Abstract
In recent years, researchers have generated a variety of mouse models in an attempt to dissect the contribution of individual genes to the complex phenotype associated with Williams syndrome (WS). The mouse genome is easily manipulated to produce animals that are copies of humans with genetic conditions, be it with null mutations, hypomorphic mutations, point mutations, or even large deletions encompassing many genes. The existing mouse models certainly seem to implicate hemizygosity for ELN, BAZ1B, CLIP2, and GTF2IRD1 in WS, and new mice with large deletions of the WS region are helping us to understand both the additive and potential combinatorial effects of hemizygosity for specific genes. However, not all genes that are haploinsufficient in humans prove to be so in mice and the effect of genetic background can also have a significant effect on the penetrance of many phenotypes. Thus although mouse models are powerful tools, the information garnered from their study must be carefully interpreted. Nevertheless, mouse models look set to provide a wealth of information about the neuroanatomy, neurophysiology and molecular pathways that underlie WS and in the future will act as essential tools for the development and testing of therapeutics.
Keywords: Williams syndrome, mouse models, behavior, gene deletion, phenotype
INTRODUCTION
Genotype–phenotype correlation in Williams syndrome (WS) relies on the identification of individuals with atypical deletions of the critical region. Due to the mechanism of unequal meiotic recombination and subsequent deletion of 7q11.23, these individuals are extremely rare, with the vast majority of deletions spanning the entire 1.5 million base pair interval [Bayes et al., 2003]. Since the discovery of the WS deletion roughly 30 individuals with atypical deletions have been identified, but attempts to correlate genotype with phenotype have been only moderately successful [Morris, 2006]. Although atypical deletion of the WS region can provide some insight into the role of specific genes in the complex phenotype, clear correlations are proving elusive for several reasons. There are only a few individuals, each with a different deletion and in many cases the exact breakpoints of each deletion have not been established. In a few cases, families with multiple affected members have been identified and these enable intra-familial comparisons between people with identical deletions and similar genetic backgrounds [Frangiskakis et al., 1996; Morris et al., 2003; Antonell et al., 2009]. Even where the deletion breakpoints have been defined, the effect on the expression of neighboring genes, which may be altered due to deletion of regulatory elements, has in most cases not been examined. Another major problem with comparing different individuals is that they have been evaluated by different physicians and have not been subject to the same battery of clinical, cognitive, and psychological testing. Finally, and perhaps the most significant problem to date has been the enormous ascertainment bias toward individuals with deletions that include ELN. Hemizygosity for ELN results in an easily distinguishable cardiovascular phenotype that immediately indicates testing for a deletion of the WS region [Ewart et al., 1993]. With the widespread use of genome-wide clinical microarrays to detect copy number changes it is possible that more varied deletions of the WS region will be identified, but to date only a single individual has been found with a deletion that does not encompass ELN [Edelmann et al., 2007].
The generation of mouse models offers a way to circumvent some of these problems, though some still remain, as discussed below. Mice can be engineered with specifically chosen genetic alterations and many mice with the identical genetic alteration can be studied. This removes the ascertainment bias seen in people with atypical deletions and also dramatically decreases the variation between individuals. Since WS is a developmental disorder, mouse models give unparalleled access to pre-natal and post-natal phenotypic characterization and also to tissue for further molecular analysis. Thus, whilst bearing in mind the caveats mentioned in the next section, mouse models of WS provide invaluable tools to study the effect of both individual and combinatorial gene disruption over a wide spectrum of analyses, from the whole animal through to the single molecule.
MICE AS MODELS OF HUMAN DISEASE
Despite the rich clinical resources available for the study of human genetic disease, animal models, and particularly mouse models, can provide valuable insight into the pathogenic mechanisms underlying these disorders. The mouse genome sequence has recently become available for comparison with that of humans and has revealed a very similar gene content [Waterston et al., 2002]. As a consequence mice exhibit many of the clinical symptoms of human disease and sophisticated phenotyping tools are available for their assessment [Rossant and McKerlie, 2001]. Powerful techniques exist for the manipulation of the mouse genome, allowing the germ-line disruption of single genes, multiple genes and even developmental stage-or tissue-specific genetic alterations [Hardouin and Nagy, 2000]. Mouse models allow access to tissues and embryonic time points that are not possible in humans, and allow the study of gene interaction since different genetic combinations can be generated simply through breeding. Finally, a mouse that displays a similar phenotype to a human disease provides an experimental model for the development and testing of new therapeutic interventions.
It must be remembered, however, that mice are not men. Their physiology, although similar, has significant differences that can mean some proteins have divergent functions and some gene disruptions may result in quite different phenotypic consequences. For instance, mice are quadripedal so some musculoskeletal symptoms will present differently than they might in bipedal humans. Mice have a higher metabolic rate, earlier reproductive age and a far shorter lifespan than humans. Mice have adapted to environments, predators and pathogens that are not an issue for people, and vice versa. It is perhaps not surprising then, that one study estimated that at least 20% of human essential genes may be non-essential in the mouse, meaning that they can be homozygously deleted and not result in lethality [Liao and Zhang, 2008]. This is likely a result of human adaptation to environment and involves genes such as those that are necessary for the extended human lifespan. At the genetic level, mice and humans also have around 300 genes that are unique to each species, making human genetic disorders involving these genes impossible to model in another organism [Waterston et al., 2002].
There are also phenotyping hurdles that must be overcome in the mouse. There are obviously limitations to the cognitive and behavioral testing that can be carried out in mice as compared to humans. There are also documented differences in testing protocols and results between research personnel and between laboratories as well as significant effects of genetic background on the expression of some phenotypic traits [van der Staay and Steckler, 2001].
SINGLE GENE MOUSE MODELS OF WILLIAMS SYNDROME
Numerous single gene knock-outs of genes from the WS region have been generated, some with the specific aim of understanding the molecular basis of WS and others as a result of unrelated research projects (Fig. 1). Of the 26 genes that are commonly deleted in WS, published mouse models currently exist for 11 (Table I). The majority of these models were generated through conventional gene targeting techniques, whereby part of the gene is replaced by a selectable marker through homologous recombination in embryonic stem cells, usually producing a non-functioning null allele [Hardouin and Nagy, 2000]. These mice allow the study of the function of each gene in isolation, both in the heterozygous (equivalent to gene dosage in WS) and the homozygous state. Although mice completely lacking a particular gene do not mirror the gene dosage found in WS, the null genotype may enhance subtle phenotypes and give additional insight into the phenotypic consequences of hemizygosity, as well as providing avenues for molecular or biochemical studies. Perhaps the greatest promise of the single gene mouse model lies in its potential to correlate specific phenotypes with an individual gene.
