition, mutations (within the 19th intron or the flanking exons) as a cause of aberrant pre-mRNA splicing were ruled out by sequencing. These results suggest that extrinsic factors, e.g., differential expression of splicing factors during cancer progression (Stickeler et al., 1999), may restrict GIVΔCT expression to poorly invasive cells. Regardless of the mechanism, splicing-mediated exclusion of GIV's C-terminal GEF motif was a strikingly conserved theme in poorly motile cancer cells.
We also found that expression of GIV's GEF motif is dysregulated in human colorectal tumors. Noninvasive tumors of early stages are largely comprised of cells that express GIVΔCT which endows cells with a proliferative advantage. With advancing clinical stages, invasive tumors are increasingly made up of cells expressing GIV-fl, which endows cells with an invasive advantage (Jiang et al., 2008). This shift in tumor composition is in keeping with studies demonstrating that phenotypic heterogeneity exists among cells within the same tumor (Giese et al., 1996; Fedotov and Iomin, 2007). Phenotypic heterogeneity has remained a challenge in treatment of carcinomas because only the actively proliferating cells are the most vulnerable to chemotherapy, whereas the nonproliferating cells that are actively invading are resistant to anticancer drugs (Mandel and Rall, 1969). Our findings suggest that alternative splicing of GIV's GEF motif probably contributes to phenotypic heterogeneity and influences early tumor growth as well as late metastatic invasion. In fact, our unpublished work indicates that tumors made up of cells expressing GIVΔCT have increased DNA microsatellite instability and tend to grow larger but are poorly invasive and carry a good prognosis, whereas those made up of cells expressing GIV-fl tend to metastasize early and are associated with poor survival.
In conclusion, we have described a novel molecular mechanism that regulates whether cells migrate or proliferate during cancer invasion. Mechanistic insights gained as to how GIV, a nonreceptor GEF, links G protein activity to growth factor receptor signaling helps define a new paradigm for how G proteins affect growth factor signaling and sheds light on the enigmatic origin of migration-proliferation dichotomy that is observed not only in cancer progression but also during epithelial wound healing and development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gordon N. Gill for anti-EGFR antibodies, scientific advice, and thoughtful comments during preparation of this manuscript; Howard Rockman for the FLAG-tagged EGFR construct; Roger Tsien (UCSD) for sharing his confocal microscope; and Paul Steinbach (UCSD) for assistance with microscopy and analysis software. We also thank the UCSD School of Medicine Light Microscopy Facility, supported in part by National Institutes of Health National (NIH) Institute of Neurological Disorders and Stroke (NINDS) grants P30 NS-047101 and DK-80506 and its director, Jennifer Meerloo. This work was funded by NIH grants CA-100768 and DK-I7780 (to M.G.F.), and Burroughs Wellcome Fund and Research Scholar Award (American Gastroenterology Association FDN) (to P. G.), who was also supplemented by NIH grant T32 DK-07202. M.G.-M. was supported by a Basque Government Postdoctoral fellowship BFI06.300 and Susan G. Komen postdoctoral fellowship KG080079. J. E. is supported by the UCSD McNair Scholarship Program.
Abbreviations used:
- EGFR
Epidermal Growth Factor Receptor
- GEF
Guanine nucleotide Exchange Factor
- PI3K
Phosphoinositide 3-kinase
- PLC
Phospholipase C
- SH2
Src Homology 2
- ERK
Extracellular signal-Regulated Kinase
- STAT5
Signal Transduced and Activator of Transcription 5
- VEGF
Vascular Endothelial Growth Factor
- PDGF
Platelet-Derived Growth Factor
- MAPK
Mitogen Activated Protein Kinase
- HRP
Horseradish Peroxidase
- EEA1
Early Endosome Antigen 1
- PM
Plasma Membrane
- GBD
G protein Binding Domain
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- YFP
yellow fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0028) on May 12, 2010.
