Abstract
Production of extended-spectrum β-lactamases (ESBL) has been reported in virtually all species of Enterobacteriaceae, which greatly complicates the therapy of infections caused by these organisms. However, the frequency of isolates producing AmpC β-lactamases, especially plasmid mediated AmpC (pAmpC), is largely unknown. These β-lactamases confer resistance to extended spectrum cephalosporins and aztreonam, a multidrug-resistant (MDR) profile. The aim of the present study was to determine the occurrence of ESBL and pAmpC β-lactamases in a hospital where MDR enterobacterial isolates recently emerged. A total of 123 consecutive enterobacterial isolates obtained from 112 patients at a university hospital in Rio de Janeiro, Brazil during March-June 2001 were included in the study. ESBL was detected by the addition of clavulanate to cephalosporin containing disks and by double diffusion. AmpC production was evaluated by a modified tridimensional test and a modified Hodge test. The presence of plasmid-mediated ampC β-lactamase genes was evaluated by multiplex-PCR. Sixty-five (53%) of 123 enterobacterial isolates were MDR, obtained from 56 patients. ESBL production was detected in 35 isolates; 5 clonal E. coli isolates exhibited high levels of chromosomal AmpC and ESBL production. However, no isolates contained pAmpC genes. Infection or colonization by MDR enterobacteria was not associated with any predominant resistant clones. A large proportion of hospital infections caused by ESBL-producing enterobacteria identified during the study period were due to sporadic infections rather than undetected outbreaks. This observation emphasizes the need to improve our detection methods for ESBL- and AmpC-producing organisms in hospitals where extended-spectrum cephalosporins are in wide use.
Keywords: multidrug-resistance, enterobacteria, AmpC beta-lactamase, ESBL, three-dimensional test, Hodge test
1. Introduction
Infections by enterobacterial isolates resistant to extended-spectrum cephalosporins or aztreonam have become a serious problem worldwide (Winokur et al., 2001; Bell et al., 2002). Increased prevalence of MDR enterobacteria is often due to intense prescription of third-generation cephalosporins or quinolones in the community and in the hospital and dissemination of these organisms by inappropriate hygienic practices (Cohen, 1992; Davin-Regli et al., 1997; Hobson et al., 1996; Holmberg et al., 1987; Meyer et al., 1993; Rice et al., 1990). In addition, MDR strains of enteric pathogens can emerge from animal reservoirs due to selection by antimicrobial agents used as growth promoters (Ramchandani et al., 2005).
Nosocomial outbreaks of MDR infections in hospitals have led to endemic occurrence of these infections, with dissemination of resistance genes, plasmids and strains of a variety of bacterial species (Ben Redjeb et al., 1988; Bradford, 2001; Philippon et al., 1989; Winokur et al., 2001). MDR enterobacteria often produce extended spectrum β-lactamases (ESBL) or overexpress AmpC β-lactamases (AmpC) (Thomson, 2001). However, many clinical microbiologists are unaware of plasmid-encoded AmpC because its phenotypic detection is difficult, and these β-lactamases can be misidentified as ESBLs (Hanson, 2003). There is confusion about the importance of such resistance mechanisms, optimal test methods, and appropriate reporting conventions. Failure to detect these β-lactamases has contributed to their uncontrolled spread and occasional therapeutic failures. The Clinical and Laboratory Standards Institute (CLSI) recommend ESBL screening and confirmation only for Escherichia coli isolates, which is an organism with constitutive or minimal AmpC chromosomal expression, and Klebsiella oxytoca, Klebsiella pneumoniae and Proteus mirabilis, which have no chromosomally encoded AmpC (CLSI/NCCLS, 2005). No standards have been published to date for the other enterobacterial species. In addition, there are no CLSI recommendations for detecting plasmid-mediated AmpC (pAmpC) in any species.
Perez-Pérez and Hanson (2002) developed a multiplex PCR assay for the detection of plasmid-encoded ampC genes that proved useful as a rapid screening tool to distinguish cefoxitin resistant non-AmpC producers from cefoxitin resistant AmpC producers. In addition, this PCR-based method can distinguish hyper-producing chromosomal AmpC E. coli isolates from E. coli isolates encoding an ‘imported’ plasmid ampC gene.
Hospital Universitário Clementino Fraga Filho (HUCFF) is a large (490 bed), tertiary care university hospital, in the city of Rio de Janeiro, Brazil. In 2000, the Committee for Control of Healthcare-Associated Infections at HUCFF detected a five-fold increase in the number of MDR enterobacterial infections, from 35 patients, in 1998, to 172 patients, in 2000. The present study was designed to provide a better understanding of the microbiological factors associated with this observation at HUCFF. Our main study objective was to determine the occurrence of ESBL and pAmpC in association with the emergence of MDR enterobacteria and whether this sudden increase in the number of MDR enterobacterial infections was due to an outbreak of limited clonal groups of ESBL/AmpC-expressing strains, or an increase of distinct strains that were selected by the widespread use of extended-spectrum cephalosporins in the hospital.
