Abstract
The activation of T lymphocytes plays a very important role in T-cell-mediated immune response. Though there are many related literatures, the changes of membrane surface nanostructures and adhesion property of T lymphocytes at different activation stages have not been reported yet. However, these investigations will help us further understand the biophysical and immunologic function of T lymphocytes in the context of activation. In the present study, the membrane architectures of peripheral blood T lymphocytes were obtained by AFM, and adhesion force of the cell membrane were measured by acquiring force–distance curves. The results indicated that the cell volume increased with the increases of activation time, whereas membrane surface adhesion force decreased, even though the local stiffness for resting and activated cells is similar. The results provided complementary and important data to further understand the variation of biophysical properties of T lymphocytes in the context of in vitro activation.
Keywords: T lymphocytes, Cell activation, Membrane nanostructures, Adhesion force
Introduction
Human peripheral blood T lymphocytes play a key role in human adaptive immunity. Though the activation process of T lymphocytes in vivo or in vitro has been well-studied immunologically and biochemically, however, whether the membrane surface nanostructures and adhesion property change in the process of T lymphocyte activation in vitro is largely unknown yet. However, the characterization of the nano-mechanical changes in the process of T lymphocytes activation in vitro has been hampered by the lack of sensitive quantitative techniques [1]. Atomic force microscopy (AFM) [2] is a powerful nano-technology tool that has been applied to observe DNA micropatterns on the polycarbonate surface [3], to fabricate the nanostructure materials [4], and to measure the adhesion force, elasticity and stiffness of sample [5-9]. The ultra-high force sensitivity of AFM and its ability to measure properties of individual cell makes the technique particularly appropriate for measuring viscoelastic changes of cell membrane. However, up to now, there are only a few reports of AFM application on T-cell-related studying. Franco-Obregón et al. [10] reported on the application of AFM to measure distinct ion channel classes on the outer nuclear envelope of human Jurkat T cell, and to determine the density of pore proteins. Wojcikiewicz et al. studied the interaction of leukocyte function-associated antigen-1 (LFA-1), expressed on Jurkat T cells, with intercellular adhesion molecules-1 and -2 using AFM, and the interaction between individual pairs of living T lymphocytes and endothelial cells [1,11-13]. The studies on biophysical properties (topography, nanostructures, adhesion force, stiffness, and others) of cells will provide fundamental insights into cellular structures and biology functions [14]. However, the variation of membrane surface nanostructures and nano-mechanical property of T lymphocytes in the context of in vitro activation remains unclear.
In the present work, we reported on the application of AFM to characterize the topography and to measure the membrane adhesion force in the process of human peripheral blood T lymphocytes upon in vitro activation. Firstly, we evaluated the effect of fixative (glutaraldehyde) and cell isolation process on the adhesion force of cell membrane, indicating the fixative resulted in the increases of adhesion force of cell membrane, whereas cell isolation process decreased the adhesion force. Then, we found that the adhesion force of lymphocytes decreased with the increasing of the activation time. Our results provide complementary and important data for further interpreting the activation time-dependent variation of membrane surface nanostructures and nano-mechanics, which may be helpful in investigating the membrane function of T lymphocytes at the nanoscale resolution.
Materials and Methods
T Lymphocyte Isolation and Preparation
Peripheral venous blood was drawn from healthy, drug-free adult donors and mixed with an anticoagulant (heparin) immediately. The isolation of T lymphocytes was conducted according to the RosetteSep Procedure: (1) 100 μL of RosetteSep®human CD3+T lymphocyte enrichment cocktail was fully mixed with 2 mL of whole blood, and incubated for 20 min at room temperature; (2) diluted with 2 mL of PBS containing 2% bovine serum albumin (BSA) gently; (3) layered the diluted solution on the top of 3 mL density medium (Ficoll), then centrifuged at 1200gfor 20 min; (4) remove the enriched cells from the density medium:blood plasma interface, and washed the enriched cells with 2% PBS (centrifuged at 425gfor 10 min) (repeated once). The isolated T lymphocytes (1.44 × 106) were cultured with culture medium RPMI 1640 for the next experiments.
