Abstract
Machado–Joseph disease (MJD) is a neurodegenerative disorder, caused by the expansion of the (CAG)n tract in the MJD gene. This encodes the protein ataxin-3, of unknown function. The mouse Mjd gene has a structure similar to that of its human counterpart and it also contains a TATA-less promoter. Its 5′ flanking region contains conserved putative binding regions for transcription factors Sp1, USF, Arnt, Max, E47, and MyoD. Upon differentiation of P19 cells, the Mjd gene promoter is preferentially activated in endodermal and mesodermal derivatives, including cardiac and skeletal myocytes; and less so in neuronal precursors. Mouse ataxin-3 is ubiquitously expressed during embryonic development and in the adult, with strong expression in regions of the CNS affected in MJD. It is particularly abundant in all types of muscle and in ciliated epithelial cells, suggesting that it may be associated with the cytoskeleton and may have an important function in cell structure and/or motility.
Keywords: Ataxin-3, Polyglutamine, Spinocerebellar ataxia, Triplet repeat, Muscle, Myocytes, MyoD, E47, Max, Arnt
Machado–Joseph disease (MJD), also known as spinocerebellar ataxia type 3 (SCA3), is a neurodegenerative disorder of late onset and the most common dominant spinocerebellar ataxia worldwide (30% among all forms) [1]. Patients present with cerebellar ataxia and progressive external ophthalmoplegia, associated in a variable degree with pyramidal signs, extrapyramidal signs (dystonia or rigidity), amyotrophy, and peripheral neuropathy [2]. Pathologically, MJD is characterized by neuronal loss in the spinocerebellar, dentate, pontine, and vestibular nuclei, the substantia nigra, the locus coeruleus, the palidoluysian complex, the motor cranial nerve and medulla anterior horn nuclei, and the dorsal root ganglia [2–4].
The MJD causative gene was mapped to chromosome 14q32.1 in 1993 [5] and cloned in 1994 [6], but its structure has only recently been elucidated [7]. Alternative splicing of MJD results in the production of different isoforms of ataxin-3 [8], which are expressed in various tissues and have been detected both in the nucleus and in the cytoplasm [7,9–11]. MJD is one of the CAG repeat/polyglutamine disorders, which includes Huntington disease (HD), spinal and bulbar muscular atrophy, dentatorubropallidoluysian atrophy (DRPLA), and other spinocerebellar ataxias, such as SCA1, 2, 6, 7, 12, and 17. Each is characterized by selective neuronal cell death in specific regions of the brain, and their causative genes do not show homology with each other, except for the polyglutamine segment itself. The minimal poly(Q) length to cause disease is variable among these disorders. The CAG repeat tract in MJD contains 10–51 triplets in healthy individuals and 55–87 in patients [12,13].
Mutant ataxin-3, like other pathogenic proteins carrying expanded poly(Q) stretches, appears to undergo a conformational change and aggregate in cells forming inclusion bodies [12]. Ubiquitinated neuronal intranuclear inclusions (NIIs) have been observed in the brains of patients [14], cultured cells [15], and transgenic mouse models [16,17]. The observation that NIIs sequestrate transcriptional activators/coactivators suggests that an interference with gene transcription might be underlying the polyglutamine pathogenic mechanism [18]. It is not clear, however, if these NIIs are the cause or a consequence of the pathogenic mechanism. The potential conformational change and protein misfolding may be of importance to pathogenesis, since increased expression of chaperones can diminish the poly(Q) toxicity [19,20], while inhibition of the proteasome pathway enhances cell death [21,22]. Another hypothesis for the mechanism of neurodegeneration is the possibility of amyloid formation, as in Alzheimer disease and other neurodegenerative disorders, but with a different subcellular location [23]. Infrared spectroscopy measurements have shown that ataxin-3, containing an expanded poly(Q) tract, also forms fibrils in vitro presenting a higher content of β sheets [24]. There is evidence that polyglutamine aggregates exhibit most of the features of amyloid [25].
The first transgenic mouse models of MJD were generated with truncated and full-length human MJD1a cDNAs (containing 79 CAGs) under the control of the L7 promoter, which specifically directs their expression in Purkinje cells [16]. Recently, YAC transgenic mouse models for MJD were also generated [17]. These mice, carrying the full-length human MJD gene with expanded CAG repeats and its own regulatory elements, showed a mild and slowly progressive cerebellar deficit, cell loss in the dentate and pontine nuclei, as well as in other regions of the cerebellum, and peripheral nerve demyelination.
The function of ataxin-3 remains unknown. Recent studies suggest that ataxin-3 protein interacts with DNA-repair proteins HHR23A and HHR23B [26] and with the major histone acetyltransferases CBP, p300, and PCAF [27]. A recent computational structure-based sequence alignment study revealed that ataxin-3 has homology to ENTH and VHS domain proteins, which are involved in membrane trafficking and regulatory adaptor functions [28].
Several protein sequences highly homologous to human ataxin-3 have been described in rat [29] and chicken [30] and found in databases for mouse, Fugu, Drosophila, Caenorhabditis elegans, and Arabidopsis thaliana [28], suggesting that this protein is highly conserved throughout evolution and has functional relevance. To gain insight into the function of ataxin-3, we isolated and characterized the mouse Mjd gene. We cloned mouse Mjd, determined its genomic structure, characterized (spatially and functionally) the promoter region using the P19 cell model, and analyzed the expression pattern of mouse ataxin-3 during embryonic development and in the adult, using immunohistochemistry and immunoblotting techniques. Our results indicate that mouse ataxin-3 is ubiquitously expressed and is developmentally regulated, with high expression in muscle and ciliated epithelial cells, as well as in regions of the central nervous system (CNS) affected in MJD.
Results
Cloning of full-length cDNA and analysis of mouse Mjd gene organization
Using one of the cDNA sequences of human MJD (pMJD1a), we searched for homologous sequences in GenBank and found one mouse cDNA (Accession No. AK008675) with 1097 bp. A full-length mouse cDNA clone with 1100 bp (pMjd1) was obtained by RT-PCR from cerebral cortex total RNA and verified by sequencing. Analysis of the nucleotide sequence revealed 88, 84, 81, 80, and 68% identity with the human MJD1-1, MJD5-1, MJD1a, MJD2-1, and H2 cDNA sequences, respectively, and 92% identity with the rat homologue cDNA. The pMjd1 cDNA contains a coding region of 1065 bp, predicted to encode a 355-amino-acid protein—mouse ataxin-3.
A multiple sequence alignment, using CLUSTAL W, of mouse ataxin-3 with rat ataxin-3 and human ataxin-3-v1, ataxin-3-v2, ataxin-3-v3 [28], and ataxin-3-v4 (see Materials and methods) showed a 94, 86, 80, 82, and 83% identity, respectively, at the amino acid level (Fig. 1). This indicates that the obtained sequence is in fact the mouse homologue to human ataxin-3.
Fig. 1.
Multiple sequence analysis of mouse and rat ataxin-3 homologues and human ataxin-3 variants (CLUSTAL W). Aligned residues that are identical or similar are shaded with a black or gray background, respectively.