REFERENCES
- Anai M., et al. A novel protein kinase B (PKB)/AKT-binding protein enhances PKB kinase activity and regulates DNA synthesis. J. Biol. Chem. 2005;280:18525–18535. doi: 10.1074/jbc.M500586200. [DOI] [PubMed] [Google Scholar]
- Athale C., Mansury Y., Deisboeck T. S. Simulating the impact of a molecular ‘decision-process’ on cellular phenotype and multicellular patterns in brain tumors. J. Theor. Biol. 2005;233:469–481. doi: 10.1016/j.jtbi.2004.10.019. [DOI] [PubMed] [Google Scholar]
- Ausprunk D. H., Folkman J. Migration and proliferation of endothelial cells in preformed and newly formed blood vessels during tumor angiogenesis. Microvasc. Res. 1977;14:53–65. doi: 10.1016/0026-2862(77)90141-8. [DOI] [PubMed] [Google Scholar]
- Bagrodia S., Chackalaparampil I., Kmiecik T. E., Shalloway D. Altered tyrosine 527 phosphorylation and mitotic activation of p60c-src. Nature. 1991;349:172–175. doi: 10.1038/349172a0. [DOI] [PubMed] [Google Scholar]
- Band V., Zajchowski D., Swisshelm K., Trask D., Kulesa V., Cohen C., Connolly J., Sager R. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990;50:7351–7357. [PubMed] [Google Scholar]
- Bernards R., Weinberg R. A. A progression puzzle. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]
- Bonneton C., Sibarita J. B., Thiery J. P. Relationship between cell migration and cell cycle during the initiation of epithelial to fibroblastoid transition. Cell Motil. Cytoskeleton. 1999;43:288–295. doi: 10.1002/(SICI)1097-0169(1999)43:4<288::AID-CM2>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Brattain M. G., Willson J.K.V., Koterba A., Patil S., Venkateswarlu S. Colorectal cancer. In: Masters J.R.W., Palsson B., editors. Human Cell Culture, Vol. 2, Cancer Cell Lines Part 2. London, United Kingdom: Kluwer Academic; 1999. pp. 293–303. [Google Scholar]
- Bresalier R. S., Hujanen E. S., Raper S. E., Roll F. J., Itzkowitz S. H., Martin G. R., Kim Y. S. An animal model for colon cancer metastasis: establishment and characterization of murine cell lines with enhanced liver-metastasizing ability. Cancer Res. 1987;47:1398–1406. [PubMed] [Google Scholar]
- Burke P., Schooler K., Wiley H. S. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell. 2001;12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Gupta K., Wells A. Cell movement elicited by epidermal growth factor receptor requires kinase and autophosphorylation but is separable from mitogenesis. J. Cell Biol. 1994a;124:547–555. doi: 10.1083/jcb.124.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Xie H., Sekar M. C., Gupta K., Wells A. Epidermal growth factor receptor-mediated cell motility: phospholipase C activity is required, but mitogen-activated protein kinase activity is not sufficient for induced cell movement. J. Cell Biol. 1994b;127:847–857. doi: 10.1083/jcb.127.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H., Kehrl J. H. Localization of Gi alpha proteins in the centrosomes and at the midbody: implication for their role in cell division. J. Cell Biol. 2007;178:245–255. doi: 10.1083/jcb.200604114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E. H., Hutcheon A. E., Joyce N. C., Zieske J. D. Synchronization of the G1/S transition in response to corneal debridement. Invest. Ophthalmol. Vis. Sci. 1999;40:1952–1958. [PubMed] [Google Scholar]
- Dhanasekaran D. N. Transducing the signals: a G protein takes a new identity. Sci. STKE. 2006;2006 doi: 10.1126/stke.3472006pe31. [DOI] [PubMed] [Google Scholar]
- De Donatis A., Comito G., Buricchi F., Vinci M. C., Parenti A., Caselli A., Camici G., Manao G., Ramponi G., Cirri P. Proliferation versus migration in platelet-derived growth factor signaling: the key role of endocytosis. J. Biol. Chem. 2008;283:19948–19956. doi: 10.1074/jbc.M709428200. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V. N., Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., Takahashi M. Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell. 2005;9:389–402. doi: 10.1016/j.devcel.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Enomoto A., Ping J., Takahashi M. Girdin, a novel actin-binding protein, and its family of proteins possess versatile functions in the Akt and Wnt signaling pathways. Ann. N Y Acad. Sci. 2006;1086:169–184. doi: 10.1196/annals.1377.016. [DOI] [PubMed] [Google Scholar]
- Fedotov S., Iomin A. Migration and proliferation dichotomy in tumor-cell invasion. Phys. Rev. Lett. 2007;98:118101. doi: 10.1103/PhysRevLett.98.118101. [DOI] [PubMed] [Google Scholar]