To understand the effects of glutaraldehyde fixative on adhesion force, we firstly performed two groups that included unfixed cells and fixed cells respectively (which did not incubate in culture medium), and the group of fixed cells was set as control I. However, because fixative could stabilize cell membrane, glutaraldehyde was still used in the following experiments to acquire the repeatable images and force–distance curves. To estimate the effects of isolation process on cells, we measured the adhesion force of cells that incubated in culture medium for 24 h (no stimulation), and the acquired data were set as control II. Then, three testing (activation) groups, which were stimulated with phorbol dibutyrate (PDB, 1 × 10−7 mol/mL; Calbiochem Co.) plus ionomycin (ION, 0.5 μg/mL; Sigma) for 6, 24, and 48 h were performed. Cells were fixed by 2.5% glutaraldehyde (Sigma) in buffer solution for 10 min and air dried before AFM measurements. The prepared samples were measured immediately by AFM.
AFM Measurements
Atomic force microscopy (Autoprobe CP Research, Veeco, USA) was performed using a commercial AFM, which was performed in the contact mode or the tapping mode in air (room temperature, humidity: 75%). The glass substrate carrying cells was mounted onto the XY stage of the AFM and the integral video camera was used to locate the cells. The curvature radius of the silicon tip is less than 10 nm, scan rate is 0.3–1 Hz.
The contact mode was for measuring adhesion force (f, pN) cell membrane. Over ten thousands force curves were acquired, each curve representing the mean value of 15 times automatic measurements by the instrument. The tapping mode was for topographical analysis. The acquired images were reproducible during repeated scanning. More than 20 cells were investigated by the same two AFM probes (the tapping-mode probe for imaging and the contact-mode probe for force acquisition) for statistic analysis. And the adhesion force measurement of all samples was carried out with the same contact mode probe in air at room temperature.
Data Processing and Statistics
The acquired images (256 × 256 pixels) were only processed with the instrument-equipped software (Image Processing Software Version 2.1, IP 2.1) to eliminate low-frequency background noise in the scanning direction or to level the images (flatten order: 0–2). The data were reported as mean ± SD, and data analysis was conducted using Origin 6.0 software. The cell stiffness was qualitatively analyzed according to reported methods [7,8,15].
Results
Topographical Changes of T Lymphocytes in Activation
Resting T lymphocytes present typical spherical shape (Fig. 1a–c), and cellular microvilli and pseudopodia are clearly seen at the edge of cell. Figure 1b is an error-signal mode image, in which the structural details like pseudopodia can be more easily distinguished. Figure 1c shows nanostructural image of cell membrane, indicating the smooth and intact membrane surface structure. Figure 1i presents a height profile generated along the broken line in Fig. 1a, and measurement indicated the size of cell is 4.5 μm in diameter and 1.87 μm in height.
Figure 1.
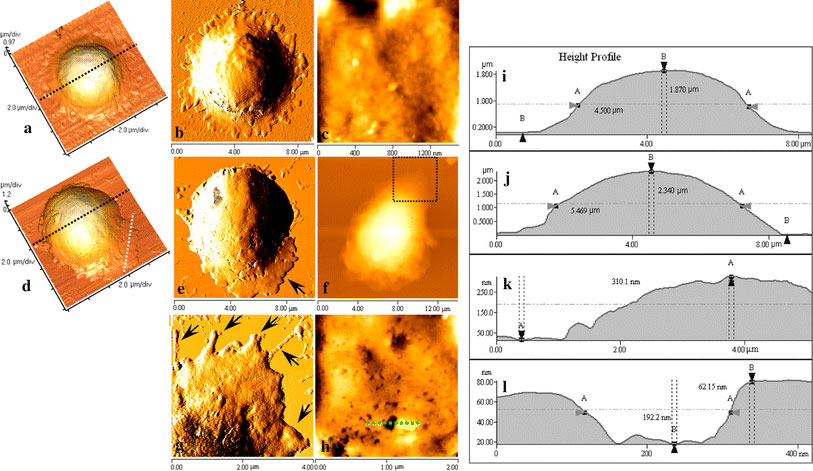