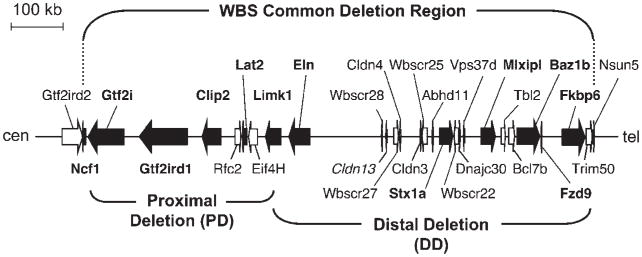
Figure 1.
The region of mouse chromosome 5G syntenic to the WS region. A schematic representation of mouse chromosome 5G is shown with genes shown as arrows in the direction of transcription. The WS commonly deleted region is shown above. Filled arrows represent genes for which single gene mouse models exist and their names are shown in bold type. The range of the two existing deletion mouse models are shown below. Note that both deletions disrupt Limk1, so that mice carrying both deletions are Limk1-null. Cldn13 is present within this region in mice, but not in humans. WSCR23, is present within an intronic region of GTF2IRD1 in humans, but not in mice.
TABLE I.
Phenotypic Features of Single Gene Mouse Models of WS
| Gene | Phenotype | References |
|---|---|---|
| Fkbp6 |
Heterozygotes: fertile, with no phenotype observed Homozygotes: males have abnormal spermatocyte pairing resulting in aspermia and infertility |
Crackower et al. [2003] |
| Fzd9 |
Heterozygotes: diminished seizure threshold, abnormal hippocampal structure Homozygotes: splenomegaly, thymic atrophy, developing B-cell depletion; diminished seizure threshold, abnormal hippocampal structure, impaired spatial learning and memory |
Ranheim et al. [2004], Zhao et al. [2005] |
| Baz1b |
Heterozygotes: mild craniofacial abnormalities, low frequency of cardiac malformations Homozygotes: high frequency of neonatal lethality, growth retardation, craniofacial abnormalities. Cardiac malformations such as ASD, VSD, and aortic coarctation |
Ashe et al. [2008], Yoshimura et al. [2009] |
| Mlxipl |
Heterozygotes: no data reported Homozygotes: decreased lipogenesis, decreased ability to metabolize glucose leading to glycogen accumulation in liver |
Iizuka et al. [2004] |
| Stx1a |
Heterozygotes: no deficits in leaning and memory, anxiety or locomotor activity observed (small number of mice tested) Homozygotes: high frequency of embryonic lethality Overexpression: hyperglycemia, impaired insulin secretion and insulin tolerance Truncation mutation: impaired insulin secretion, altered synaptic plasticity |
Fujiwara et al. [2006], Ohara-Imaizumi et al. [2007], McRory et al. [2008] |
| Eln |
Heterozygotes: hypertension, decreased aortic compliance and mild cardiac hypertrophy Homozygotes: perinatal embryonic lethality due to aortic obstruction by SMC proliferation |
Li et al. [1998a,b] |
| Limk1 |
Heterozygotes: no data reported Homozygotes: abnormal dendrite spine morphology, altered hipocampal function, mild deficit in spatial learning and memory |
Meng et al. [2002] |
| Lat2 |
Heterozygotes: no data reported Homozygotes: abnormal T-cell activation and mast cell response resulting in an autoimmune syndrome |
Volna et al. [2004], Zhu et al. [2004, 2006] |
| Clip2 |
Heterozygotes: mild growth deficiency, hippocampal dysfunction, deficits in motor coordination Homozygotes: mild growth deficiency, hippocampal dysfunction, deficits in motor coordination |
Hoogenraad et al. [2002] |
| Gtf2ird1 |
Heterozygotes: mild growth deficiency, hypersociability, learning, and memory deficits Homozygotes: mild growth deficiency, craniofacial abnormalities, hypersociability, learning, and memory deficits |
Tassabehji et al. [2005], Young et al. [2008], Palmer et al. [2010] |
| Gtf2i |
Heterozygotes: no data reported Homozygotes: early embryonic lethality |
Enkhmandakh et al. [2009] |
ASD, atrial-septal defect; VSD, ventrical-septal defect; SMC, smooth muscle cell.
DELETION MOUSE MODELS
Single gene mouse models can provide valuable information about the function of an individual gene, but they are not true genetic models of WS, which is by definition a contiguous gene deletion disorder. To address this issue, Li et al. [2008] took advantage of the high conservation of gene content and order across the WS region with its syntenic region on mouse chromosome 5G (Fig. 1). They used Cre-loxP recombination between separately targeted endpoints in embryonic stem cells to generate mice with deletions spanning either the proximal or the distal part of the region commonly deleted in people with WS. One deletion (DD) spanned Trim50–Limk1 (the equivalent of the proximal end of the deletion in people with WS) and the other (PD) spanned Limk1–Gtf2i (the equivalent of the distal end of the deletion in people with WS). Together(P/D), these deletions encompassed the entire common WS deletion, although because of the nature of the targeting, mice heterozygous for the two deletions were actually null for Limk1.
GENOTYPE–PHENOTYPE CORRELATION IN MOUSE MODELS OF WS
Cardiovascular Disease
Cardiovascular disease is one of the defining clinical characteristics of WS and ELN was the first gene to be linked with WS etiology. Although the pathology is not identical to that seen in humans, a knock-out mouse model has demonstrated a critical role for elastin in the regulation of vascular morphogenesis [Li et al., 1998a]. Mice lacking elastin die shortly after birth due to aortic obstruction caused by smooth muscle cell proliferation in the arterial wall [Li et al., 1998a]. Heterozygous mice are viable but produce ~50% less Eln mRNA and protein, are hypertensive, exhibit thinner elastic lamellae, more lamellar units, decreased aortic compliance, and mild cardiac hypertrophy [Li et al., 1998b]. The Eln mouse model does not exhibit supravalvular aortic stenosis (SVAS), the most common cardiovascular manifestation seen in WS, however a transgenic mouse carrying a human version of ELN on a bacterial artificial chromosome resulted in thickening of the wall of the ascending aorta when combined with heterozygosity for the mouse Eln gene [Hirano et al., 2007]. This suggests a fundamental difference in the function of the mouse and human gene in the developing aorta but also provides a more clinically relevant model for the study of therapeutic intervention for SVAS.