A probe of 400 bp (nucleotides 1 to 400 of the cDNA), obtained by PCR, was used to screen a BAC Mouse II library. One positive BAC clone was identified and sequenced. Comparisons of the mouse cDNA clone sequence with the BAC clone sequence enabled us to determine the exon/intron structure of the mouse Mjd gene (Table 1). This gene of approximately 36 kb, localized on chromosome 12, is very similar to its human counterpart, in both number and sizes of exons and introns. Mjd is composed of 11 exons and 10 introns, carrying a (CAG)6 repeat on exon 10 (Table 1). Thus, the genomic structure of mouse Mjd is highly homologous to that of human MJD, which is localized on chromosome 12, in a region syntenic to the human chromosome 14.
Table 1.
Exon–intron boundaries of the mouse Mjd gene
| Exon/intron | Length (bp) | Acceptor site | Donor site | |
|---|---|---|---|---|
| Exon | Intron | |||
| Exon 1 | 67 | CCACGAGAAA/gtgagtgtgg | ||
| Intron 1 | 10098 | |||
| Exon 2 | 165 | gaaacactag/CAAGAAGGCT | ATTTTTACAG/gtactgattt | |
| Intron 2 | 226 | |||
| Exon 3 | 45 | aattgaacag/CAGCCTTCTG | CTCTATTCAA/gtaagtagtc | |
| Intron 3 | 2084 | |||
| Exon 4 | 86 | cttttgacag/GTTATAAGCA | TTGATCCTAT/gtaagatttt | |
| Intron 4 | 367 | |||
| Exon 5 | 67 | tttcttctag/AAACGAAAGA | AGGCAAGCAG/gtaatatatc | |
| Intron 5 | 3198 | |||
| Exon 6 | 88 | tttttcctag/TGGTTTAACT | CAGCAAGAAG/gtaaataaga | |
| Intron 6 | 4648 | |||
| Exon 7 | 133 | ctctttttag/GTTATTCTAT | AAGAGCAGAG/gtaaaactac | |
| Intron 7 | 3192 | |||
| Exon 8 | 167 | tgttgtttag/TGCCCTCAAA | AGTATGCAAG/gtttggacat | |
| Intron 8 | 1183 | |||
| Exon 9 | 97 | ttgtttttag/GTAGTTCCAG | ACTTTGAAAA/gtaaagtagt | |
| Intron 9 | 6386 | |||
| Exon 10 | 98 | aatgtttcag/GCAACAGCAG | AGCGACCAAG/gtttgctttc | |
| Intron 10 | 3279 | |||
| Exon 11 | 97 | tctttcccag/GAGGCGACGC | ||
Sequence analysis of the 5′ flanking region
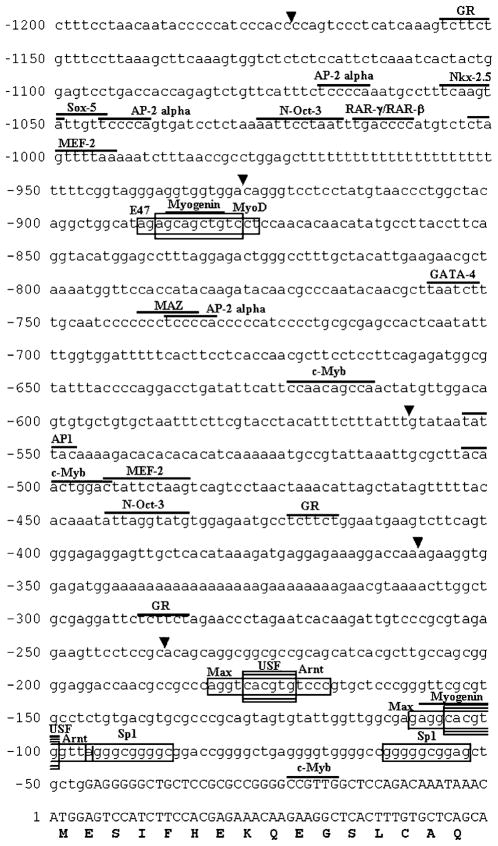
Analysis of the 5′ flanking region of Mjd revealed that its promoter consists of a sequence lacking TATA boxes or CCAAT boxes, similar to the human counterpart (Fig. 2). The G+C-rich region extends from −307 to −8 bp (GC content, 68%). Analysis of the 1173-bp region upstream of the start codon of both mouse and human genes for conservation of putative transcription factor binding regions using the TRES/TRANSFAC program revealed the presence of putative Sp1, upstream stimulatory factor (USF), aryl hydrocarbon receptor nuclear translocator (Arnt), Myc-associated factor X (Max), E47 and MyoD regulatory element binding sites, and also GC box elements in the gene promoters of both species (see supplementary data). Potential transcription factor binding sites for GATA-4, Nkx-2.5, myogenin, myocyte-specific enhancer factor-2 (MEF-2), c-Myb, activating enhancer-binding protein-2α (AP-2α), retinoic acid receptor-γ/retinoid X receptor-β (RAR-γ/RXR-β), N-Oct-3, SRY-related HMG-box-5 (Sox-5), glucocorticoid receptor (GR), activating protein-1 (AP1), and Myc-associated zinc-finger protein (MAZ) were also found in the 5′ flanking region of mouse Mjd (see supplementary data). These data suggest that the Mjd gene may be developmentally regulated.
Fig. 2.
Nucleotide sequence of the 5′ flanking region of the mouse Mjd gene. The translation start site is numbered as +1. Uppercase letters represent exon sequences and untranscribed sequences are represented by lowercase letters. The coding region of the first exon is shown as codon triplets. For the detection of promoter activity, an arrowhead indicates the start point of each construct. Consensus binding sequences of the conserved transcription factor binding sites are boxed; other potential transcription factor binding sites are indicated by a line.
Binding of MyoD to the Mjd promoter
To confirm the binding of the transcription factor MyoD, whose target region is conserved in mouse and human, we expressed and purified a recombinant mouse MyoD-His tag protein and analyzed its binding to the −928 bp fragment of the 5′ flanking region of the Mjd gene by electrophoretic mobility-shift assay (EMSA). We analyzed the binding of various quantities of the recombinant protein (2.5, 5, and 15 μg) to the DNA and verified that the binding, as revealed by gel retardation assay, increased proportionally with the amount of protein added (Fig. 3). To verify that the binding of the recombinant protein to the −928 bp fragment was specific, we performed a competition assay using two different quantities of oligonucleotides corresponding to the MyoD binding region of the Mjd gene promoter (localized at position −888 bp) and verified that in the presence of these oligonucleotides the binding of the recombinant protein to the promoter fragment decreased proportionally to the amount of competing oligonucleotides (Fig. 3). This result suggests that the Mjd gene promoter is indeed regulated by MyoD, which is a transcriptional activator of muscle-specific genes.
Fig. 3.
Binding of MyoD to the Mjd gene promoter. EMSAs were performed with 2.5 (lane 2), 5 (lane 3), or 15 μg (lane 4) of purified recombinant MyoD incubated with the −928 bp fragment of the promoter. For the competition assay 2.5 μg of purified recombinant MyoD was incubated with 2 (lane 6) or 4 μg (lane 7) of competitor double-stranded oligonucleotides followed by the addition of the −928 bp fragment. Lanes 1 and 5 correspond to the −928 bp fragment of the promoter.