a–cRepresentative AFM topographical images of resting T lymphocyte.a3-D image of a resting T lymphocyte;berror-signal mode image ofa, the size of cell is 4.5 μm in diameter and 1.87 μm in height as shown in height profile (i), and cellular pseudopodia at the edge of cell could be clearly seen (b);cnanostructural image.d–hRepresentative AFM images of activated T lymphocytes.d3-D image of an activated T lymphocyte;eerror-signal mode image ofd, the size of cell is 5.469 μm in diameter and 2.34 μm in height as shown in height profile (j). The lamellipodia-like protrusion was also clearly visible (white dotted lineindandarrowine, whose height is 310.1 nm as shown in height profile (k).gAn enlarged error-signal mode image of the square frame inf;hnanostructural image possessing many concaves and the size of the largest concave is 192.2 nm in diameter and 62.15 nm in depth, which is shown by profile (l) that generated along the broken line inh
Figure 1d–h indicates topographies and nanostructures of activated T lymphocytes, and the lamellipodia-like protrusion that is found to have a height of about 310 nm (profile 1k) is shown by white dotted line in Fig. 1d and the black arrow in Fig. 1e. Figure 1g is an enlarged view (error-signal mode) of the square frame in Fig. 1f, and cellular pseudopodia are shown by black arrows. Figure 1h displays a representative nanostructural image of membrane surface, displaying a large number of concaves or membrane pores, whose average size is 40.73 ± 10.95 nm in diameter; and the largest concave is about 200 nm in diameter and 62 nm in depth, as shown in height profile Fig. 1l. Figure 1j presents a height profile generated along the black broken line in Fig. 1d, showing the size of cell is 5.469 μm in diameter and 2.34 μm in height.
To quantify the topographical difference between resting and activated CD3+T lymphocytes, a statistical analysis was performed as shown in Fig. 2, including the changes of average roughness (Ra) and particle mean height of surface nanostructure, cell height, cell diameter and cell volume. The results demonstrate that cell volume double increases from 42.87 ± 0.84 μm3(resting) to 94.24 ± 8.81 μm3(activation) (Fig. 2c), which is in accordance with the increases of cell height and cell diameter (Fig. 2b). When the measurements were conducted on nanoscale images, the results indicated that both average roughness (Ra) and the mean height of membrane surface particles increased after activation (Fig. 2a).
Figure 2.
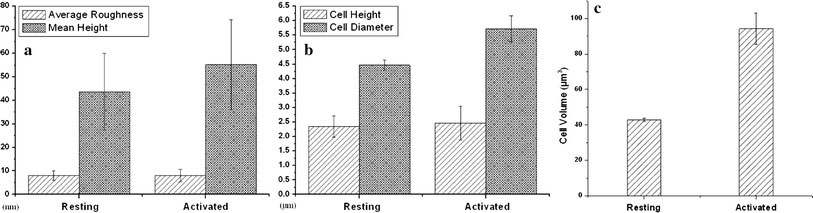
Histograms of statistical results.a,bThe average roughness and cell height of the resting and the activated groups are approximately equal. However, mean height of particles of surface nanostructure, cell diameter and cell volume in the activated group are larger than those in the resting group (a–c)
Adhesion Force Changes of T Lymphocytes in Activation
To compare the difference in adhesion property of cell membrane between resting and activated T lymphocytes, surface adhesion force was measured by acquiring force–distance curves. Figure 3a–f presents representative force–distance curves. To analyze the effects of fixative on cellular mechanical properties, we firstly measured adhesion force of fixed and unfixed lymphocytes (Fig. 3a, b). The results indicated that the fixative results in the increases of adhesion force of T cell membrane (Fig. 3g).
Figure 3.