Alongside cardiovascular disease a lower, but still significant frequency of structural cardiac defects is also found in people with WS, though they have not been reported in families with isolated SVAS suggesting they are unrelated to ELN haploinsufficiency. Abnormalities include aortic coarctation, atrial and ventricular septal defects, mitral valve insufficiency or prolapse, bicuspid aortic valve and tetralogy of Fallot [Eronen et al., 2002; Del Pasqua et al., 2009]. A recent knock-out mouse model of Baz1b, a gene in the proximal part of the WS deletion region, may explain the occurrence of these developmental heart defects. BAZ1B is a shared subunit of 2 distinct chromatin remodeling complexes; WICH for DNA repair and WINAC for transcriptional control. Mice lacking BAZ1B showed major heart abnormalities and died shortly after birth, but heterozygous mice showed a range and frequency of developmental heart defects similar to those seen in people with WS [Yoshimura et al., 2009]. These mice also had altered expression of genes known to be important for proper heart development, such as Gja5 and Irx3, suggesting that BAZ1B is required for the normal function of cardiac transcriptional regulators.
Interestingly, the WINAC protein complex has also been shown to interact directly with the vitamin D receptor (VDR) where it mediates VDR recruitment and binding to target sites in the promoters of genes regulated by VDR, including key enzymes regulating vitamin D biosynthesis and catabolism [Kitagawa et al., 2003]. Heterozygous Baz1b mice showed elevated serum calcium levels that were consistent with the transient hypercalcemia seen in infants and children with WS suggesting that hemizygosity for BAZ1B may explain this phenomenon [Morris et al., 1988; Yoshimura et al., 2009].
Craniofacial Development
There has been much debate about the genetic etiology of the characteristic facial features associated with WS. The WS facial gestalt is a unique combination of alterations to both the soft tissue of the face and the underlying skull. The characteristic face of WS can be easily measured and defined using three-dimensional imaging to generate dense surface models of the face and identify discriminating features that support clinical diagnosis [Hammond et al., 2005]. Skull analysis, however, is far less specific to WS with only a few common features present [Mass and Belostoky, 1993; Axelsson et al., 2005]. People with smaller atypical deletions of the WS region show varying degrees of dysmorphology but without careful, standardized analysis of facial features in each individual, it is hard to draw firm conclusions about the contributions of specific genes to facial morphogenesis [Morris, 2006].
There are two genes that have been postulated to be involved in the craniofacial features of WS based on existing mouse models, Gtf2ird1 and Baz1b. Gtf2ird1 was proposed as a key gene in facial morphogenesis based on the finding of a characteristic facial appearance which included periorbital fullness and a short snout, in Gtf2ird1Tg(Alb1-Myc)166.8Sst-null mice [Tassabehji et al., 2005]. Some of the mice also had more severe craniofacial abnormalities with misaligned jaws and a twisted snout. Evidence that GTF2IRD1 may regulate the developmental gene goosecoid, which is known to be important for craniofacial morphogenesis, suggested that hemizygosity for GTF2IRD1 could play a significant role in the WS facial phenotype [Rivera-Perez et al., 1995; Yamada et al., 1995; Tassabehji et al., 2005]. The heterozygous mice, however, showed normal craniofacial development, as did other Gtf2ird1-null mice, suggesting that this phenotype may be influenced by strain effects in the different mice, all of which were maintained on different genetic backgrounds genetic effects in the different mice, all of which were maintained on different strains [Palmer et al., 2007; Young et al., 2008]. Dramatic strain differences have been observed for mouse models of other syndromic disorders with distinctive craniofacial features, such as Smith–Magenis syndrome (SMS). The comparison of different mouse models for SMS also uncovered differences in the penetrance of the craniofacial phenotype based on the type of mutation generated in the mouse model, with large deletions having a more severe effect than point mutations in the single causative gene, Rai1 [Yan et al., 2007]. There may be similar effects in the Gtf2ird1Tg(Alb1-Myc)166.8Sst-null mouse with a craniofacial phenotype since in this model the Gtf2ird1 locus is interrupted by the insertion of between 5 and 10 tandem copies of a 4.6 kb transgene, potentially disrupting the regulation or expression of other genes within the region [Durkin et al., 2001].
Baz1b was implicated in craniofacial development through a mouse model generated by random chemical mutagenesis [Ashe et al., 2008]. This mouse line was found to have a substitution of a highly conserved amino acid, which resulted in an unstable protein, the majority of which was subsequently degraded. Mice homozygous for this mutation died within the first week of life, similar to the Baz1b knock-out mice described in the previous section. These mice had skulls that were significantly different from WT skulls primarily due to a reduction of the parietal and nasal bones as well as a relative hypoplasia of the lower jaw. Heterozygotes also had slightly narrower and shorter craniums compared to wild-type mice as well as reduced size of the posterior region of the lower jaw. Baz1b is expressed strongly in all the major facial primordia from early in embryogenesis including the cranial neural crest-derived mesenchyme that drives facial morphogenesis. From these results it is possible that BAZ1B is important for cranial development, which is altered in people with WS who have a significantly shorter cranial base [Mass and Belostoky, 1993; Axelsson et al., 2005].
Perhaps the most informative WS mouse models as far as craniofacial development is concerned are the deletion models. Here the mice recapitulate the genetic etiology of WS and so might be expected also to recapitulate the classic facies. Indeed, the most striking abnormalities in craniofacial development are observed for mice lacking the entire WS syntenic region [Li et al., 2008]. These mice have a significantly shorter cranial base with some narrowing of the posterior part of the skull, which is present to a lesser degree in mice lacking only the human proximal region spanning Trim50–Limk1. No differences in cranial morphology are seen in mice lacking the human distal region (Limk1–Gtf2i), in contrast to reports of individuals with atypical deletions which seem to point to genes at the distal end being more involved in the craniofacial phenotype [Tassabehji et al., 2005; Morris, 2006; Antonell et al., 2009; Ferrero et al., 2010]. These conflicting results highlight the complexity of both mammalian craniofacial development and WS. Although many of the developmental pathways in mammals are conserved, there are bound to be subtle differences in the roles of individual genes. The almost complete penetrance of the WS facial gestalt in humans suggests clear differences in the function of some genes from the region that are important for craniofacial morphogenesis [Morris et al., 1988].