Promoter activity of the 5′ flanking region
To characterize the regions regulating the transcription activity of the mouse Mjd gene, we constructed a series of plasmids containing different lengths of the 5′ flanking region of Mjd fused to the promoterless CAT gene in pCAT3-Basic (Promega): p-237MjdCAT, p-358MjdCAT, p-559Mjd CAT, p-928MjdCAT, and p-1173Mjd CAT. Transient transfection assays were performed using undifferentiated mouse embryonic carcinoma P19 cells. CAT activity was normalized for transfection efficiency, with reference to the β-galactosidase activity derived from the cotransfected internal control plasmid. As shown in Fig. 4A, we observed minimal activity of the promoter with the p-1173MjdCAT construct and maximal activity with either p-237MjdCAT or p-358MjdCAT plasmid, which presented a 94- and a 128-fold increase over the empty pCAT3-Basic plasmid, respectively. The fragments with 237 and 358 bp presented a 1.4-fold increase over the 1173-bp segment of the 5′ flanking region of the gene. We observed a decrease in the promoter activity for the fragments larger than −358 bp, which suggests the existence of repressor-binding sequences in these regions.
Fig. 4.
Functional analysis of the mouse Mjd gene promoter. (A) A series of constructs containing different sizes of the 5′ flanking region of the Mjd gene was transfected into P19 cells. CAT activity is represented in arbitrary units, normalized for β-galactosidase activity and for total protein amount. (B) Activation of the mouse Mjd promoter during RA- or DMSO-induced differentiation of P19 cells. Undifferentiated P19 were incubated with medium or with DMSO- or RA-supplemented medium, on bacterial-grade petri dishes, for 3 days, then dissociated, transfected with p-237MjdCAT and p-1173MjdCAT constructs, and further grown on tissue culture plates. Cells were harvested at day 3 after replating (day 6 after induction of differentiation). Data are expressed as the fold increase in CAT activity compared with transfections using empty CAT plasmid, pCAT3-Basic. The results are the averages of three independent transfections ± SEM (error bars).
These results suggest that the −237 bp region of the cloned 5′ region of the Mjd gene is sufficient to direct maximal transcription in P19 cells.
Activation of the mouse Mjd gene promoter during differentiation of P19 cells
We next examined the behavior of the isolated promoter sequence in terms of transcriptional activation during the differentiation of P19 cells onto three different types of cells: neuronal precursors, cardiac/skeletal myocytes, and endoderm cells [31,32]. To promote cellular differentiation, P19 cells were cultured as aggregates in the presence of medium or all-trans-retinoic acid (RA)- or dimethyl sulfoxide (DMSO)-supplemented medium to obtain endoderm-like cells (untreated), neuron-like cells, or cardiac/skeletal myocytes, respectively. A significantly higher activity of the Mjd gene promoter was observed in cardiac and skeletal myocytes transfected with constructs p-237MjdCAT and p-1173MjdCAT, which presented a 244.4- and a 231.5-fold increase in gene activity, respectively, over cells transfected with the empty pCAT3-Basic plasmid (Fig. 4B). Endoderm-like cells presented a 164.7- and 144.3-fold increase over the empty-vector-transfected cells, when transfected with constructs p-237MjdCAT and p-1173MjdCAT, respectively. In contrast, neuron-like cells transfected with the same constructs, p-237MjdCAT and p-1173MjdCAT, showed a 19.3- and a 23.0-fold increase, respectively, relative to cells transfected with the empty vector (Fig. 4B). These data indicate that at early stages of embryogenesis, the Mjd gene is preferentially activated in endodermal (DMSO-treated and untreated) and mesodermal (DMSO-treated) derivatives, including cardiac and skeletal muscle, and relatively less in neuronal precursors.
Mouse ataxin-3 expression in adult mouse
The expression pattern of mouse ataxin-3 was studied by immunoblotting analysis of several tissues from adult wild-type mice, using the anti-ataxin-3-specific antiserum that cross-reacted with endogenous mouse ataxin-3. Tissue samples from the spinal cord, brain stem, cerebellum, cerebral cortex, heart, skeletal muscle, testis, kidney, lung, spleen, pancreas, liver, and stomach all presented an immunoreactive band of 42 kDa that corresponded to full-length mouse ataxin-3 and another band of approximately 160 kDa, potentially corresponding to an SDS-resistant tetramer (Fig. 5).
Fig. 5.
Mouse ataxin-3 expression in wild-type C57Bl/6 mouse tissues. Protein extracts were resolved on 10% SDS– polyacrylamide gels. Immunoblotting reveals immunoreactive bands of 42 and 160 kDa present in all tissues analyzed. Heart, skeletal muscle, and testis presented another, lower molecular mass of 38 kDa, while skeletal muscle presented an additional band of 40 kDa.
Cardiac and skeletal muscle expressed an additional and more prominent lower molecular mass species of 38 kDa that is also present in human counterpart tissues [9]. This species and another of higher molecular mass were also present in testis. Skeletal muscle presented additionally another immunoreactive band of 40 kDa (Fig. 5). These species could either represent different splice variants of the mouse Mjd gene (potentially corresponding to human H2 cDNA variant described by Ichikawa and colleagues [7]) or be the result of proteolysis. Their conservation in mouse and human suggests that they could be of functional relevance.
To assess the regional and cellular distribution of mouse ataxin-3, we performed immunohistochemistry studies, using the anti-ataxin-3 antiserum, in peripheral tissues and organs and in the CNS of wild-type mice. Mouse ataxin-3 seemed to be ubiquitously distributed and to have a cytoplasmic localization, as evidenced in immunostained sections (Figs. 6, 7, and 8). The protein was highly expressed in skeletal, cardiac, and smooth muscle (Figs. 6A–6D). The flagellum of the spermatozoa stained intensely (Fig. 6K), and Leydig cells were also reactive to the antibody (Fig. 6H).
Fig. 6.
Mouse ataxin-3 immunoreactivity in adult mouse tissues. Bright-field micrographs of (A) transversal and (B) longitudinal sections of skeletal muscle, (C) cardiac muscle, (D) smooth muscle, (E) pancreas, (F) control for staining, without the primary antibody, in pancreas section, (G) spleen, (H) testis, and (I) liver. (A – D) High immunoreactivity was found in all types of muscles analyzed. (E) In the pancreas, staining was ubiquitous and could be clearly observed in the islets of Langerhans, though it was also present in the exocrine component. (G) In the spleen, labeling was found in both the white and the red pulp. (H) In the testis, the protein was also widely distributed, both within and outside the seminiferous tubules; in the tubules, it was found on spermatogonia, spermatocytes, spermatids, and spermatozoa and appeared to be absent on Sertoli cells; high reactivity was also found in the surrounding tunica propria (D). (I) In the liver, mouse ataxin-3 immunoreactivity was widely distributed throughout the section. (J) High magnification of two hepatocytes evidencing the more intense perimembrane staining, and (K) high magnification of spermatozoa. The arrows point to, in (A, B), a nucleus of muscular cell, showing no anti-mouse ataxin-3 immunoreactivity; in (D), the smooth muscle layer in the tunica propria of the testis; and in (K), the flagellum of a spermatozoon. The black bar on the lower right corner is equivalent to 50 μm. h, hepatocyte; il, islet of Langerhans; lc, Leydig cells; pa, pancreatic acinar cells; rp, red pulp; sc, Sertoli cell; sp, spermatocytes; st, spermatids; sz, spermatozoa; v, venule; wp, white pulp.