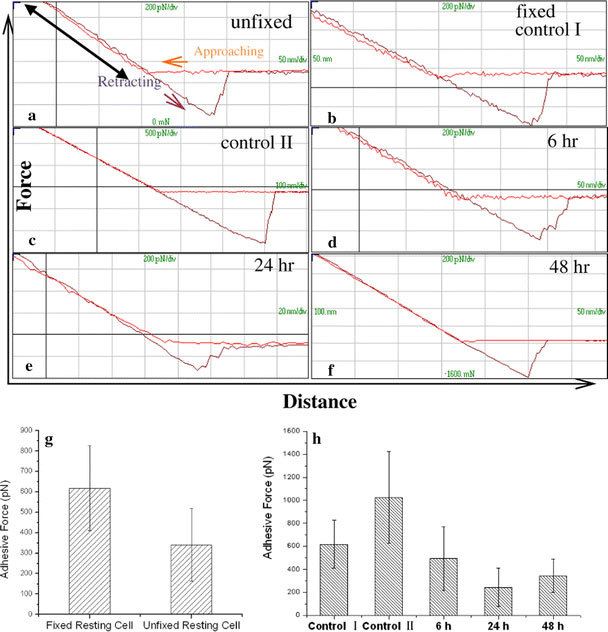
Representative force–distance curves.aForce curve of unfixed cells.bForce curve of fixed cells, which were measured immediately after isolation.cForce curve of fixed cells (control II) that incubated in culture medium for 24 h after isolation.d–fForce curves of fixed T cells, which were activated for 6, 24 and 48 h, respectively. According to the slope variation of approaching portion of force curve (black double-head arrowina), the changes of cell stiffness can be preliminary determined.gHistograms of statistical results of surface adhesion force, showing the fixed cells have a greater adhesion force than unfixed cells.hComparison of adhesion force of control groups and testing groups, indicating the adhesion force of non-incubated cells (control I) is smaller than that of incubated cells (control II); and the cell surface adhesion force decreases with the increases of activation time
On the other hand, AFM observation exhibits that the topography of T lymphocytes could be easily affected by the isolation processes (such as centrifugation, washing with PBS). Therefore, to evaluate how the isolation process alters the adhesion property of cell membrane, the difference of adhesion force between control I (non-incubated) and control II (incubated) (Fig. 3b, c) were then analyzed. The results clearly indicated that the adhesion force increased from 618 ± 207.28 pN (control I group) to 1025 ± 399.84 pN (control II group) (Fig. 3h), implying isolation processes lowered the membrane surface adhesion force.
Figure 3d, e, and f presents representative force–distance curves of T lymphocytes that were stimulated with PDB plus ION for 6, 24 and 48 h, respectively. The statistical analysis (Fig. 3h) suggests that the adhesion force of cell membrane decreased with the increases of stimulation time.
Discussion
AFM is not only a surface imaging technique, but also a sensitive force spectrometer. It has emerged as a powerful tool to measure the changes of mechanical property of cell membrane [5,6,8,16-19], cell stiffness [8,17], cell viscoelasticity [20,21], and to measure the interaction between cells [1], by which one could get some valuable information about the biophysical changes of the activated lymphocytes. AFM-based force spectroscopy is also particularly well suited for research in cell adhesion [19], and can stretch cells thereby allowing measurement of their rheological properties [17].
AFM observation indicates that cellular topography changed after PDB plus ION treatment for 24 h (Fig. 2), for example, the cell volume increased due to the cell activation. Cellular pseudopodia and lamellipodia-like protrusion of activated cells become more obvious and abundant, and the nanostructures of nano-concaves or membrane pores formed on the membrane surface of activated cell are readily seen (Fig. 1h); however, the cell membrane of resting T lymphocytes maintains integrity (Fig. 1c). The topographical and nanostructural changes (such as formation of membrane concaves/pores) might correlate with cytoskeleton rearrangement and/or more mass exchange of activated T cells than resting T cells.
Furthermore, the comparison of adhesion force between fixed cells and unfixed cells reveals that fixative can result in the increases of adhesion force of cell membrane, which is in accordance with the previous literature result [22]. On the other hand, the isolation process also affects the mechanical properties of T lymphocytes, inducing the decreases of the adhesion property of cell membrane. This result implies that the isolation process may affect the membrane biological function of T lymphocytes. Furthermore, as for testing groups, the measured adhesion force is clearly smaller than that of both control I and control II groups; after stimulated by PDB plus ION, the adhesion force decreases with the increases of stimulation time (Fig. 3h), and reaches the lowest at the stimulation time of 24 h, whereas the cell stiffness does not change obviously according to the qualitative analysis of approaching branch of force–distance curves.