2. Materials and Methods
2.1. Study design
The study period was from March to June, 2001. Patients were retrospectively selected by review of records at the microbiology laboratory and the patient hospital database. A total of 56 patients with MDR enterobacteria and 56 with non-MDR enterobacteria were included in the study. The institution’s ethics committee approved the study.
2.2. Microbiological methods
2.2.1. Study population and bacterial isolates
A total of 128 enterobacterial isolates were obtained from 117 patients consecutively admitted to HUCFF between March and June, 2001. Patients included in the study were those who had the bacterial isolate confirmed as an Enterobacteriaceae species and obtained at least 72h after admission. Only one isolate of each bacterial species was selected per patient, preferably, one obtained from a normally sterile site. Five isolates from five (7.5%) patients were excluded: two were from patients whose records were not found, and three were from patients with more than one isolate of the same bacterial species. Therefore, 123 enterobacterial isolates obtained from 112 patients infected (90, 80.4%) or colonized (22, 19.6%) by these agents were included in the study.
Bacterial species were identified by the Vitek system GNI card (bioMérieux, Marcy l’Etoile, France) and conventional biochemical tests (Farmer III, 2003; Winn Jr. et al., 2006). Antimicrobial susceptibility was evaluated by the disk diffusion method (CLSI/NCCLS, 2000) for the following agents: amikacin, aztreonam, cefepime, cefotaxime, ceftazidime, cefoxitin, cephalothin, ciprofloxacin, gentamicin, imipenem, piperacillin-tazobactam, and trimethoprim-sulfamethoxazole. The category of intermediate susceptibility was analyzed together with the resistant strains.
2.2.2. Tests for β-lactamase production
ESBL screening was based on disk diffusion results for aztreonam, ceftazidime and cefotaxime and confirmed by standard ceftazidime and cefotaxime disks combined with clavulanic acid (10μg) (CLSI/NCCLS, 2005). In addition, we evaluated all isolates by the double-disk diffusion method with disks containing cefepime, cefotaxime, ceftazidime, and aztreonam placed 25mm apart (center to center) to a disk containing a β-lactamase inhibitor (amoxicillin-clavulanic acid) (Jarlier et al., 1988).
Ceftazidime-resistant and -intermediate isolates were also evaluated for metallo-β-lactamase production by a disk approximation test with disks containing ceftazidime and 2-mercaptopropionic acid (Arakawa et al., 2000).
AmpC production was evaluated for isolates belonging to species with no chromosomally encoded AmpC type β-lactamase (K. pneumoniae and P. mirabilis) or with constitutive or minimal AmpC chromosomal expression (E. coli). AmpC detection methods included the Hodge test and the tridimensional test modified from Yong and colleagues (2002) and Coudron and colleagues (2000), respectively. Indicator strains were E. coli ATCC 25922 and E. coli ATCC 35218. To test P. mirabilis isolates, we used MacConkey agar plates to suppress swarming (including those of unlysed cells in the tridimensional test), which could interfere with interpretation of results in both Hodge and tridimensional tests.
Briefly, for the Hodge test, the surface of two Mueller-Hinton agar plates were inoculated with each one of indicator strains and a 30 μg cefoxitin disk was placed at the center of the plate. Two or three colonies of the overnight-cultured test strains were picked and heavily streaked outward from the cefoxitin disk with a bacteriologic loop. After 18-h incubation, the radius of the inhibition zone of the indicator organism, and the decreased radius of the inhibition zone along the growth of test strain were measured; the difference was expressed in millimeters as a distorted inhibition zone (Figure 1A).
Figure 1.