Although these mouse models do not clarify the genetic origin of the craniofacial phenotype in WS, they do not rule out a role for Baz1b or Gtf2ird1 in the phenotype. Careful selection of background strains may help dissect the role of GTF2IRD1 and may also modulate the phenotype of the deletion mice. Baz1b is a tantalizing candidate since it shows a heterozygous phenotype both when mutated and when disrupted in the context of the larger deletion. Interestingly, the mouse model deleted for the human distal region (PD, Fig. 1), though containing two intact copies of Baz1b, showed reduced expression of this gene [Li et al., 2008]. This may indicate long-range regulation of Baz1b by elements or genes situated within the distal region of the common WS deletion, raising the possibility of different thresholds of dosage sensitivity that may rely both on the extent of the deletion and expression of the non-deleted gene copy.
Endocrine Abnormalities
In the past few years it has emerged that the majority of adults with WS suffer from diabetes mellitus or pre-clinical glucose intolerance [Pober et al., 2010]. Mouse models with either decreased or increased levels of syntaxin 1A, which plays a role in membrane vesicle fusion and pancreatic β-cell exocytosis of insulin granules, show significant alterations in glucose metabolism due to abnormal insulin secretion [Lam et al., 2005; Ohara-Imaizumi et al., 2007], STX1A is therefore an excellent functional candidate for the high frequency of glucose dysregulation seen in WS. Another potential player in the regulation of glucose metabolism in WS is MLXIPL, a carbohydrate response element binding protein. MLXIPL is involved in the regulation of expression of carbohydrate responsive enzymes in the liver, which in turn control glucose metabolism and the synthesis of fatty acids and triglycerides [Iizuka and Horikawa, 2008]. Mlxipl-null mice showed elevated plasma glucose levels, suggesting that this gene may contribute to the phenotype in humans, however these mice also had reduced fatty acid synthesis and fat deposition which is in direct contrast to people with WS who tend to have a high percent body fat [Cherniske et al., 2004; Iizuka et al., 2004; Pober et al., 2010].
Cognition and Behavior
Several of the current mouse models of WS point to the possible involvement of specific genes in the cognitive and behavioral phenotypes of WS. LIMK1 and CLIP2 both regulate the cytoskeleton, LIMK1 through actin dynamics and CLIP2 through the microtubule network, and mouse models of both genes exhibited some degree of hippocampal dysfunction as evidenced by deficits in contextual fear conditioning and altered synaptic plasticity [Hoogenraad et al., 2002, 2004; Meng et al., 2002]. Both mouse models had normal brain volumes and neither had major structural brain abnormalities, but the Limk1-null mice did show altered dendritic spine morphology in pyramidal neurons, a phenomenon that has previously been associated with other genetic disorders involving intellectual disability, such as Down, fragile X and Rubinstein–Taybi syndromes [Kaufmann and Moser, 2000]. No analysis of the Limk1 heterozygotes was reported, so at this time it is unclear whether hemizygosity for LIMK1 might result in similar neuronal abnormalities in humans. Clip2 hetrozygotes and homozygotes showed impaired motor coordination on some tasks, but no differences in anxiety or amygdala function, suggesting that CLIP2 may contribute to coordination problems in WS but not to the characteristic behavioral profile.
The Gtf2i genes have emerged as prime candidates for involvement in the WS cognitive and behavioral profile through the study of individuals with atypical deletions. Although the Gtf2i mouse model has not yet undergone any assessment, the Gtf2ird1 mouse showed several phenotypes that overlap with WS [Young et al., 2008]. Both heterozygous and homozygous mice exhibited increased social interaction, reduced aggression and anxiety and impaired amygdala-based learning and memory, which correlates with the high sociability, lack of social anxiety and disinhibition seen in individuals with WS. In contrast to the Clip2 and Limk1 mice, their hippocampal function appeared to be intact and they had no problems with spatial tasks. Serotonin metabolism was also altered in the frontal cortex of these mice, and subsequent studies have demonstrated selectively enhanced serotonin receptor 1A-mediated responses in layer V pyramidal neurons of the pre-frontal cortex, suggesting altered neurophysiology [Proulx et al., 2010].
Other genes that produce changes in learning and behavior when disrupted in mice are Fzd9 and Stx1a. Though the role of these genes in WS is less compelling based on individuals with atypical deletions, they may still contribute to the WS phenotype. Fzd9 hemizygosity caused mild changes in hippocampal structure, whilst loss of Fzd9 produced changes in hippocampal structure and function [Zhao et al., 2005]. Hemizygosity for Stx1a did not produce any obvious behavioral or cognitive phenotype, but mice homozygous for a truncated form of STX1A also had altered synaptic plasticity, related to hippocampal dysfunction [Fujiwara et al., 2006; McRory et al., 2008].
A key question that has not yet been answered is whether the hemizygous deletion of two different genes together can result in an additive or a combinatorial effect on brain development and function. This is particularly relevant for Limk1 and Clip2, which both have major roles in the regulation of cytoskeletal integrity and remodeling, as well as for the Gtf2i genes that may co-regulate some of the same downstream target genes as well as perhaps regulating each other’s function [Tussie-Luna et al., 2001; Hoogenraad et al., 2004; Jackson et al., 2005]. At this time neither of these double heterozygous mice have been generated, but they will hopefully be analyzed in the future.