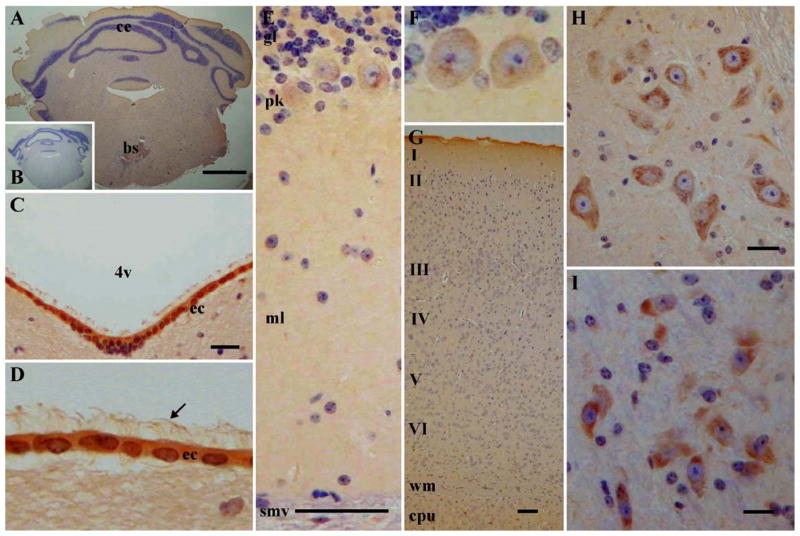
Fig. 7.
Ataxin-3 is ubiquitously expressed in the central nervous system of the adult mouse. (A, B) Low-magnification photomicrographs of a coronal section of the cerebellum and brain stem (approximate level 5.68 mm anterior to bregma) [46]. (A) Mouse ataxin-3 immunoreactivity and (B) control of staining, without the primary antibody. Mouse ataxin-3 staining is prominent in ependymal cells, as evidenced (C) in the cells in the fourth ventricle and at high magnification in (D), also showing the labeled cilia. (E) Low magnification of the anti-ataxin-3 labeling, in the different layers of the cerebellum, evidencing the cytoplasmic labeling in the Purkinje cells, also shown in higher magnification in (F). (G) Low magnification of immunoreactivity in cerebral cortex layers. (H) High magnification of the neuronal immunolabeling in the brain stem and (I) in the substantia nigra. The arrow points to, in (D), the labeled cilia of the ependymal cells. The black bar on the lower right corner is equivalent in (A) to 1 mm, in (G) to 200 μm, and in (C, E, H, I) to 50 μm. I – VI, cortical layers I – VI; 4v, fourth ventricle; bs, brain stem; ce, cerebellum; cpu, caudate putamen; ec, ependymal cells; gl, granule cell layer; ml, molecular cell layer; pk, Purkinje cells; smv, superior medullary velum; wm, white matter.
Fig. 8.
Ataxin-3 immunoreactivity during mouse embryonic development. Sagittal micrographs of mouse embryos at (A) embryonic day 11.5 (E11.5), (B) E12.5, (C) E13.5, (D) E14.5, (E) E15.5, and (F) E16.5. (F) At E16.5, the developing cardiac, skeletal, and smooth muscle were visibly labeled for mouse ataxin-3; immunoreactivity was also found in the respiratory and digestive tracts, especially marking the nonkeratinizing stratified squamous epithelia of the oral cavity and nasopharynx and the intestinal villi, in the apical portion of enterocytes, as well as in the adrenal glands, the pancreas, and the salivary glands (sublingual and submaxillary). (G) Control for staining, without the primary antibody, in a sagittal section of an E13.5 mouse embryo. (H –N) High-magnification images of E16.5 embryo. The black bar in the lower right corner is equivalent in (A, B) to 1 mm, in (C– G) to 2 mm, and in (H– N) to 50 μm. Intense ataxin-3 staining is evidenced (H) in the cochlear and (I) in the olfactory epithelia. Ataxin-3 is marked in the developing (J) kidney, (K) lung, (L) stomach, (M) thymus (both in the medulla and in the cortex), and (N) ossification areas (the periosteum was stained, as well as the cytoplasm of osteoprogenitor cells, whereas larger and more cuboidal osteoblasts appeared to exhibit no staining). bs, brain stem; cc, costal cartilage; ce, cerebellar primordium; cp, cortical plate; h, heart; i, intestine; k, kidney; l, lung; li, liver; s, stomach; sc, spinal cord; sm, submaxillary gland; t, thymus.
Anti-mouse ataxin-3 immunoreactivity was found ubiquitously in the CNS (Fig. 7A) and particularly localized in the cytoplasm of both neurons and glial cells. Marked labeling was found in the cytoplasm of neurons in the brain stem (Fig. 7H) and in the substantia nigra (Fig. 7I). In the substantia nigra, mouse ataxin-3 was expressed both in the pars compacta and in the pars reticulata. In the cerebellum, immunoreactivity was found in both the granular and the molecular layer (Fig. 7E); in the Purkinje cells, mouse ataxin-3 appeared to be located mainly in the cytoplasm, as suggested by the absence of nuclear staining (Fig. 7F). As shown in Fig. 7G, staining for mouse ataxin-3 was found in all layers of the cerebral cortex. Myelinated fibers of the white matter were also marked, although the staining was less pronounced. Likewise, in fiber areas, such as the corpus callosum and the anterior commissure, the staining was less intense. Immunolabeling was very intense in the ependymal cells, as observed in the lateral, third, and fourth ventricles (Fig. 7C); the cilia of these cells were also labeled (Fig. 7D).
Expression of mouse ataxin-3 during embryonic development
To determine the pattern of expression of mouse ataxin-3 during mouse embryonic development, we performed immunohistochemistry using anti-ataxin-3 antiserum in sagittal sections of embryos, at embryonic day (E) 11.5, E12.5, E13.5, E14.5, E15.5, and E16.5. Mouse ataxin-3 was continuously expressed in several tissues, including the CNS, from the earliest age analyzed (E11.5) and exhibited a ubiquitous distribution (Figs. 8A–8F). Ciliated epithelia, like the olfactory (Fig. 8I) and cochlear epithelia (Fig. 8H), were intensely stained.
Discussion
In this study, we determined the entire genomic structure of the mouse Mjd gene, characterized the functional promoter activity for the 5′ flanking region in transient transfection assays, and investigated the expression of the gene in mouse embryonic carcinoma P19 cells and its developmental regulation in mice. Our results showed that the mouse Mjd gene, a gene of 36 kb, has an organization very similar to that of human MJD: both the number and the sizes of exons and introns—11 exons and 10 introns—are identical [7]. The Mjd gene contains a CAG repeat localized in exon 10, as occurs in the human gene. The Mjd cDNA has a high sequence similarity to its rat and human counterparts. At the amino acid level, mouse ataxin-3 revealed 94 and 82–86% identity with rat and human proteins, respectively. The (CAG)n repeat in the mouse Mjd gene, as well as in the rat gene [29], is shorter than in its human counterpart, containing 6 triplet units compared to the range of 10–51 repeats in the normal human population [12,13]. Reduced length of polyglutamine tracts and high conservation at the amino acid level have also been observed in mouse homologues of other genes involved in human polyglutamine disorders, such as SCA1 [33], SCA2 [34], SCA7 [35], and HD [36,37]. This suggests that the polyglutamine stretch may have no relevance for function in the wild-type protein and that the expansion of repeats may be characteristic of primates, since no natural models of expanded poly(Q) are known in rodents.
