Abstract
Gold nanoparticles (GNPs) are widely used to detect DNA. We studied the effect of pH on the assembly/disassembly of single-stranded DNA functionalized GNPs. Based on the different binding affinities of DNA to GNPs, we present a simple and fast way that uses HCl to drive the assembly of GNPs for detection of DNA sequences with single nucleotide differences. The assembly is reversible and can be switched by changing the solution pH. No covalent modification of DNA or GNP surface is needed. Oligonucleotide derived from human p53 gene with one-base substitution can be distinguished by a color change of the GNPs solution or a significant difference of the maximum absorption wavelength (λmax), compared with wildtype sequences. This method enables detection of 10 picomole quantities of target DNA.
Keywords: Single-stranded DNA, HCl, Gold nanoparticles, p53, Mutation
Introduction
Gold nanoparticles (GNPs) coupled with biomolecules are of great current interest because of their biomedical applications. GNPs have been used to detect DNA with high sensitivity and selectivity [1]. Mutations, single nucleotide polymorphisms (SNPs), chromosomal translocations, gene expression, and pathogens from clinical samples can be easily detected [2-5]. The first reports on GNPs–DNA complex were published in the August issue of Nature in 1996. Two groups of CA Mirkin and AP Alivisatos described the organization of GNPs with DNA oligonucleotides [6,7]. Mirkin et al. mixed two batches of 13-nm gold particles, each attached by thiolated non-complementary oligonucleotides. When a complementary oligonucleotide duplex was added to the solution, the nanoparticles assembled into aggregates, which provoked a red-to-blue color change accomplished by a red-shift of the surface plasmon band. The disadvantage of this method is the requirements of two sets of oligonucleotide probes and thiol-modification of the probe DNA, which is expensive and time-consuming [8].
The optical property of DNA-linked gold nanoparticle assemblies was developed for sequence-specific DNA detection. Li and Rothberg invented another non-cross-linking method, where GNPs aggregation was induced by an increasing salt concentration. This method took advantage of the preferential nonspecific binding of single-stranded oligonucleotides over double-stranded ones to the surface of GNPs and devised a label-free way, which requires no covalent modification of the DNA. The presence of non-complementary probes could prevent aggregation and the solution remains red; while complementary probes could not prevent gold nanoprobe aggregation resulting in a color change from red to blue. This simple assay has been applied to clinical samples of genomic DNA that screen for SNPs associated with long QT syndrome [9,10].
Adsorption of single-stranded DNA (ssDNA) on GNPs can effectively stabilize GNPs against salt-induced aggregation [11]. GNPs synthesized by citrate reduction of HAuCl4 are stable in solution, while electrostatic repulsion among citrate ion-coated GNPs prevents them from aggregation. But irreversible aggregation occurs when salt is added in the solution. To our knowledge, it is still unclear whether ssDNA can stabilize GNPs against HCl. In this article, we have studied the effect of pH on the assembly/disassembly of ssDNA functionalized GNPs (ssDNA–GNPs). Differentiation of single nucleotide mutation of oligonucleotide is achieved by only one-step reaction. The target sequences harbor 12 of the most frequent point mutations in exon 5, 7, and 8 of human p53 gene (Table 1).
Table 1.
Target oligonucleotide sequences
| ssDNA | No. | Sequence (5′-3′) |
|---|---|---|
| 175wd |
|
GGCACCCGCGTCCGCGCCATGGCCATCTAC |
| 175G–A |
1 |
GGCACCCGCGTCCCGCCATGGCCATCTAC |
| 245wd |
|
TGCATGGGCGGCATGAACCGGAGGCCCATC |
| 245G–T |
2 |
TGCATGGGCGCATGAACCGGAGGCCCATC |
| 245G–A |
3 |
TGCATGGGCGCATGAACCGGAGGCCCATC |
| 248wd |
|
GCATGAACCGGAGGCCCATCCTCACCATCA |
| 248G–A |
4 |
GCATGAACCGAGGCCCATCCTCACCATCA |
| 248G–T |
5 |
GCATGAACCGAGGCCCATCCTCACCATCA |
| 248C–T |
6 |
GCATGAACGGAGGCCCATCCTCACCATCA |
| 249wd |
|
GCATGAACCGGAGGCCCATCCTCACCATCA |
| 249G1–T |
7 |
GCATGAACCGGAGCCCATCCTCACCATCA |
| 249G2–T |
8 |
GCATGAACCGGAGCCCATCCTCACCATCA |
| 249G-C |
9 |
GCATGAACCGGAGCCCATCCTCACCATCA |
| 273wd |
|
GAACAGCTTTGAGGTGCGTGTTTGTGCCTG |
| 273G–T |
10 |
GAACAGCTTTGAGGTGCTGTTTGTGCCTG |
| 273G–A |
11 |
GAACAGCTTTGAGGTGCTGTTTGTGCCTG |
| 282wd |
|
TCCTGGGAGAGACCGGCGCACAGAGGAAGA |
| 282C–T | 12 | TCCTGGGAGAGACGGCGCACAGAGGAAGA |
The underline corresponds to the single base mismatch comparing with the wildtype (wd). 12 of the most frequent point mutations in exon 5, 7, and 8 of human p53 gene are included. 248wd and 249wd are the same sequences. The mutant sequences are numbered from 1 to 12
Experimental
Materials
HAuCl4, sodium tris-citrate, NaOH and HCl were purchased from Xilong Chemical (Guangdong, China). All reagents were of analytical grade. Single-stranded DNA (ssDNA) was synthesized by Shanghai Sangon Biological and Engineering Company with the following sequences (Table 1). Milli-Q water (18.2 MΩ) was used in all of the experimental processes.