The human immune system mainly includes cell-mediated immune system and humoral immune system. T lymphocytes play a key role in cell-mediated immune response, and the activation investigation of T lymphocytes in vitro can help researchers interpret the function of the whole immune system. Because the activation process of T cells is a key stage in T-cell mediated immune response, the investigation of biophysical changes of T cells could lead to further understanding of the mechanism of immune response. Phorbol dibutyrate (PDB), an effective T cell mitogen and an activator of protein kinase C (PKC), can enter cells and activate T lymphocytes. Ionomycin (ION) is a Ca2+ ionophore and used in research to raise the intracellular level of Ca2+ and in research on Ca2+ transport across biological membranes (http://en.wikipedia.org/wiki/Ionomycin). Therefore, PDB plus ION can play the role of the “two-signal” of T cell activation. In the process of activation and proliferation of T lymphocytes, the variation of both cellular topography and membrane biophysical properties might correlate with the changes of biological function of T lymphocytes, such as the phosphorylation of signaling molecules, changes of cell polarity, and Ca2+ release [23-26]. Moreover, both PDB and ION are strong pharmaceutical reagents that can quickly upregulate CD69 expression as early as 4 h after stimulation [27], and following the expression of CD25 and CD71, therefore, the expression of these activation markers and the mitosis of cells might altogether contribute to the changes of cellular topography and the decreases of membrane adhesion force. In this work, the measurement results are useful to further understand the relationship between cellular topography or membrane mechanical property and the function of T lymphocytes in immune response, which provide the complementary data on studying T cell in vitro activation.
Conclusions
In the present work, the characterization of cellular topography and measurement of membrane adhesion force in the process of activation and proliferation of T lymphocytes are reported. After stimulated with PDB plus ION for 24 h, the cell volume of T lymphocytes increased onefold; the adhesion force, however, decreased approximately to one-fifth control II. As the activation time increased (6, 24, and 48 h), the adhesion force of lymphocytes decreased, and it was the smallest at the 24 h stimulation time, but the cell stiffness does not alter obviously. The variation in membrane nanostructures adhesion force between resting cells and activated cells might closely correlate with the stimulus-induced changes in immunologic function of T lymphocytes. Taken together, this investigation provides complementary and important data to further interpret the relationship between immune function and the biophysical properties of T lymphocytes.
Contributor Information
Yangzhe Wu, Email: jerry_wyzhe@hotmail.com.
Jiye Cai, Email: tjycai@jnu.edu.cn.
Acknowledgements
This work was supported by the general project of NSFC (No. 60578025 and No. 30540420311) (J. C.), the general project of NSFC (No. 30572199) and the major project of NSFC (No. 30230350) (X. H.).
References
- Zhang X, Wojcikiewicz EP, Moy VT. Exp. 2006. p. 1306. COI number [1:CAS:528:DC%2BD28XptlSgsLY%3D] [DOI] [PMC free article] [PubMed]