Analysis of the 1173-bp regulating region upstream of mouse Mjd revealed the presence of putative binding sites for the transcription factors Sp1, USF, Max, Arnt, E47 and MyoD, which were conserved in the 5′ flanking region of the human gene, and the presence of potential binding sites for GATA-4, Nkx-2.5, myogenin, MEF-2, c-Myb, AP-2α, RAR-γ/RXR-β, N-Oct-3, Sox-5, GR, AP1, and MAZ. Similar to the human MJD promoter gene, no TATA or CCAAT boxes were found on the 5′ flanking region of Mjd. This and the presence of both constitutive and tissue-specific transcription factor regulatory sites suggest that the Mjd promoter is unusual, since it presents characteristics of both housekeeping and regulated genes. Interestingly, the results of the functional studies of this TATA-less promoter region correlate very well with the predicted roles of these putative transcription factor binding regions. Analysis of the promoter activity, using a series of constructs containing different lengths of the 5′ upstream region of the Mjd gene and CAT as a reporter gene, showed minimal activity of the promoter for the −1173 bp fragment, whereas fragments −237 and −358 bp presented maximal activity. The progressive decrease in activity for fragments larger than −358 bp suggest the existence of repressor binding sites in these regions. Sequence analysis of segments −237 and −358 bp revealed the repeated presence of similar regulatory regions on both fragments, except for the presence on the −358 bp segment of a putative GR binding region at position −290 bp. The similarity of results for the two constructs may relate to the absence of glucocorticoid exposure in the P19 system used in this study. The −237 bp region, containing putative binding sites for several ubiquitous transcription factors, such as Sp1, USF, Max, and Arnt, that are conserved in the human promoter, seems to be sufficient to direct maximal transcription in the P19 cells.
Having identified the minimal promoter region and potential regulatory elements, we then studied the Mjd promoter activity, in both RA-induced (neuronal) and DMSO-induced (cardiac/skeletal myocytes) differentiated P19 cells [31,32]. Cells transfected with constructs p-237MjdCAT and p-1173MjdCAT exhibited an increase in promoter activity of the Mjd gene when treated with DMSO—there was a 1.5- and 1.6-fold higher activity, respectively, relative to untreated cells. This is in agreement with the presence on the promoter of several putative binding regions for muscle-specific transcription factors, including myogenin, MyoD/E47, MEF-2, GATA-4, and Nkx-2.5. Of these, the MyoD binding site is also present in the human promoter. We confirmed the binding of the transcription factor MyoD to the mouse Mjd promoter by gel shift assay. These results are confirmed by the immunoblotting results that showed a high expression of the Mjd gene product—mouse ataxin-3—in cardiac and skeletal muscle. These tissues expressed smaller isoforms of the protein, 38 and 40 kDa in size, which are conserved in humans [9], suggesting the functional relevance of these isoforms. The expression of mouse-ataxin-3 in cardiac, skeletal, and smooth muscle was also confirmed by immunohistochemistry, both in adult mouse and during development, showing a cytoplasmic localization.
Untreated preaggregated P19 cells that correspond to endoderm-like cells [38] presented a relatively high activity of both Mjd promoter regions (−237 and −1173 bp), suggesting that the Mjd gene could be of functional relevance from the first stages of cell differentiation.
Neuron-like cells presenting morphologic and metabolic characteristics of normal neurons were obtained by RA-induced P19 differentiation. Interestingly, transient transfection of these cells with constructs p-237MjdCAT and p-1173MjdCAT originated an activity 8.5- and 6.3-fold weaker, respectively, than the untreated cells. The weaker activity of the minimal promoter (−237 bp) could be explained by the presence of two conserved putative binding sites for Max, at positions −109 and −182 bp. The Max and Mad genes are known to be up-regulated during RA-induced differentiation [39], thus Max/Mad heterodimers could bind and act as repressors of transcription. Simultaneously, down-regulation of the activator gene c-Myc, which is later expressed at high level (at day 14 of differentiation) [39], could also contribute to the observed decrease in promoter activity. Alternatively, a decrease in transcription may occur through the AhR/Arnt pathway, potentially acting at the conserved −105 and −178 bp binding sites. In fact, aryl hydrocarbon receptor (AhR) has been shown to be down-regulated in RA-treated cells [40].
Despite the presence on the p-1173MjdCAT construct of several potential binding regions for AP-2α, N-Oct-3, and RAR-γ/RXR-β, which are activators for neuron-specific genes, CAT activity on RA-differentiated cells transfected with this construct was rather similar to that obtained for the minimal promoter construct. It is possible that the potential activation of transcription by these factors is not sufficient to overcome the transcription repression of Mjd by the Max/Mad pathway, at least at this stage (day 6) of RA-induced differentiation. This does not exclude that at later stages of embryonic development, which are not mimicked by P19 cells, these transcription factors could eventually become activated. The presence of additional neuronal-specific enhancers of transcription upstream of the −1173 bp promoter region of Mjd also cannot be excluded and could account for the high expression of mouse ataxin-3 observed in the CNS during both adult and embryonic stages of the mouse.
In the CNS of the adult mouse, mouse ataxin-3 was ubiquitously expressed and was localized in the cytoplasm of both neurons and glial cells. Relevant reactivity was found in the brain stem (mainly in neurons), in the substantia nigra, and in both the granular and the molecular layer of the cerebellum. However, a very intense expression was also observed in other types of cells of the CNS, which are not affected in MJD. We suggest that the pathogenic mechanism in MJD may be independent of the function of ataxin-3, being due not only to a prominent expression of the mutant protein in the affected regions but also to other factors that contribute to a selective neuronal loss.
The study of the expression pattern of mouse ataxin-3 during embryonic development revealed that it is expressed since the early stages and confirmed the ubiquitous expression observed in adult mice.
The intense immunoreactivity of mouse ataxin-3 in ependymal cells, in the flagellum of the spermatozoa, and in ciliated olfactory and cochlear epithelia suggests that this protein may have an important function in cell structure and/or motility. Additionally, the presence in the promoter region of conserved binding sites for muscle-specific transcription factors, and the high activity of the Mjd promoter in DMSO-treated cells, in conjunction with the very intense expression of mouse ataxin-3 in cardiac, skeletal, and smooth muscle, suggests that the function of Mjd may be relevant in these tissues. These results can also be related to the structural modeling of ataxin-3, which has a distant homology to adaptins [28]. Adaptins are functionally related to and involved in the cytoskeleton machinery, vesicular trafficking, and clathrin-dependent endo- and exocytosis [28]. Further studies are necessary to determine whether mouse ataxin-3 may be a cytoskeleton-associated protein that could be important for motility and for cellular transport.
In summary, our studies indicate that MJD is highly conserved between species, within coding and 5′ regulatory regions, and its regulated expression during development suggests that ataxin-3 may have an important biological function. Given this conservation, the study of mouse Mjd may provide fundamental insights into the function of human ataxin-3.
Materials and methods
Computational methods
We applied the BLAST program [41] to search for the homologue cDNA sequence in mouse under Accession No. AK008675. All protein sequences were taken from the TrEMBL database. The accession numbers of human variants ataxin-3-v1, ataxin-3-v2/v3 as denoted in [28], and ataxin-3-v4 corresponding to H2 in [7]; rat ataxin-3;and mouse ataxin-3 are Q96TC4, Q96TC3, Q96TC3, BAB18798, O35815, and Q9CVD2, respectively. Multiple sequence alignment was constructed by means of CLUSTAL W [42]. Comparison of MJD sequence to the homologous 5′ upstream region in mouse Mjd was performed to uncover evolutionarily conserved regions using TRES/TRANSFAC with a score of 95% [43]. Conserved regions in Mjd were then scanned for the presence of potential transcription factor binding regions using TESS [44].