Multiple Models of Gtf2ird1 With Different Phenotypes
In most cases a single mouse model for any particular gene exists. In the case of Gtf2ird1, several models exist, each generated in a different manner and each with it own phenotypic traits. The stark differences between these models underline the effect of mutation and genetic background. There are three Gtf2ird1 models that show milder phenotypes, and have significant overlap with the clinical and behavioral aspects of WS. As discussed earlier, the Gtf2ird1 insertional model (Gtf2ird1Tg(Alb1-Myc)166.8Sst) exhibited craniofacial abnormalities, while two other knockout models (Gtf2ird1tm1Lro and Gtf2ird1tm1Hrd) exhibited behavioral phenotypes [Tassabehji et al., 2005; Young et al., 2008; Palmer et al., 2010]. A fourth model (Gtf2ird1Gt(XE465)Byg) showed a far more severe phenotype in both the heterozygous and homozygous state [Enkhmandakh et al., 2009]. Phenotypic features of the homozygous mice included severe neural tube and vascular defects, including abnormal vascularization of the yolk sac, which was postulated to contribute to the embryonic lethality seen in this model. Even heterozygous Gtf2ird1Gt(XE465)Byg mice showed severe skeletal abnormalities including kyphosis and loss of bones in the skull as well as hypopigmentation and hydrocephalus. The dramatically different phenotype observed in this mouse model is likely due not to genetic background effects, but to the nature of the Gtf2ird1 mutation. This model was generated from an embryonic stem cell line that has an insertion of a LacZ gene-trap cassette in intron 22 of the gene. The gene-trap technology allows for the expression of an in-frame fusion protein from the trapped locus, which in this case would consist of most of the GTF2IRD1 protein, fused with LacZ. The fusion protein would include multiple DNA-binding domains, as well as the dimerization domain, but would lack a nuclear localization signal. This fusion protein may be able to bind its DNA targets or interact with its regular protein partners, but would presumably not be able to carry out its normal function in the nucleus, perhaps acting in a dominant negative fashion. A role for GTF2I in the cytoplasm was recently identified, where it functions as a negative regulator of agonist-induced calcium entry, making it possible that GTF2IRD1 may also have a cytoplasmic function [Caraveo et al., 2006]. In addition, GTF2IRD1 and GTF2I are reported to counter-regulate the same gene targets, and GTF2IRD1 has been proposed to repress GTF2I transcriptional activation through nuclear exclusion [Tussie-Luna et al., 2001; Jackson et al., 2005]. The striking similarity between the phenotype of the Gtf2ird1Gt(XE465)Byg mice and another gene-trap line, Gtf2iGt(XE029)Byg, suggests that the Gtf2ird1Gt(XE465)Byg mutant may in fact be disrupting GTF2I function [Enkhmandakh et al., 2009].
Correlation of Genetic Mouse Model With WS
It was hoped that a mouse model with deletion of the entire region commonly deleted in WS, would recapitulate the full spectrum of the WS phenotype. In fact, these mice do show many of the classic symptoms of WS, but also lack some of the key characteristics (Table II) [Li et al., 2008]. In particular, heightened anxiety and increased sociability are present in the deletion mice, as are cardiovascular symptoms, growth retardation and motor coordination deficits. Other core features of WS, such as the soft tissue features of the face and hypersensitivity to sound are not present and visuospatial ability has not been explored in this model, although additional tests of spatial learning and memory may yet uncover deficits. Other similarities with WS include reduced whole brain volume and an increase in the density of neuronal packing in layer V of the somatosensory cortex, which may correlate with an increase in neuronal packing density observed in layer IV of the visual cortex in WS [Galaburda et al., 2002].
TABLE II.
Comparison of Phenotypes in Mice and Humans With Deletions of the WS Region
| WS common deletiona | WS deletion mouseb |
Human deletion of distal regionc (CLDN3–GTF2I) |
Mouse deletion of proximal regionb,d (Gtf2i–Limk1) |
|---|---|---|---|
| Cognition | |||
| Developmental delay/learning impairment | YES | YES | — |
| Weakness in spatial skills | n.d. | YESe | n.d. |
| Relative strength in expressive language | n.d. | YESe | n.d. |
| Brain morphology | |||
| Reduced brain volume | YES | n.d. | YES |
| Increased neuronal density | YES | n.d. | YES |
| Behavior | |||
| Excessively social | YES | YES | YES |
| Attention deficit hyperactivity disorder | — | YESe | — |
| Hypersensitivity to sound | — | YES | YES |
| Anxiety | YES | YESe | YES |
| Craniofacial | |||
| Broad forehead | — | YES | — |
| Bitemporal narrowing | — | YES | — |
| Periorbital fullness | — | YES | — |
| Stellate iris | n.d. | YES | n.d. |
| Short cranial base | YES | n.d. | — |
| Small, widely spaced teeth | n.d. | n.d. | n.d. |
| Other | |||
| Low birth weight | n.d. | YES | n.d. |
| Growth deficiency | Males | YES | Males (mild) |
| Impaired glucose tolerance | n.d. | n.d. | n.d. |
| Reduced visual acuity | n.d. | n.d. | n.d. |
| Hypercalcemia | n.d. | — | n.d. |
| Cardiovascular abnormalities | YES | YES | — |
| Motor co-ordination problem | YES | YES | Mild |
WS, Williams syndrome; n.d., not determined.
Three cases reported [Botta et al., 1999; Heller et al., 2003].
WS proximal region in mouse is equivalent to WS distal region in human.
In a single case where the individual was old enough to be evaluated.
FUTURE DIRECTIONS
The ever-increasing number of mouse models for WS is helping to unravel the contribution of individual genes to the features of WS and provide a rich resource for complex molecular, biochemical and genetic studies that are not possible in humans. Genome-wide analysis of gene or protein expression has already begun in many of these mouse models to identify molecules and pathways that may be either directly or indirectly altered in WS due to the reduced expression of specific genes. These mice will also important for in-depth invasive studies of the brain that will help to determine the underlying neurophysiology of the cognitive and behavioral aspects of WS.
Behavioral and physiological analyses have already shown that many of the common WS phenotypes are able to be recapitulated in the mouse, although some aspects of WS will be hard or even impossible to model in a mouse: phobias are unlikely to be present in a mouse model and language studies are not feasible. One area that is highly characteristic of WS but has received little attention in the mouse is visuospatial ability. Although spatial learning and memory may be readily assessed in mice using paradigms such as the Morris water maze and the Barnes maze, most of the mice, including the deletion models, have not been tested [Holmes et al., 2002]. Attempts to define the contribution of specific genes to the visuospatial deficits in WS through the study of atypical deletions have been inconclusive and the mouse models provide valuable tools with which to assess the individual and combinatorial role that genes such as Limk1, Clip2, and the Gtf2i genes play in normal and atypical visuospatial cognition.