- Frame M. C. Newest findings on the oldest oncogene; how activated src does it. J. Cell Sci. 2004;117:989–998. doi: 10.1242/jcs.01111. [DOI] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., Farquhar M. G. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc. Natl. Acad. Sci. USA. 2009;106:3178–3183. doi: 10.1073/pnas.0900294106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Marcos M., Ghosh P., Ear J., Farquhar M. G. A structural determinant that renders Gαi3 sensitive to activation by GIV/Girdin is required to promote cell migration. J. Biol. Cell. 2010;285:12765–12777. doi: 10.1074/jbc.M109.045161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylarde P. M., Sarkany I. Cell migration and DNA synthesis in organ culture of human skin. Br. J. Dermatol. 1975;92:375–380. doi: 10.1111/j.1365-2133.1975.tb03096.x. [DOI] [PubMed] [Google Scholar]
- Gerhardt H., et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J. Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P., Garcia-Marcos M., Bornheimer S. J., Farquhar M. G. Activation of Galphai3 triggers cell migration via regulation of GIV. J. Cell Biol. 2008;182:381–393. doi: 10.1083/jcb.200712066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese A., Loo M. A., Tran N., Haskett D., Coons S. W., Berens M. E. Dichotomy of astrocytoma migration and proliferation. Int. J. Cancer. 1996;67:275–282. doi: 10.1002/(SICI)1097-0215(19960717)67:2<275::AID-IJC20>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Gill G. N., Kawamoto T., Cochet C., Le A., Sato J. D., Masui H., McLeod C., Mendelsohn J. Monoclonal anti-epidermal growth factor receptor antibodies which are inhibitors of epidermal growth factor binding and antagonists of epidermal growth factor binding and antagonists of epidermal growth factor-stimulated tyrosine protein kinase activity. J. Biol. Chem. 1984;259:7755–7760. [PubMed] [Google Scholar]
- Hans F., Dimitrov S. Histone H3 phosphorylation and cell division. Oncogene. 2001;20:3021–3027. doi: 10.1038/sj.onc.1204326. [DOI] [PubMed] [Google Scholar]
- Haugh J. M. Localization of receptor-mediated signal transduction pathways: the inside story. Mol. Interv. 2002;2:292–307. doi: 10.1124/mi.2.5.292. [DOI] [PubMed] [Google Scholar]
- Hermouet S., Merendino J. J., Jr, Gutkind J. S., Spiegel A. M. Activating and inactivating mutations of the alpha subunit of Gi2 protein have opposite effects on proliferation of NIH 3T3 cells. Proc. Natl. Acad. Sci. USA. 1991;88:10455–10459. doi: 10.1073/pnas.88.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlin J., Rosenkvist J., Andersson T. TNK2 preserves epidermal growth factor receptor expression on the cell surface and enhances migration and invasion of human breast cancer cells. Breast Cancer Res. 2008;10:R36. doi: 10.1186/bcr2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer A. K., Tran K. T., Griffith L., Wells A. Cell surface restriction of EGFR by a tenascin cytotactin-encoded EGF-like repeat is preferential for motility-related signaling. J. Cell Physiol. 2008;214:504–512. doi: 10.1002/jcp.21232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janes K. A., Lauffenburger D. A. A biological approach to computational models of proteomic networks. Curr. Opin. Chem. Biol. 2006;10:73–80. doi: 10.1016/j.cbpa.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Jiang P., Enomoto A., Jijiwa M., Kato T., Hasegawa T., Ishida M., Sato T., Asai N., Murakumo Y., Takahashi M. An actin-binding protein Girdin regulates the motility of breast cancer cells. Cancer Res. 2008;68:1310–1318. doi: 10.1158/0008-5472.CAN-07-5111. [DOI] [PubMed] [Google Scholar]
- Kitamura T., et al. Regulation of VEGF-mediated angiogenesis by the Akt/PKB substrate Girdin. Nat. Cell Biol. 2008;10:329–337. doi: 10.1038/ncb1695. [DOI] [PubMed] [Google Scholar]
- Kloth M. T., Laughlin K. K., Biscardi J. S., Boerner J. L., Parsons S. J., Silva C. M. STAT5b, a mediator of synergism between c-src and the epidermal growth factor receptor. J. Biol. Chem. 2003;278:1671–1679. doi: 10.1074/jbc.M207289200. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H., Niesman I., Fischer T., DeVries L., Farquhar M. G. Identification and characterization of GIV, a novel Galpha i/s-interacting protein found on COPI, endoplasmic reticulum-Golgi transport vesicles. J. Biol. Chem. 2005;280:22012–22020. doi: 10.1074/jbc.M501833200. [DOI] [PubMed] [Google Scholar]