Cloning of the full-length Mjd and MyoD cDNAs by RT-PCR
Total RNA was isolated from adult C57Bl/6 mouse brain and skeletal muscle, using Trizol reagent (Gibco BRL). Five micrograms of total RNA was used to perform reverse transcription using the SuperScript First-Strand Synthesis System for RT-PCR (Gibco BRL) with an oligo(dT) primer. To amplify the Mjd cDNA, PCR was carried out using the Expand High Fidelity System (Roche) with primers mmMJD1 and mmMJD2 (see below) using mouse brain total RNA as template. To amplify the MyoD cDNA, PCR was carried out using the same system with primers MyoD1 and MyoD2 (see below) using mouse skeletal muscle total RNA as template. The PCR conditions consisted of 1 cycle of 2 min at 94°C, followed by 32 cycles of 1 min at 94°C, 1 min at 62°C, and 1 min at 72°C, and ending with 5 min at 72°C. The PCR products corresponding to Mjd and to MyoD cDNAs were purified using GFX PCR DNA and the Gel Band Purification Kit (Amersham Pharmacia Biotech) and cloned into the pGEM-T Easy vector (Promega) (pMjd1) and into the pDONR207 vector, respectively, using the Gateway system (Invitrogen Life Technologies) (pDON207MyoD).
PCR
The primers used for RT-PCR were mmMJD1 (5′-GAGGGGCTGCTCCGCGCCGGGGCCGT), mmMJD2 (5′-TCTGCTTTCAAGTTGTCTTTAGCAG), MyoD1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATATGGAGCTTCTATCGCCGCCACTCCG), and MyoD2 (5′-GGGGACCACTTTGTCCAAGAAAGCTGGGTCTCAAAGCACCTGATAAATCGCATTG). The probe used for screening the BAC library was amplified by PCR, using the Mjd cDNA (pMjd1) as template, with primers mmMJD1 and mmMJD3 (5′-GTAAACCAGTGTTCTTTATAATTGCA), and performed with the same cyclic parameters used for RT-PCR. PCRs for the construction of CAT reporter plasmids were all performed using the Expand High Fidelity System (Roche) and performed under the following conditions: 1 cycle of 5 min at 95°C, followed by 35 cycles of 1 min at 95°C, 1 min at 55°C, 1 min at 72°C, and ending with 5 min at 72°C. For the insertion of the restriction enzyme sites we used the reverse primer PMjd-1BglIIR (5′-GCACAGATCTGTTTATTTGTCTGGAG) in conjunction with one of the forward primers: PMjd-237KpnIF (5′-CGAGGGTACCACAGCAGGCGGCGCC), PMjd-358KpnIF (5′-GCACGGTACCAGAAGGTGGAGATGG), PMjd-559KpnIF (5′-GGCTGGTACCGTATAATATTACAAAAG), PMjd-928KpnIF (5′-CGAGGGTACCCAGGGTCCTCCTATGTAAC), or PMjd-1173KpnIF (5′-GGCTGGTACCCCAGTCCCTCATCAAAGT).
Isolation of a genomic BAC clone
The genomic BAC clone was obtained from Incyte Genomics, Inc. A BAC Mouse II library was screened by direct hybridization, using a probe of 400 bp, constructed by PCR, using primers mmMJD1 and mmMJD3. These primers were designed on the basis of the cloned Mjd cDNA sequence. The positive BAC clone was grown overnight, at 37°C, in 2XYT medium containing the appropriate antibiotic (25 μg/ml chloramphenicol). BAC DNA was isolated using the Qiagen Large-Construct Kit (Qiagen).
Cell culture and differentiation
The P19 clone was obtained from the American Type Collection. Cells were cultured (plating 106 cells in 75-cm3 tissue culture flasks, every 2 days) in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% fetal bovine serum (FBS), in a 5% CO2 humidified chamber at 37°C. To promote cellular differentiation, cells were cultured as aggregates, in bacterial petri dishes, in medium supplemented with 1 μM RA (Sigma) for neuronal differentiation or with 1% DMSO for cardiac/skeletal myocyte differentiation. Cells (1 × 105/ml culture) were plated into bacterial petri dishes (day 0) and incubated with medium (no treatment) or with RA- or DMSO-supplemented medium, for 2 days. After 2 days, floating aggregates were collected and washed with medium and replated into bacterial petri dishes for 1 additional day. At day 3, the aggregates were harvested; dissociated by treatment with 0.25% trypsin, 1 mM EDTA (Gibco BRL); and then plated in DMEM, 10% FBS in 60-mm diameter tissue-culture-grade dishes. At day 4, cells were transfected with constructs p-237MjdCAT and p-1173MjdCAT. Cell viability was assessed by trypan blue exclusion.
Construction of CAT reporter and MyoD-His tag expression plasmids
To obtain different lengths of the 5′ flanking region of Mjd, PCRs were performed using the BAC clone 27521 as the template. We used the primer PMjd-1BglIIR to insert the 3′ restriction enzyme site for BglII (at the position −1 bp) into all fragments, in conjunction with one of the primers PMjd-237KpnIF, PMjd-358KpnIF, PMjd-559KpnIF, PMjd-928KpnIF, or PMjd-1173KpnIF to insert the 5′ restriction enzyme site for KpnI, at the respective lengths of the 5′ region −237, −358, −559, −928, or −1173 bp. These PCR products were digested with BglII and KpnI and subcloned into BglII—KpnI-digested pCAT3-Basic vector (Promega). The resultant constructs were p-237MjdCAT, p-358MjdCAT, p-559MjdCAT, p-928MjdCAT, and p-1173MjdCAT. The recombinant mouse MyoD-His tag protein expression plasmid (pDEST17MyoD) was obtained by recombination of the pDONR207MyoD with the pDEST17 vector using the Gateway system (Invitrogen Life Technologies). All constructs were confirmed by automatic sequencing.
Expression and purification of recombinant MyoD-His tag protein
The pDEST17MyoD construct was transformed in Escherichia coli BL21SI bacteria. Culture was grown to OD600 of 0.5 L of LBON medium plus ampicillin (100 μg/ml). The expression was induced by the addition of NaCl to a final concentration of 0.3 M and culture was grown for 30 min. Cells were harvested, resuspended in 15 ml of phosphate buffer 1×, imidazole 10 mM, lysozyme 66 μg/ml, Triton X-100 0.1%, Complete EDTA-free protease inhibitor cocktail tablet (Roche) and incubated on ice for 1 h. The lysate was sonicated on ice eight times for 30 s each time and then centrifuged for 15 min at 12,000 rpm at 4°C. The supernatant was collected and the recombinant protein was purified using the HisTrap Kit (Amersham Pharmacia Biotech) using the manufacturer’s conditions.