Perhaps the most exciting prospect for mouse models is the ability to develop and test new therapeutic interventions. It has already been demonstrated that some of the serious cardiovascular abnormalities found in the Eln-null mice can be alleviated by the introduction of a human ELN gene, suggesting that although not identical, the underlying mechanisms of disease are somewhat similar in humans and mice [Hirano et al., 2007]. This provides a sound base for the pre-clinical testing of pharmaceutical therapies aimed at reducing hypertension, smooth muscle cell proliferation and vascular stenoses, which are the main causes of mortality in WS. In the future, mouse models will likely be just as important for the development and testing of new therapies to combat anxiety, disinhibition, visuospatial deficits, and even intellectual disability.
Acknowledgments
Grant sponsor: Canadian Institutes of Health Research.
Footnotes
Dr. Osborne’s Research aims to understand the molecular basis of Williams syndrome and her laboratory utilizes both clinical samples and animal models to probe the relationship between genes at 7q11.23 and cognition, language and behavior.
References
- Antonell A, Del Campo M, Magano LF, Kaufmann L, Martinez de la Iglesia J, Gallastegui F, Flores R, Schweigmann U, Fauth C, Kotzot D, Perez-Jurado LA. Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams syndrome neurocognitive profile. J Med Genet. 2009 doi: 10.1136/jmg.2009.071712. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Ashe A, Morgan DK, Whitelaw NC, Bruxner TJ, Vickaryous NK, Cox LL, Butterfield NC, Wicking C, Blewitt ME, Wilkins SJ, Anderson GJ, Cox TC, Whitelaw E. A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development. Genome Biol. 2008;9:R182. doi: 10.1186/gb-2008-9-12-r182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson S, Kjaer I, Heiberg A, Bjornland T, Storhaug K. Neurocranial morphology and growth in Williams syndrome. Eur J Orthod. 2005;27:32–47. doi: 10.1093/ejo/cjh065. [DOI] [PubMed] [Google Scholar]
- Bayes M, Magano LF, Rivera N, Flores R, Perez Jurado LA. Mutational mechanisms of Williams syndrome deletions. Am J Hum Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta A, Novelli G, Mari A, Novelli A, Sabani M, Korenberg J, Osborne LR, Digilio MC, Giannotti A, Dallapiccola B. Detection of an atypical 7q11.23 deletion in Williams syndrome patients which does not include the STX1A and FZD3 genes. J Med Genet. 1999;36:478–480. [PMC free article] [PubMed] [Google Scholar]
- Caraveo G, van Rossum DB, Patterson RL, Snyder SH, Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science. 2006;314:122–125. doi: 10.1126/science.1127815. [DOI] [PubMed] [Google Scholar]
- Cherniske EM, Carpenter TO, Klaiman C, Young E, Bregman J, Insogna K, Schultz RT, Pober BR. Multisystem study of 20 older adults with Williams syndrome. Am J Med Genet Part A. 2004;131A:255–264. doi: 10.1002/ajmg.a.30400. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R, Mo R, Hui CC, Nieves E, Cohen PE, Osborne LR, Wada T, Kunieda T, Moens PB, Penninger JM. Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science. 2003;300:1291–1295. doi: 10.1126/science.1083022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Pasqua A, Rinelli G, Toscano A, Iacobelli R, Digilio C, Marino B, Saffirio C, Mondillo S, Pasquini L, Sanders SP, de Zorzi A. New findings concerning cardiovascular manifestations emerging from long-term follow-up of 150 patients with the Williams-Beuren-Beuren syndrome. Cardiol Young. 2009;19:563–567. doi: 10.1017/S1047951109990837. [DOI] [PubMed] [Google Scholar]
- Durkin ME, Keck-Waggoner CL, Popescu NC, Thorgeirsson SS. Integration of a c-myc transgene results in disruption of the mouse Gtf2ird1 gene, the homologue of the human GTF2IRD1 gene hemizygously deleted in Williams syndrome. Genomics. 2001;73:20–27. doi: 10.1006/geno.2001.6507. [DOI] [PubMed] [Google Scholar]
- Edelmann L, Prosnitz A, Pardo S, Bhatt J, Cohen N, Lauriat T, Ouchanov L, Jimenez Gonzalez P, Manghi ER, Bondy P, Esquivel M, Monge S, Fallas M, Splendore A, Francke U, Burton BK, McInnes LA. An atypical deletion of the Williams syndrome interval implicates genes associated with defective visuospatial processing and autism. J Med Genet. 2007;44:136–143. doi: 10.1136/jmg.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkhmandakh B, Makeyev AV, Erdenechimeg L, Ruddle FH, Chimge NO, Tussie-Luna MI, Roy AL, Bayarsaihan D. Essential functions of the Williams syndrome-associated TFII-I genes in embryonic development. Proc Natl Acad Sci USA. 2009;106:181–186. doi: 10.1073/pnas.0811531106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eronen M, Peippo M, Hiippala A, Raatikka M, Arvio M, Johansson R, Kahkonen M. Cardiovascular manifestations in 75 patients with Williams syndrome. J Med Genet. 2002;39:554–558. doi: 10.1136/jmg.39.8.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart AK, Morris CA, Atkinson D, Jin W, Sternes K, Spallone P, Stock AD, Leppert M, Keating MT. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet. 1993;5:11–16. doi: 10.1038/ng0993-11. [DOI] [PubMed] [Google Scholar]