GNPs were prepared in water as follows. An aqueous solution of sodium tris-citrate (5 mL, 1%) was mixed with HAuCl4(1 mL, 1%) solution in a conical flask and sealed with parafilm. The reaction solution was first boiled in a 700-watt microwave oven on high power for 1 min, and then kept heating on medium power for 5 min.
UV–Vis Spectroscopy
GNPs (10 nM) were mixed with 50 pmol ssDNA and incubated at room temperature for 10 min. The pH of aqueous solutions was adjusted by adding 1 M HCl or 1 M NaOH. 50 μL cuvettes (Beckman) were used to hold the samples, and all UV–visible spectra were collected on a Beckman H800 UV–vis spectrophotometer with 1 nm resolution.
Zeta Potentials and Particle Size Measurement
The determination of zeta potential was carried out using Zetasizer Nano-ZS (Malvern Instruments Ltd. UK). The zeta potentials of GNPs and GNPs binding with ssDNA at different pH values were measured. Each data point for zeta potential was an average of at least 15 measurements.
Transmission Electron Microscopy (TEM)
Samples for TEM characterization were prepared by placing a drop of sample solution on a carbon coated copper grid and dried at 37 °C. TEM measurements were made in a JEM-2100 transmission electron microscopy operated at an accelerating voltage of 200 kV.
Results and Discussion
Tumor suppressor gene p53 (MIM 191170) is the most frequently mutated gene in human cancer. Detection of p53 abnormalities is very important for cancer diagnosis and early therapy [12,13]. In the present work we developed a simple colorimetric assay using unmodified GNPs to detect 12 of the most frequent point mutations in exon 5, 7, and 8 of human p53 gene. The target mutant ssDNA with one-base substitution can be differentiated clearly from the wildtype oligonucleotide.
We improved the conventional method to synthesize GNPs by citrate reduction of HAuCl4 under microwave [14,15]. The average particle size was 13 nm in diameter measured by TEM. The surface plasmon band (λmax) was 520 nm. The initial colloidal solution of GNPs or ssDNA-coated GNPs was wine red at pH 7.0. A characteristic blue color of naked GNPs was shown in alkaline condition (pH > 12.0). With addition of HCl or NaOH (pH 2.0–12.0), GNPs solution turned light pinkish, at pH < 2.0, the solution first became blue, then turned colorless instantaneously, and, finally, a black precipitation settled to the bottom. If GNPs were incubated with single-stranded DNA, addition of HCl caused a color change to blue or purple, while addition of NaOH (pH 7.0–12.0) did not change the solution color. The absorption peak red-shifted to different long-wave-length regions according to different DNA sequences (pH < 4.0 or pH > 12.0). And the redshift was accompanied by the attenuation of its intensity, which clearly indicated the occurrence of aggregation.