Electrophoretic mobility-shift assay
The −928 bp fragment of the Mjd gene promoter, generated by KpnI/BglII restriction of the p-928MjdCAT, was used for EMSA. For the competition assay, single-stranded sense and antisense MyoD oligonucleotides were synthesized based on both the sequence in the 5′ region of the Mjd gene and the binding matrix for MyoD with Accession No. M00184 (TRANSFAC database). The oligonucleotides MyoD-sense (5′-AGCAGCTGTC) and MyoD-antisense (5′-GACAGCTGCT) were annealed by incubation at 30°C for 1 min and by slow cooling to room temperature. DNA–protein binding assays were performed at room temperature for 30 min, always using the same amount of DNA (300 ng of the −928 bp fragment) and different quantities of recombinant mouse MyoD protein in a final volume of 40 μl of TE. In the competition assay, the double-stranded oligonucleotides (2 or 4 μg) were incubated with the recombinant protein for 30 min before the incubation with the DNA. The mixtures were loaded in a 1.5% agarose gel and electrophoresed at room temperature.
Cell transfection and CAT assay
To analyze activities of the CAT reporter plasmids in undifferentiated, untreated (day 3), RA-treated (day 3), and DMSO-treated (day 3) P19 cells, these were seeded at 2 × 105, 7.5 × 105, 7.5 × 105, and 3 × 106 cells, respectively, per 60-mm-diameter tissue-culture-grade dishes in DMEM, 10% FBS 24 h prior to transfection. Transient transfection was carried out in serum-free medium (Opti-MEM; Life Technologies), using 10 μl of Lipofectamine2000 Reagent (Life Technologies), to introduce 1.5 μg of reporter construct and 0.5 μg of pSV-β-galactosidase control vector (Promega). After transfection for 5 h, medium was replaced with DMEM, 10% FBS. Cells were harvested 48 h after transfection and washed three times with phosphate-buffered saline (PBS), and then cell extracts were prepared in 1× reporter lysis buffer (Roche). Cell extracts were assayed for CAT activity, using the CAT ELISA kit (Roche). Measurements were performed in duplicate, averaged, and then normalized to the β-galactosidase activity, to correct for transfection efficiency, and to the total protein amount. β-Galactosidase activity was measured using the β-Galactosidase Enzyme Activity System (Promega), and the total protein amount was assessed using the Lowry Protein assay kit (Sigma).
Immunoblotting
Proteins were extracted from different tissues of adult C57Bl/6 mice, using standard methods [45]. Total protein quantification was assessed using the Lowry Protein assay kit (Sigma). Proteins of spinal cord, brain stem, cerebellum, cerebral cortex, heart, skeletal muscle, testis, kidney, lung, spleen, pancreas, liver, and stomach tissues (30 μg) were resolved on a 10% SDS–polyacrylamide gel and transferred onto nitrocellulose membrane (Amersham Biotechnologies). The anti-human ataxin-3-specific antiserum (kindly provided by Henry Paulson) was used (1:2000). For detection of the immunocomplexes formed, the secondary antibody peroxidase-conjugated goat anti-rabbit IgG (Stressgen) (1:4000) or goat anti-mouse IgG (Jackson Immunoresearch Laboratories) (1:1000) was used. Staining intensity was developed with the chemiluminescence system (Roche).
Immunohistochemistry
Adult male C57Bl/6 mice were perfused transcardially, under anesthesia, with fixative solution (4% paraformaldehyde, in 0.1 M phosphate buffer, pH 7.4), for 10 min. The encephalon and peripheral tissues (skeletal muscle, cardiac muscle, pancreas, spleen, liver, and testis) were postfixed for 2 h in the same fixative. Mouse embryos were isolated from pregnant C57Bl/6 female mice, at E11.5, E12.5, E13.5, E14.5, E15.5, and E16.5. The day of plug detection was considered to be E0.5 (embryonic day 0.5). Embryos were washed with ice-cold PBS and fixed in 10% buffered formalin, for at least 24 h, at 4°C. After fixation, the samples (adult tissues and organs and embryos) were paraffin embedded. Nominal 5-mm tissue sections were cut and mounted onto positively charged slides (X-Tra Slides), deparaffinized on xylene, and rehydrated by passages through graded ethanol series. In formalin-fixed material, heating was performed to allow antigen recovery. Nonspecific binding was blocked with normal goat serum (Vectastain Elite ABC kit; Vector Laboratories). Sections were incubated overnight in primary anti-ataxin-3 antiserum diluted (1:2000) in PBS. Antibody labeling was visualized using the secondary anti-rabbit antibody by the avidin–biotin conjugation method (Vectastain Elite ABC kit; Vector Laboratories) and 3,3-diaminobenzidine (Vector Laboratories). Sections were counterstained with hematoxylin. Dehydration of sections was assessed through graded ethanols and xylene. Slides were mounted with Histomount. Positive controls were processed in parallel. Sections were analyzed using an Olympus BX-50 microscope, equipped with a Camedia C-2000Z Olympus digital camera. High-quality (1600 × 1200 pixel) images were captured to a PC equipped with the Olympus DP-Soft 3.0 software. Brain sections were referenced to [46].
Acknowledgments
We thank Mark Tessaro, Laura Montermini, Margarida Duarte, João Sousa, and Teresa Summavielle for their cooperation in this study and Sandra Macedo Ribeiro for critical review of the manuscript. We also thank Henry Paulson for providing of the anti-human ataxin-3 antiserum. This work was supported by the FSE/FEDER and the Fundacão para a Ciência e Tecnologia (FCT) (Project POCTI/MGI 33759/99) and the Fundacão Luso-Americana para o Desenvolvimento (Proc. 3.L./A.II/P.582/99) and by a grant from the Canadian Institutes of Health Research (CIHR, Grant MOP44045) to M.M.S. M.M.S. is the recipient of a CIHR New Investigator award. M.C.C. is the recipient of a Ph.D. scholarship from FCT, (BD/9759/2003). Printing of color figures was supported by Bioportugal, Bonsai Technologies, and Vidrolab.
Footnotes
Supplementary data for this article may be found on ScienceDirect.