- Ferrero GB, Howald C, Micale L, Biamino E, Augello B, Fusco C, Turturo MG, Forzano S, Reymond A, Merla G. An atypical 7q11.23 deletion in a normal IQ Williams syndrome patient. Eur J Hum Genet. 2010;18:33–38. doi: 10.1038/ejhg.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, Proschel C, Gutowski NJ, Noble M, Atkinson DL, Odelberg SJ, Keating MT. LIM-kinase1 hemizygosity implicated in impaired visuospatial constructive cognition. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Mishima T, Kofuji T, Chiba T, Tanaka K, Yamamoto A, Akagawa K. Analysis of knock-out mice to determine the role of HPC-1/syntaxin 1A in expressing synaptic plasticity. J Neurosci. 2006;26:5767–5776. doi: 10.1523/JNEUROSCI.0289-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaburda AM, Holinger DP, Bellugi U, Sherman GF. Williams syndrome: Neuronal size and neuronal-packing density in primary visual cortex. Arch Neurol. 2002;59:1461–1467. doi: 10.1001/archneur.59.9.1461. [DOI] [PubMed] [Google Scholar]
- Hammond P, Hutton TJ, Allanson JE, Buxton B, Campbell LE, Clayton-Smith J, Donnai D, Karmiloff-Smith A, Metcalfe K, Murphy KC, Patton M, Pober B, Prescott K, Scambler P, Shaw A, Smith AC, Stevens AF, Temple IK, Hennekam R, Tassabehji M. Discriminating power of localized three-dimensional facial morphology. Am J Hum Genet. 2005;77:999–1010. doi: 10.1086/498396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardouin SN, Nagy A. Mouse models for human disease. Clin Genet. 2000;57:237–244. doi: 10.1034/j.1399-0004.2000.570401.x. [DOI] [PubMed] [Google Scholar]
- Heller R, Rauch A, Luttgen S, Schroder B, Winterpacht A. Partial deletion of the critical 1.5 Mb interval in Williams syndrome. J Med Genet. 2003;40:e99. doi: 10.1136/jmg.40.8.e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano E, Knutsen RH, Sugitani H, Ciliberto CH, Mecham RP. Functional rescue of elastin insufficiency in mice by the human elastin gene: Implications for mouse models of human disease. Circ Res. 2007;101:523–531. doi: 10.1161/CIRCRESAHA.107.153510. [DOI] [PubMed] [Google Scholar]
- Holmes A, Wrenn CC, Harris AP, Thayer KE, Crawley JN. Behavioral profiles of inbred strains on novel olfactory, spatial and emotional tests for reference memory in mice. Genes Brain Behav. 2002;1:55–69. doi: 10.1046/j.1601-1848.2001.00005.x. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Koekkoek B, Akhmanova A, Krugers H, Dortland B, Miedema M, van Alphen A, Kistler WM, Jaegle M, Koutsourakis M, Van Camp N, Verhoye M, van der Linden A, Kaverina I, Grosveld F, De Zeeuw CI, Galjart N. Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet. 2002;32:116–127. doi: 10.1038/ng954. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A, Galjart N, De Zeeuw CI. LIMK1 and CLIP-115: Linking cytoskeletal defects to Williams syndrome. Bioessays. 2004;26:141–150. doi: 10.1002/bies.10402. [DOI] [PubMed] [Google Scholar]
- Iizuka K, Horikawa Y. ChREBP: A glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr J. 2008;55:617–624. doi: 10.1507/endocrj.k07e-110. [DOI] [PubMed] [Google Scholar]
- Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101:7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TA, Taylor HE, Sharma D, Desiderio S, Danoff SK. Vascular endothelial growth factor receptor-2: Counter-regulation by the transcription factors, TFII-I and TFII-IRD1. J Biol Chem. 2005;280:29856–29863. doi: 10.1074/jbc.M500335200. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, Matsui D, Ogawa S, Unno K, Okubo M, Tokita A, Nakagawa T, Ito T, Ishimi Y, Nagasawa H, Matsumoto T, Yanagisawa J, Kato S. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- Lam PP, Leung YM, Sheu L, Ellis J, Tsushima RG, Osborne LR, Gaisano HY. Transgenic mouse overexpressing syntaxin-1A as a diabetes model. Diabetes. 2005;54:2744–2754. doi: 10.2337/diabetes.54.9.2744. [DOI] [PubMed] [Google Scholar]
- Li DY, Brooke B, Davis EC, Mecham RP, Sorensen LK, Boak BB, Eichwald E, Keating MT. Elastin is an essential determinant of arterial morphogenesis. Nature. 1998a;393:276–280. doi: 10.1038/30522. [DOI] [PubMed] [Google Scholar]
- Li DY, Faury G, Taylor DG, Davis EC, Boyle WA, Mecham RP, Stenzel P, Boak B, Keating MT. Novel arterial pathology in mice and humans hemizygous for elastin. J Clin Invest. 1998b;102:1783–1787. doi: 10.1172/JCI4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HH, Roy M, Kuscuoglu U, Spencer CM, Halm B, Harrison KC, Bayle JH, Splendore A, Ding F, Meltzer LA, Wright E, Paylor R, Deisseroth K, Francke U. Induced chromosome deletions cause hypersociability and other features of Williams syndrome in mice. EMBO Mol Med. 2008;1:50–65. doi: 10.1002/emmm.200900003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc Natl Acad Sci USA. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E, Belostoky L. Craniofacial morphology of children with Williams syndrome. Cleft Palate Craniofac J. 1993;30:343–349. doi: 10.1597/1545-1569_1993_030_0343_cmocww_2.3.co_2. [DOI] [PubMed] [Google Scholar]
- McRory JE, Rehak R, Simms B, Doering CJ, Chen L, Hermosilla T, Duke C, Dyck R, Zamponi GW. Syntaxin 1A is required for normal in utero development. Biochem Biophys Res Commun. 2008;375:372–377. doi: 10.1016/j.bbrc.2008.08.031. [DOI] [PubMed] [Google Scholar]
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, Jia Z. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- Morris CA. Genotype-phenotype correlations in Williams syndrome. In: Morris CA, Lenhoff HM, Wang PP, editors. Williams syndrome. Baltimore, MD: Johns Hopkins University Press; 2006. pp. 59–82. [Google Scholar]
- Morris CA, Demsey SA, Leonard CO, Dilts C, Blackburn BL. Natural history of Williams syndrome: Physical characteristics. J Pediatr. 1988;113:318–326. doi: 10.1016/s0022-3476(88)80272-5. [DOI] [PubMed] [Google Scholar]