Since the isoelectric point (IP) of ssDNA is about 4.0–4.5, ssDNA is negatively charged at pH above the IP, and can be easily adsorbed on the surface of GNPs. Such electrostatic interaction is mainly caused by functional groups (amines and carbonyls) of the exposed bases, which bind DNA to GNPs [9,16]. Stabilization of GNPs and ssDNA–GNPs complex at different pH values (1.0–14.0) was investigated with UV–vis spectroscopy and zeta potential analysis. The effect of pH on the zeta potentials is shown in Fig. 1a. The zeta potentials of GNPs and ssDNA–GNPs at all pH values are negative. Gold particles dispersed in water are negatively charged because the nanoparticles are coated by citrate ions. Zeta potentials of DNA-binded GNPs are more negative than those of GNPs at the same pH, indicating that ssDNA–GNPs complex are more stable than GNPs. In general, zeta potentials of both GNPs and ssDNA–GNPs decrease with increasing pH (pH 1.0–12.0). At low pH the particles have less potential due to the increased chemical potential of H+ ions in solution. Between pH 4.0 and pH 8.0, the zeta potential changes a little. At pH 12.0, the zeta potentials of GNPs and ssDNA–GNPs reach the minimum values below −30 mv. However, at pH > 12.0, the zeta potentials dramatically increase. This is in accordance with the result of UV–vis spectrum (Fig. 1b). In this case, the colloidal particles aggregate to large agglomerates, which is the reason for the significant increase of λmax at high pH (>12.0). Between pH 2.0 and pH 12.0, λmax of GNPs almost keeps at the same level, while at pH 1.0, the intensity of the adsorption peak decreases nearly to zero (not shown in Fig. 1b). But it is quite different for ssDNA–stablilized GNPs, a redshift of λmax appears at pH under 4.0 (λmax = 542 nm at pH 1.0).
Figure 1.
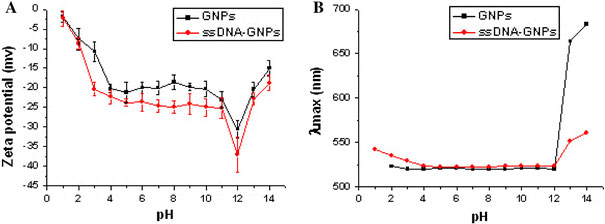
Zeta potentials (a) and λmax(b) of GNPs and ssDNA–GNPs at different solution pH values. ssDNA: 273G–T
Further study was performed to investigate whether HCl-driven GNPs aggregation could differentiate ssDNA with single-base substitution. We tested altogether 17 sequences (Table 1), including 5 wildtype oligonucleotides and 12 respective mutant oligonucleotides. Of the 12 single base-pair substitutions, 6 were transitions (C→T, G→A) and 6 were transversions (G→T, G→C). Upon addition of 1 M HCl to 50 μL GNPs solution incubated with a wildtype (wd) ssDNA 248WT, a dramatic color change from red to blue was observed within a few seconds, while solution containing the mutant (mt) ssDNA 248C–T became dark purple. The UV–vis spectra showed a significant shift in the surface plasmon resonance (SPR) from λmax = 520 nm to 644 nm for 248WT,while the adsorption peak shifted to 599 nm for 248C–T (Fig. 2). Thus oligonucleotides with one-base substitution could be distinguished clearly by a significant difference of λmaxof GNPs–wd ssDNA (644 nm) and GNPs–mt ssDNA (599 nm). We compared λmaxof 12 groups (Fig. 3), statistically significant differences were observed between wildtype and mutant sequences of all groups (P < 0.05, Student’st-test for paired data). We suggest that ssDNA with a single-base substitution should have different binding strength to GNPs, leading to different stabilization efficiency against HCl, which is confirmed by the UV–vis spectra (Fig. 2). This method enables detection of 10 picomole quantities of target DNA. Under alkaline conditions, there are no significant difference of λmaxbetween GNPs–wd ssDNA and GNPs–mt ssDNA.
Figure 2.
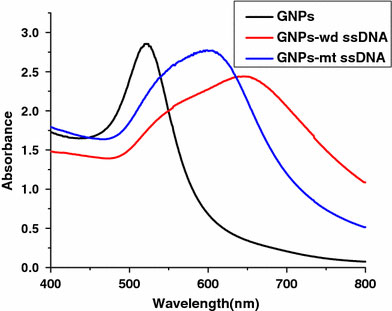
UV–vis spectra of GNPs solutions mixed with wildtype (wd) and mutant (mt) ssDNA upon addition of HCl (pH 1.0). ssDNA: 248C–T
Figure 3.