- Lelievre V., Muller J. M., Falcon J. Adenosine modulates cell proliferation in human colonic carcinoma. II. Differential behavior of HT29, DLD-1, Caco-2 and SW403 cell lines. Eur. J. Pharmacol. 1998;341:299–308. doi: 10.1016/s0014-2999(97)01463-5. [DOI] [PubMed] [Google Scholar]
- Levkowitz G., Waterman H., Zamir E., Kam Z., Oved S., Langdon W. Y., Beguinot L., Geiger B., Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrell L. M., Daaka Y., Lefkowitz R. J. Regulation of tyrosine kinase cascades by G-protein-coupled receptors. Curr. Opin. Cell Biol. 1999;11:177–183. doi: 10.1016/s0955-0674(99)80023-4. [DOI] [PubMed] [Google Scholar]
- Mandel H. G., Rall D. P. The present status of cancer chemotherapy–a summary of papers delivered at the Cherry Hill Conference on “a critical evaluation of cancer chemotherapy”. Cancer Res. 1969;29:2478–2485. [PubMed] [Google Scholar]
- Murphy J. E., Padilla B. E., Hasdemir B., Cottrell G. S., Bunnett N. W. Endosomes: a legitimate platform for the signaling train. Proc. Natl. Acad. Sci. USA. 2009;106:17615–17622. doi: 10.1073/pnas.0906541106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma T., et al. Beta-arrestin-mediated beta1-adrenergic receptor transactivation of the EGFR confers cardioprotection. J. Clin. Invest. 2007;117:2445–2458. doi: 10.1172/JCI31901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman L., Franzen L., Rudolph U., Birnbaumer L., Hornquist E. H. Regression of Peyer's patches in G alpha i2 deficient mice prior to colitis is associated with reduced expression of Bcl-2 and increased apoptosis. Gut. 2002;51:392–397. doi: 10.1136/gut.51.3.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao M., Iglehart J. D., Pardee A. B. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007;67:5293–5299. doi: 10.1158/0008-5472.CAN-07-0877. [DOI] [PubMed] [Google Scholar]
- Razi M., Futter C. E. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol. Biol. Cell. 2006;17:3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijken P. J., Hage W. J., van Bergen en Henegouwen P. M., Verkleij A. J., Boonstra J. Epidermal growth factor induces rapid reorganization of the actin microfilament system in human A431 cells. J. Cell Sci. 1991;100:491–499. doi: 10.1242/jcs.100.3.491. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- Simpson F., Martin S., Evans T. M., Kerr M., James D. E., Parton R. G., Teasdale R. D., Wicking C. A novel hook-related protein family and the characterization of hook-related protein 1. Traffic. 2005;6:442–458. doi: 10.1111/j.1600-0854.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Srebrow A., Kornblihtt A. R. The connection between splicing and cancer. J. Cell Sci. 2006;119:2635–2641. doi: 10.1242/jcs.03053. [DOI] [PubMed] [Google Scholar]
- Stickeler E., Kittrell F., Medina D., Berget S. M. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene. 1999;18:3574–3582. doi: 10.1038/sj.onc.1202671. [DOI] [PubMed] [Google Scholar]
- Tetreault M. P., Chailler P., Beaulieu J. F., Rivard N., Menard D. Epidermal growth factor receptor-dependent PI3K-activation promotes restitution of wounded human gastric epithelial monolayers. J. Cell Physiol. 2007;214:545–557. doi: 10.1002/jcp.21239. [DOI] [PubMed] [Google Scholar]
- Tice D. A., Biscardi J. S., Nickles A. L., Parsons S. J. Mechanism of biological synergy between cellular Src and epidermal growth factor receptor. Proc. Natl. Acad. Sci. USA. 1999;96:1415–1420. doi: 10.1073/pnas.96.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware M. F., Tice D. A., Parsons S. J., Lauffenburger D. A. Overexpression of cellular Src in fibroblasts enhances endocytic internalization of epidermal growth factor receptor. J. Biol. Chem. 1997;272:30185–30190. doi: 10.1074/jbc.272.48.30185. [DOI] [PubMed] [Google Scholar]
- Waters C., Pyne S., Pyne N. J. The role of G-protein coupled receptors and associated proteins in receptor tyrosine kinase signal transduction. Semin. Cell Dev. Biol. 2004;15:309–323. doi: 10.1016/j.semcdb.2003.12.020. [DOI] [PubMed] [Google Scholar]
- Weber G. F. Molecular mechanisms of metastasis. Cancer Lett. 2008;270:181–190. doi: 10.1016/j.canlet.2008.04.030. [DOI] [PubMed] [Google Scholar]
- Wells A., Kassis J., Solava J., Turner T., Lauffenburger D. A. Growth factor-induced cell motility in tumor invasion. Acta Oncol. 2002;41:124–130. doi: 10.1080/028418602753669481. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.