- Binnig G, Quate CF, Gerber C. Phys. 1986. p. 930. Bibcode number [1986PhRvL..56..930B] [DOI] [PubMed]
- Wang Z, Li RX. Nanoscale Res. 2007. p. 69. COI number [1:CAS:528:DC%2BD2sXktVartrc%3D] [DOI]
- Jian SR, Juang JY. Nanoscale Res. 2008. p. 249. COI number [1:CAS:528:DC%2BD1cXhsVyhtrzJ]; Bibcode number [2008NRL.....3..249J] [DOI]
- Butt HJ, Cappella B, Kappl M. Surf. 2005. p. 1. COI number [1:CAS:528:DC%2BD2MXht1ahsrrK]; Bibcode number [2005SurSR..59....1B] [DOI]
- Waar K, van der Mei HC, Harmsen HJM, de Vries J, Atema-Smit J, Degener JE, Busscher HJ. Microbiology-Sgm. 2005. p. 2459. COI number [1:CAS:528:DC%2BD2MXmvV2ls74%3D] [DOI] [PubMed]
- Cross SE, Jin YS, Rao J, Gimzewski JK. Nat. 2007. p. 780. COI number [1:CAS:528:DC%2BD2sXhtlyktL7L]; Bibcode number [2007NatNa...2..780C] [DOI] [PubMed]
- Lam WA, Rosenbluth MJ, Fletcher DA. Blood. 2007. p. 3505. COI number [1:CAS:528:DC%2BD2sXksFWhs74%3D] [DOI] [PMC free article] [PubMed]
- Parot P, Dufrene YF, Hinterdorfer P, Le Grimellec C, Navajas D, Pellequer JL, Scheuring S. J. 2007. p. 418. COI number [1:CAS:528:DC%2BD1cXhsFOkt78%3D] [DOI] [PubMed]
- Franco-Obregon A, Wang HW, Clapham DE. Biophys. 2000. p. 202. COI number [1:CAS:528:DC%2BD3cXks1Klu7o%3D] [DOI] [PMC free article] [PubMed]
- Wojcikiewicz EP, Abdulreda MH, Zhang X, Moy VT. Biomacromolecules. 2006. p. 3188. COI number [1:CAS:528:DC%2BD28XhtVajt7zO] [DOI] [PMC free article] [PubMed]
- Zhang X, Chen A, De Leon D, Li H, Noiri E, Elitok S, Moy VT, Goligorsky MS. J. 2003. p. 46. [DOI] [PubMed]
- Zhang X, Wojcikiewicz E, Moy VT. Biophys. 2002. p. 2270. COI number [1:CAS:528:DC%2BD38XnslSrt7s%3D]; Bibcode number [2002BpJ....83.2270Z] [DOI] [PMC free article] [PubMed]
- Heinz WF, Hoh JH. Trends Biotechnol. 1999. p. 143. COI number [1:CAS:528:DyaK1MXlt1KjtLo%3D] [DOI] [PubMed]
- Strasser S, Zink A, Kada G, Hinterdorfer P, Peschel O, Heckl WM, Nerlich AG, Thalhammer S. Forensic Sci. 2007. p. 8. [DOI] [PubMed]
- Bowen WR, Lovitt RW, Wright CJ. J. 2001. p. 54. COI number [1:CAS:528:DC%2BD3MXjtFeqsr8%3D] [DOI] [PubMed]
- Canetta E, Duperray A, Leyrat A, Verdier C. Biorheology. 2005. p. 321. COI number [1:CAS:528:DC%2BD2MXht1entrrE] [PMC free article] [PubMed]
- Lulevich V, Zink T, Chen HY, Liu FT, Liu GY. Langmuir. 2006. p. 8151. COI number [1:CAS:528:DC%2BD28XmvFWhtro%3D] [DOI] [PubMed]
- Wojcikiewicz EP, Zhang X, Chen A, Moy VT. J. 2003. p. 2531. COI number [1:CAS:528:DC%2BD3sXltFemtr0%3D] [DOI] [PMC free article] [PubMed]
- Fritzsche W. Microsc. 1999. p. 357. COI number [1:STN:280:DyaK1M7ovVGqtQ%3D%3D] [DOI] [PubMed]
- Fritzsche W, Henderson E. Ultramicroscopy. 1997. p. 191. COI number [1:CAS:528:DyaK2sXmvFWmtbk%3D] [DOI] [PubMed]
- Burks GA, Velegol SB, Paramonova E, Lindenmuth BE, Feick JD, Logan BE. Langmuir. 2003. p. 2366. COI number [1:CAS:528:DC%2BD3sXnsVKisA%3D%3D] [DOI]
- Burkhardt JK, Carrizosa E, Shaffer MH. Annu. 2008. p. 233. COI number [1:CAS:528:DC%2BD1cXltlWksbo%3D] [DOI] [PubMed]
- Dadsetan S, Zakharova L, Molinski TF, Fomina AF. J. 2008. p. 12512. COI number [1:CAS:528:DC%2BD1cXltVyqsLc%3D] [DOI] [PubMed]
- Feske S. Nat. 2007. p. 690. COI number [1:CAS:528:DC%2BD2sXpsV2qtLc%3D] [DOI] [PubMed]
- Negulescu PA, Krasieva TB, Khan A, Kerschbaum HH, Cahalan MD. Immunity. 1996. p. 421. COI number [1:CAS:528:DyaK28Xjt1GhtL4%3D] [DOI] [PubMed]
- Xu L Liu Y He X Cell 20052295.COI number [1:CAS:528:DC%2BD2MXhtlSitrvK]16274628