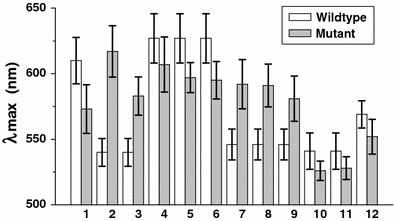
Comparison of λmaxof ssDNA–GNPs upon addition of HCl. The sequence numbers are indicated in Table 1. Each experiment was repeated at least five times.P < 0.05, Student’st-test for paired data
It is interesting that this HCl-driven GNPs assembly is reversible. When the pH was changed to 7.0 by addition of 1 M NaOH, the blue solution turned red. By addition of HCl, it changed to blue again. Figure 4shows the reversibility of the color change in alternating additions of aliquots of H+and OH−. Figure 5shows the changes in the absorption maximum, as the solution pH is cycled between 7.0 and 1.0 by the alternating addition of 1 M HCl or 1 M NaOH. UV–vis absorbance was measured after each pH change, which showed a transition between 520 nm and 650 nm. This phenomenon is very different from that of GNPs aggregation triggered by HCl without ssDNA protection, which is irreversible. However, the color of the GNPs solution becomes weaker (Fig. 4) and the difference of λmaxgradually decreases (Fig. 5) when the number of pH cycles is increased, which is caused presumably by dilution of the solution and degradation of DNA by repeated addition of strong acid and alkali. This suggests that the assembly and disassembly of the GNPs is partly reversible.
Figure 4.

Reversible color change of GNPs solution. ssDNA: 273G–T
Figure 5.
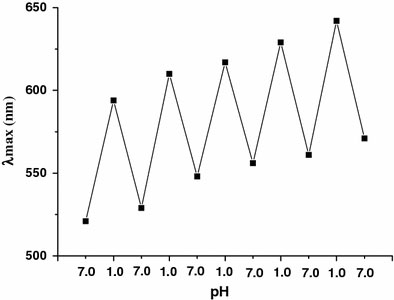
The absorption maximum for the ssDNA–GNPs conjugates as the solution pH is cycled between 7.0 and 1.0 by adding 1 M NaOH or 1 M HCl. ssDNA: 273G–T
TEM images of GNPs under acidic conditions are shown in Fig. 6. The GNPs are transformed into interconnected chainlike superstructures under acidic condition (pH 1.0). But when protected by ssDNA, GNPs assemble to big aggregate, individual round nanoparticles are clearly visible in the aggregate. A previous report demonstrated that when in contact with the HCl solution, Au atoms located at the surfaces of the particles dissolved in HCl solution, forming AuCl4− ions [17]. We suggest that GNPs fuse into a network structure because their surfaces are partly dissolved in HCl for their high chemical activity. Previous research demonstrated that ssDNA has a protective effect on GNPs against aggregation in a salt solution for the highly charged phosphate backbone of ssDNA sustains the electrostatic repulsion among ssDNA-adsorbed GNPs [18]. Our data confirm that ssDNA can also effectively stabilize GNPs against HCl-induced particles’ fusion. Addition of H+ may neutralize the negative charges of phosphate groups of DNA, leading to GNPs’ aggregation. The result of Fig. 6b is consistent with that of Fig. 5, for aggregated ssDNA–GNPs (Fig. 6b) caused a significant increase in absorbance at pH 1.0 (Fig. 5).
Figure 6.
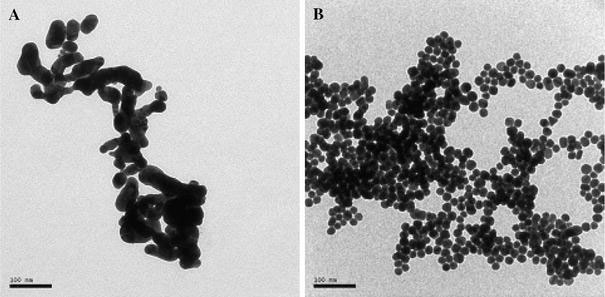
TEM image ofaGNPs + HCl (pH 1.0),bssDNA–GNPs + HCl (pH 1.0). ssDNA: 273G–T. Scale bar: 100 nm
Conclusions
This work demonstrated that adsorption of ssDNA on GNPs could effectively stabilize the colloid against HCl-induced fusion. In addition, we presented an easy and inexpensive way for single-base mismatch detection. Based on the electrostatic interaction of DNA and GNPs upon addition of HCl, ssDNA sequence can be easily distinguished from sequence with single-base mismatch by measuring λmaxof the two systems. Comparing with other nanoparticle-based DNA detection assays, our method has the following advantages: (1) no need of complicated modification of GNPs or DNA, (2) no additional DNA probes are required, (3) no need of signal amplification or temperature control. Only three components exist in our system: GNPs, target oligonucleotide, and HCl. We successfully applied this method to detect 12 point mutations derived from human p53 gene. Since this methodology is limited to a single color change, two individual reactions are required for comparison-a wildtype sequence and a mutant sequence. This colorimetric method should have the potential for genetic tests.