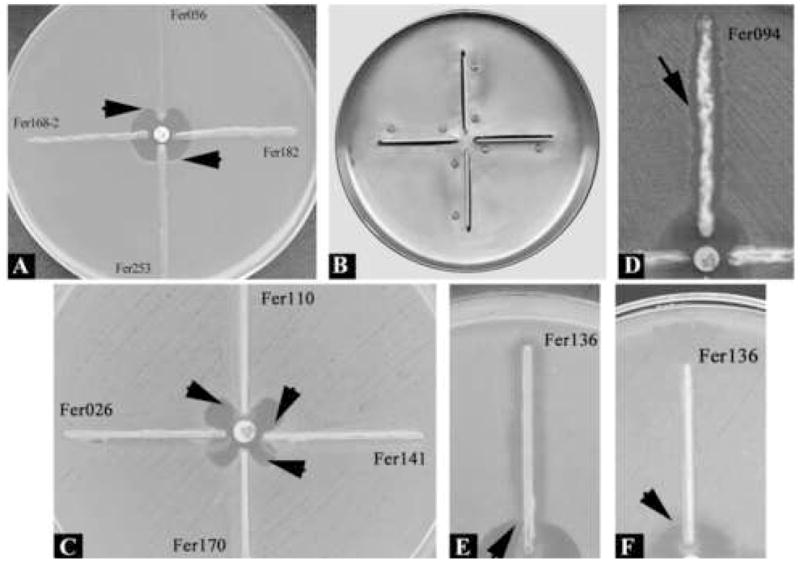
Hodge and tridimensional test patterns. (A) Hodge test (upper arrow): distorted inhibition zone (2mm) of the indicator organism by a positive isolate; (lower arrow) distorted inhibition zone (1mm) of the indicator organism by an indeterminate isolate; right and left: no effect on the zone of the indicator organism by negative isolates. (B) Metal apparatus specially prepared for molding Müeller-Hinton agar plates for AmpC detection. (C) Tridimensional test. Enhanced growth (6 or 7mm) of the surface organism E. coli ATCC 25922 is seen near agar slits (arrows) that contain extracts of E. coli (Fer110, Fer141, and Fer170) AmpC positive isolates; left: enhanced growth (3mm) of surface E. coli ATCC 25922 is seen near agar slits that contain extract of E. coli (Fer026) test isolate with indeterminate result; the extract of non-AmpC-producing E. coli isolate Fer136 inhibited the growth of surface E. coli ATCC 25922. On MacConkey agar, the growth of P. mirabilis isolate Fer094 inhibited the growth of surface E. coli ATCC 25922 (D) (arrow), but did not interfere with the growth of surface E. coli ATCC 35218 (not shown). The extract of E. coli isolate Fer136 inhibited the growth of surface E. coli ATCC 25922 (E) (arrow), but did not interfere with the growth of surface organism E. coli ATCC 35218 (F).
For the tridimensional test, a loopful of overnight growth of test organism was transferred to a sterile microcentrifuge tube. The technique was standardized so as to obtain 40–50mg of bacterial wet weight for each sample. Bacterial growth was suspended in 150μL of 0.1 M phosphate buffer (pH 7.0) and crude enzyme extracts were obtained by freeze–thawing five times. Four 5cm long linear slits were molded in the agar by a metal apparatus specially prepared for AmpC detection in Mueller-Hinton agar plates (Figure 1B), which were then inoculated with one of the two indicator strains. A 30μg cefoxitin disk was placed on the center of the inoculated agar. Each slit was loaded with the enzyme extract for a test strain in 10μL increments until the well was filled to the top. Approximately 60–80μL of the extract were loaded into each well avoiding overfilling. The plates were kept upright for 5–10 min until the solution dried, and were incubated at 35°C overnight. Any enhanced growth of the indicator strain decreasing the radius of the cefoxitin inhibition zone at the endpoint of the slit was measured, and three different types of results were recorded: a clear distortion of the cefoxitin inhibition zone (>5mm) was interpreted as a positive result and the isolate was considered AmpC producer; if no distortion was observed, the result was interpreted as negative and the isolate was considered as non-AmpC producer; the observation of a minimal distortion (> 0 and ≤ 3mm) was interpreted as a dubious result and AmpC production was considered indeterminate (Figure 1C).
All isolates were tested by a multiplex PCR for detection of family-specific plasmid-mediated ampC β-lactamase genes (Pérez-Pérez et al., 2002). Negative controls were PCR mixtures with the addition of water in place of template DNA. A positive control DNA preparation kindly provided by Dr Nancy Hanson was used to test each set of ampC-specific primers (Pérez-Pérez et al., 2002).
One cefoxitin resistant M. morganii isolate (Fer056) from the present study collection was selected as a positive control for the AmpC phenotypic tests. This strain gave clear positive Hodge and tridimensional test results. The ~405bp amplicon obtained from this isolate in the ampC multiplex PCR was purified by the QIAquick PCR Purification kit (Courtaboeuf, France) and sequenced in both directions at the UC Berkeley DNA Sequencing Facility, University of California, Berkeley, CA, USA. Nucleotide sequences were compared by BLAST, based at the National Center of Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov). A ~400bp nucleotide sequence 100% identical to the M. morganii chromosomal AmpC blaDHA-1 gene (accession # AJ237702) was obtained.