- Morris CA, Mervis CB, Hobart HH, Gregg RG, Bertrand J, Ensing GJ, Sommer A, Moore CA, Hopkin RJ, Spallone PA, Keating MT, Osborne L, Kimberley KW, Stock AD. GTF2I hemizygosity implicated in mental retardation in Williams syndrome: Genotype-phenotype analysis of five families with deletions in the Williams syndrome region. Am J Med Genet Part A. 2003;123A:45–59. doi: 10.1002/ajmg.a.20496. [DOI] [PubMed] [Google Scholar]
- Ohara-Imaizumi M, Fujiwara T, Nakamichi Y, Okamura T, Akimoto Y, Kawai J, Matsushima S, Kawakami H, Watanabe T, Akagawa K, Nagamatsu S. Imaging analysis reveals mechanistic differences between first- and second-phase insulin exocytosis. J Cell Biol. 2007;177:695–705. doi: 10.1083/jcb.200608132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SJ, Tay ES, Santucci N, Cuc Bach TT, Hook J, Lemckert FA, Jamieson RV, Gunnning PW, Hardeman EC. Expression of Gtf2ird1, the Williams syndrome-associated gene, during mouse development. Gene Expr Patterns. 2007;7:396–404. doi: 10.1016/j.modgep.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Santucci N, Widagdo J, Bontempo SJ, Tay ES, Hook J, Lemckert F, Gunning PW, Hardeman EC. Negative auto-regulation of GTF2IRD1 in Williams syndrome via a novel DNA binding mechanism. J Biol Chem. 2010;285:4715–4724. doi: 10.1074/jbc.M109.086660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober BR, Wang E, Caprio S, Petersen K, Brandt C, Stanley T, Osborne LR, Dzuria J, Gulanski B. High prevalence of diabetes and pre-diabetes in adults with Williams syndrome. Am J Med Genet Part C. 2010 doi: 10.1002/ajmg.c.30261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx E, Young EJ, Osborne LR, Lambe EK. Enhanced prefrontal serotonin 5-HT1A currents in a mouse model of Williams syndrome with low innate anxiety. J Neurodevelop Disord. 2010 doi: 10.1007/s11689-010-9044-5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranheim EA, Kwan HC, Reya T, Wang YK, Weissman IL, Francke U. Frizzled 9 knockout mice have abnormal B cell development. Blood. 2004;105:2487–2494. doi: 10.1182/blood-2004-06-2334. [DOI] [PubMed] [Google Scholar]
- Rivera-Perez JA, Mallo M, Gendron-Maguire M, Gridley T, Behringer RR. Goosecoid is not an essential component of the mouse gastrula organizer but is required for craniofacial and rib development. Development. 1995;121:3005–3012. doi: 10.1242/dev.121.9.3005. [DOI] [PubMed] [Google Scholar]
- Rossant J, McKerlie C. Mouse-based phenogenomics for modelling human disease. Trends Mol Med. 2001;7:502–507. doi: 10.1016/s1471-4914(01)02164-5. [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Hammond P, Karmiloff-Smith A, Thompson P, Thorgeirsson SS, Durkin ME, Popescu NC, Hutton T, Metcalfe K, Rucka A, Stewart H, Read AP, Maconochie M, Donnai D. GTF2IRD1 in craniofacial development of humans and mice. Science. 2005;310:1184–1187. doi: 10.1126/science.1116142. [DOI] [PubMed] [Google Scholar]
- Tussie-Luna MI, Bayarsaihan D, Ruddle FH, Roy AL. Repression of TFII-I-dependent transcription by nuclear exclusion. Proc Natl Acad Sci USA. 2001;98:7789–7794. doi: 10.1073/pnas.141222298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Staay FJ, Steckler T. Behavioural phenotyping of mouse mutants. Behav Brain Res. 2001;125:3–12. doi: 10.1016/s0166-4328(01)00278-9. [DOI] [PubMed] [Google Scholar]
- Volna P, Lebduska P, Draberova L, Simova S, Heneberg P, Boubelik M, Bugajev V, Malissen B, Wilson BS, Horejsi V, Malissen M, Draber P. Negative regulation of mast cell signaling and function by the adaptor LAB/NTAL. J Exp Med. 2004;200:1001–1013. doi: 10.1084/jem.20041213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O’Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yamada G, Mansouri A, Torres M, Stuart ET, Blum M, Schultz M, De Robertis EM, Gruss P. Targeted mutation of the murine goosecoid gene results in craniofacial defects and neonatal death. Development. 1995;121:2917–2922. doi: 10.1242/dev.121.9.2917. [DOI] [PubMed] [Google Scholar]
- Yan J, Bi W, Lupski JR. Penetrance of craniofacial anomalies in mouse models of Smith-Magenis syndrome is modified by genomic sequence surrounding Rai1: Not all null alleles are alike. Am J Hum Genet. 2007;80:518–525. doi: 10.1086/512043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, Yamaoka I, Yonezawa M, Kondo T, Furutani Y, Yagi H, Yoshinaga S, Masuda T, Fukuda T, Yamamoto Y, Ebihara K, Li DY, Matsuoka R, Takeuchi JK, Matsumoto T, Kato S. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci USA. 2009;106:9280–9285. doi: 10.1073/pnas.0901184106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Young EJ, Lipina T, Tam E, Mandel A, Clapcote SJ, Bechard AR, Chambers J, Mount HT, Fletcher PJ, Roder JC, Osborne LR. Reduced fear and aggression and altered serotonin metabolism in Gtf2ird1-targeted mice. Genes Brain Behav. 2008;7:224–234. doi: 10.1111/j.1601-183X.2007.00343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Aviles C, Abel RA, Almli CR, McQuillen P, Pleasure SJ. Hippocampal and visuospatial learning defects in mice with a deletion of frizzled 9, a gene in the Williams syndrome deletion interval. Development. 2005;132:2917–2927. doi: 10.1242/dev.01871. [DOI] [PubMed] [Google Scholar]
- Zhu M, Liu Y, Koonpaew S, Granillo O, Zhang W. Positive and negative regulation of FcεRI-mediated signaling by the adaptor protein LAB/NTAL. J Exp Med. 2004;200:991–1000. doi: 10.1084/jem.20041223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Koonpaew S, Liu Y, Shen S, Denning T, Dzhagalov I, Rhee I, Zhang W. Negative regulation of T cell activation and autoimmunity by the transmembrane adaptor protein LAB. Immunity. 2006;25:757–768. doi: 10.1016/j.immuni.2006.08.025. [DOI] [PubMed] [Google Scholar]