Contributor Information
Liping Sun, Email: sunliping@xmu.edu.cn.
Qiqing Zhang, Email: zhangqiq@xmu.edu.cn.
Acknowledgments
This work is supported by the Science and Technology Innovation Project of Fujian Province for Young Scientific Researchers, China (No. 2006F3128) and the Open Fund of State Key Laboratory for Physical Chemistry of Solid Surfaces, Xiamen University (No. 200602).
References
- Daniel MC, Astruc D. Chem. 2004. p. 293. COI number [1:CAS:528:DC%2BD3sXpvFGlur0%3D] [DOI] [PubMed]
- Baptista P, Doria G, Henriques D, Pereira E, Franco R. J. 2005. p. 111. COI number [1:CAS:528:DC%2BD2MXpslCht7g%3D] [DOI] [PubMed]
- Kalogianni DP, Bravou V, Christopoulos TK, Ioannou PC, Zoumbos NC. Nucleic Acids Res. 2007. p. e23. COI number [1:CAS:528:DC%2BD2sXjslOgsL4%3D] [DOI] [PMC free article] [PubMed]
- Martins R, Baptista P, Raniero L, Doria G, Silva L, Franco R, Fortunato E. Appl. 2007. p. 023903. [DOI]
- Doria G, Franco R, Baptista P. IET Nanobiotechnol. 2007. p. 53. COI number [1:CAS:528:DC%2BD2sXhtVKhtLfN] [DOI] [PubMed]
- Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996. p. 607. COI number [1:CAS:528:DyaK28XltVWqsrk%3D]; Bibcode number [1996Natur.382..607M] [DOI] [PubMed]
- Alivisatos AP, Johnsson KP, Peng X, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Nature. 1996. p. 609. COI number [1:CAS:528:DyaK28XltVWqsrY%3D]; Bibcode number [1996Natur.382..609A] [DOI] [PubMed]
- Demers LM, Mirkin CA, Mucic RC, Reynolds Iii RA, Letsinger RL, Elghanian R, Viswanadham G. Anal. 2000. p. 5535. COI number [1:CAS:528:DC%2BD3cXnsVals70%3D] [DOI] [PubMed]
- Li HX, Rothberg LJ. J. 2004. p. 10958. COI number [1:CAS:528:DC%2BD2cXmsFCrsb8%3D] [DOI] [PubMed]
- Li H, Rothberg L. Proc. Natl. Acad. Sci. USA. 2004. p. 14036. COI number [1:CAS:528:DC%2BD2cXosVylsrY%3D]; Bibcode number [2004PNAS..10114036L] [DOI] [PMC free article] [PubMed]
- Storhoff JJ, Elghanian R, Mirkin CA, Letsinger RL. Langmuir. 2002. p. 6666. COI number [1:CAS:528:DC%2BD38XltlCntbg%3D] [DOI]
- Hollstein M, Sidransky D, Vogelstein B, Harris CC. Science. 1991. p. 49. COI number [1:CAS:528:DyaK3MXks1Gmurg%3D]; Bibcode number [1991Sci...253...49H] [DOI] [PubMed]
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Cancer Res. 1994. p. 4855. COI number [1:CAS:528:DyaK2cXlslKgsLw%3D] [PubMed]
- Geraheart LA, Ploehn HJ, Murphy CJ. J. Phys. Chem. B. 2001. p. 12609. COI number [1:CAS:528:DC%2BD3MXot1elt78%3D] [DOI]
- Ken F, Cheng LY, Ko CFH, Chu TC. Mater. 2004. p. 373. COI number [1:CAS:528:DC%2BD3sXpt1Sgs7o%3D] [DOI]
- Jang NH. Bull. 2002. p. 1790. COI number [1:CAS:528:DC%2BD3sXis1Onsw%3D%3D] [DOI]
- Shi HZ, Bi HJ, Yao BD, Zhang LD. Appl. 2000. p. 276. COI number [1:CAS:528:DC%2BD3cXkvFWqsbw%3D]; Bibcode number [2000ApSS..161..276S] [DOI]
- Yang J, Lee JY, Too HP, Chow GM, Gan LM. Chem. 2006. p. 304. COI number [1:CAS:528:DC%2BD28XjvVantLg%3D]; Bibcode number [2006CP....323..304Y] [DOI]