References
- 1.Margolis RL. The spinocerebellar ataxias: order emerges from chaos. Curr Neurol Neurosci Rep. 2002;2:447–456. doi: 10.1007/s11910-002-0072-8. [DOI] [PubMed] [Google Scholar]
- 2.Coutinho P, Andrade C. Autosomal dominant system degeneration in Portuguese families of the Azores Islands: a new genetic disorder involving cerebellar, pyramidal, extrapyramidal and spinal cord motor functions. Neurology. 1978;28:703–709. doi: 10.1212/wnl.28.7.703. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg RN. Machado–Joseph disease: an autosomal dominant motor system degeneration. Mov Disord. 1992;3:193–203. doi: 10.1002/mds.870070302. [DOI] [PubMed] [Google Scholar]
- 4.Dürr A, et al. Spinocerebellar ataxia 3 and Machado– Joseph disease: clinical, molecular, and neuropathological features. Ann Neurol. 1996;39:490–499. doi: 10.1002/ana.410390411. [DOI] [PubMed] [Google Scholar]
- 5.Takiyama Y, et al. The gene for the Machado – Joseph disease is mapped to human chromosome 14q. Nat Genet. 1993;4:300–304. doi: 10.1038/ng0793-300. [DOI] [PubMed] [Google Scholar]
- 6.Kawaguchi Y, et al. CAG expansions in a novel gene for Machado–Joseph disease at chromosome 14q32.1. Nat Genet. 1994;8:221–228. doi: 10.1038/ng1194-221. [DOI] [PubMed] [Google Scholar]
- 7.Ichikawa Y, et al. The genomic structure and expression of MJD, the Machado– Joseph disease gene. J Hum Genet. 2001;46:413–422. doi: 10.1007/s100380170060. [DOI] [PubMed] [Google Scholar]
- 8.Goto J, et al. Machado– Joseph disease gene products carrying different carboxyl termini. Neurosci Res. 1997;28:373–377. doi: 10.1016/s0168-0102(97)00056-4. [DOI] [PubMed] [Google Scholar]
- 9.Paulson HL, et al. Machado – Joseph disease gene product is a cytoplasmic protein widely expressed in brain. Ann Neurol. 1997;41:453–462. doi: 10.1002/ana.410410408. [DOI] [PubMed] [Google Scholar]
- 10.Trottier Y, et al. Heterogeneous intracellular localization and expression of ataxin-3. Neurobiol Dis. 1998;5:335–347. doi: 10.1006/nbdi.1998.0208. [DOI] [PubMed] [Google Scholar]
- 11.Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cummings CJ, Zoghbi HY. Fourteen and counting: unraveling tri-nucleotide repeat diseases. Hum Mol Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 13.Maciel P, et al. Improvement in the molecular diagnosis of Machado–Joseph disease. Arch Neurol. 2001;58:1821–1827. doi: 10.1001/archneur.58.11.1821. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt T, et al. An isoform of ataxin-3 accumulates in the nucleus of neuronal cells in affected brain regions of SCA3 patients. Brain Pathol. 1998;8:669–679. doi: 10.1111/j.1750-3639.1998.tb00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evert BO, et al. High level expression of expanded full-length ataxin-3 in vitro causes cell death and formation of intranuclear inclusions in neuronal cells. Hum Mol Genet. 1999;8:1169–1176. doi: 10.1093/hmg/8.7.1169. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda H, et al. Expanded polyglutamine in the Machado – Joseph disease protein induces cell death in vitro and in vivo. Nat Genet. 1996;13:196–202. doi: 10.1038/ng0696-196. [DOI] [PubMed] [Google Scholar]
- 17.Cemal CK, et al. YAC transgenic mice carrying pathological alleles of the MJD1 locus exhibit a mild and slowly progressive cerebellar deficit. Hum Mol Genet. 2002;11:1075–1094. doi: 10.1093/hmg/11.9.1075. [DOI] [PubMed] [Google Scholar]
- 18.Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 19.Chan HY, Warrick JM, Gray-Board GL, Paulson HL, Bonini NM. Mechanisms of chaperone suppression of polyglutamine disease: selectivity, synergy and modulation of protein solubility in Drosophila. Hum Mol Genet. 2000;9:2811–2820. doi: 10.1093/hmg/9.19.2811. [DOI] [PubMed] [Google Scholar]
- 20.Paulson HL, Bonini NM, Roth KA. Polyglutamine disease and neuronal cell death. Proc Natl Acad Sci USA. 2000;97:12957–12958. doi: 10.1073/pnas.210395797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chai Y, Koppenhafer SL, Shoesmith SJ, Perez MK, Paulson HL. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet. 1999;8:673–682. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt T, et al. Protein surveillance machinery in brains with spinocerebellar ataxia type 3: redistribution and differential recruitment of 26S proteasome subunits and chaperones to neuronal intranuclear inclusions. Ann Neurol. 2002;51:302–310. doi: 10.1002/ana.10101. [DOI] [PubMed] [Google Scholar]
- 23.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–1995. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 24.Bevivino AE, Loll PJ. An expanded glutamine repeat destabilizes native ataxin-3 structure and mediates formation of parallel β-fibrils. Proc Natl Acad Sci USA. 2001;98:11955–11960. doi: 10.1073/pnas.211305198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Berthelier V, Hamilton JB, O’Nuallain B, Wetzel R. Amyloid-like features of polyglutamine aggregates and their assembly kinetics. Biochemistry. 2002;41:7391–7399. doi: 10.1021/bi011772q. [DOI] [PubMed] [Google Scholar]
- 26.Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N. Ataxin-3, the MJD1 product, interacts with two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- 27.Li F, Macfarlan T, Pittman RN, Chakravarti D. Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J Biol Chem. 2002;277:45004–45012. doi: 10.1074/jbc.M205259200. [DOI] [PubMed] [Google Scholar]
- 28.Albrecht M, et al. Structural modeling of ataxin-3 reveals distant homology to adaptins. Proteins. 2003;50:355–370. doi: 10.1002/prot.10280. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt I, Brattig T, Gossen M, Riess O. Characterization of the rat spinocerebellar ataxia type 3 gene. Neurogenetics. 1997;1:103–112. doi: 10.1007/s100480050015. [DOI] [PubMed] [Google Scholar]
- 30.Linhartova I, et al. Conserved domains and lack of evidence for polyglutamine length polymorphism in the chicken homolog of the Machado– Joseph disease gene product ataxin-3. Biochim Biophys Acta. 1999;1444:299–305. doi: 10.1016/s0167-4781(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 31.McBurney MW, Jones-Villeneuve EM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- 32.Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnis VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol. 1982;94:253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banfi S, et al. Cloning and developmental expression analysis of the murine homolog of the spinocerebellar ataxia type 1 gene (Sca1) Hum Mol Genet. 1996;5:33–40. doi: 10.1093/hmg/5.1.33. [DOI] [PubMed] [Google Scholar]
- 34.Nechiporuk T, et al. The mouse SCA2 gene: cDNA sequence, alternative splicing and protein expression. Hum Mol Genet. 1998;7:1301–1309. doi: 10.1093/hmg/7.8.1301. [DOI] [PubMed] [Google Scholar]
- 35.Strom AL, et al. Cloning and expression analysis of the murine homolog of the spinocerebellar ataxia type 7 (SCA7) gene. Gene. 2002;285:91–99. doi: 10.1016/s0378-1119(02)00399-2. [DOI] [PubMed] [Google Scholar]
- 36.Lin B, et al. Sequence of the murine Huntington disease gene: evidence for conservation, alternative splicing and polymorphism in a triplet (CCG) repeat. Hum Mol Genet. 1994;3:85–92. doi: 10.1093/hmg/3.1.85. [DOI] [PubMed] [Google Scholar]
- 37.Barnes GT, et al. Mouse Huntington’s disease gene homolog (Hdh) Somatic Cell Mol Genet. 1994;20:87–97. doi: 10.1007/BF02290678. [DOI] [PubMed] [Google Scholar]
- 38.Skerjank IS. Cardiac and skeletal muscle development in P19 embryonal carcinoma cells. Trends Cardiovasc Med. 1999;9:139–143. doi: 10.1016/s1050-1738(99)00017-1. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar SA, Sharma RP. Modulation of c-myc, max, and mad gene expression during neural differentiation of embryonic stem cells by all-trans-retinoic acid. Gene Exp. 2002;10:125–135. [PMC free article] [PubMed] [Google Scholar]
- 40.Williams BY, Brommer S, Czarnetzki BM, Rosenbach T. The differentiation-related upregulation of aryl hydrocarbon receptor transcript levels is suppressed by retinoic acid. Biochem Biophys Res Commun. 1995;209:706–711. doi: 10.1006/bbrc.1995.1556. [DOI] [PubMed] [Google Scholar]
- 41.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 42.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wingender E, et al. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schug J, Overton GC. Technical Rep. CBIL-TR-1997-1001-v0.0. Computational Biology and Informatics Laboratory, School of Medicine, Univ. of Pennsylvania; Philadelphia: 1997. TESS: Transcription Element Search Software on the WWW′. [Google Scholar]
- 45.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 46.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Academic Press; San Diego: 1997. [Google Scholar]