2.3. Strain typing
Genetic relatedness of isolates was evaluated by enterobacterial repetitive intergenic consensus-PCR with primer ERIC2 (ERIC2-PCR) (Versalovic et al., 1991). DNA was extracted by thermal lyses (Alves et al., 2006). PCR mixtures were prepared in a final volume of 25μL. Each reaction contained 75mM Tris-HCl (pH 9.0); 50mM KCl; 0.1mM each deoxynucleoside triphosphate; 3mM MgCl2; 0.3μM ERIC2 primer; and 1.5U of Taq DNA polymerase (BIOTOOLS, Madrid, Spain). Template DNA (3μL) was added to 22μL of the master mixture. A negative control without template DNA was included in each experiment. The reaction mixtures were subjected to amplification that consisted of an initial denaturation step at 94°C for 2 min, followed by 40 cycles of DNA denaturation at 94°C for 30s, primer annealing at 54°C for 1 min, primer extension at 72°C for 4 min, and a final extension step at 72°C for 1 min. Amplification products were electrophoresed in 1.5% agarose gels, stained with ethidium bromide, and photographed under UV light. Results were analyzed by visual inspection and with the software GelComparII, version 4.01 (Applied Maths, Kortrijk, Belgium) by the Dice index and the Unweighted Pair Group Method with Arithmetic Averages (UPGMA).
2.4. Statistical analysis
Univariate analysis was carried out by Chi-square test or by Fisher’s exact test for categorical variables. Student’s t-test was used to compare means. Tests were two-tailed, and a P value ≤ 0.05 was defined as statistically significant. Data were stored and analyzed with Epi Info version 3.2.
3. Results
3.1. Bacterial isolates and antimicrobial susceptibility
The distribution of enterobacterial species within the group of MDR isolates and non-MDR isolates was uneven. As shown in Table 1, non-MDR isolates included an increased number of E. coli (p < 0.001) and P. mirabilis (p = 0.04), while MDR isolates had more E. cloacae (p < 0.001) and S. marcescens (p = 0.04). However, patients with MDR-enterobacteria, compared to patients with non-MDR enterobacteria, had no significant differences with regard to demographic features: male patients were 62.5% and 44.6% (p = 0.09), respectively; and median age of patients was 59 (range 19–93) and 57 (range 14–86) (p = 0.6), respectively.
Table 1.
Distribution of enterobacterial species within the group of multidrug resistant (MDR) isolates and non-MDR isolates
| Bacterial species (number) | Number (%) of isolates |
|
|---|---|---|
| MDR | Non-MDR | |
| E. coli (42) | 12 (28.6) | 30 (71.4) |
| E. cloacae (18) | 18 (100) | 0 |
| K. pneumoniae (17) | 8 (47.1) | 9 (52.9) |
| E. aerogenes (13) | 9 (69.2) | 4 (30.8) |
| P. mirabilis (12) | 3 (25.0) | 9 (75.0) |
| S. marcescens (8) | 7 (87.5) | 1 (12.5) |
| M. morganii (7) | 5 (71.4) | 2 (28.6) |
| C. freundii (2) | 2 (100) | 0 |
| C. koseri (2) | 1 (50.0) | 1 (50.0) |
| P. agglomerans (2) | 0 | 2 (100) |
| Total (123) | 65 (52.8) | 58 (47.2) |
Urine was the most frequent source of Enterobacteriaceae isolation (52 isolates, 42.3%), followed by blood (37, 30.1%), lower respiratory tract (9, 7.3%), surgical site (8, 6.5%), normally sterile secretions and tissues (12, 9.8%), catheter tip (4, 3.3%), and other sources (1, 0.8%).
All isolates were susceptible to imipenem, except for a M. morganii isolate, which was susceptible only to piperacilin-tazobactam and intermediately susceptible to trimethoprim-sulfamethoxazole. Eleven (8.9%) isolates revealed susceptibility exclusively to imipenem. Resistance to extended spectrum cephalosporins or aztreonam (MDR isolates), including all ESBL producers, was observed for 65 (52.8%) enterobacterial isolates, obtained from the 56 patients. Six ESBL-producing isolates did not show resistance to extended spectrum cephalosporins or aztreonam by the disk diffusion method. Resistance to cefoxitin was observed for 62 (50.4%) isolates. Other resistance rates were: 78.9% (cephalotin), 41.5% (gentamicin), 52.0% (trimethoprim-sulfamethoxazole), 40.7% (ciprofloxacin), 35.0% (amikacin), 27.6% (piperacilin-tazobactam), and 20.3% (cefepime). Co-resistance to at least six antimicrobial agents was observed in 46 (37.4%) of the 123 isolates.
3.2. Tests for detection of Mβla and ESBL
Mbla production was not observed for any isolate. Of the 123 Enterobacteriaceae isolates, 74 (60.2%) had a positive screening test for ESBL and 35 (28.5%) were confirmed as ESBL producers (Table 2). However, confirmation of ESBL production was significantly more frequent among the group of E. coli, K. pneumoniae and P. mirabilis (21/30, 70%) than among the other enterobacterial species (14/44, 32%, p = 0.003). Therefore, a high rate of false-positive results (68%) was obtained for species with an easily inducible chromosomal ampC gene. A total of 20 (16.3%) isolates were positive by both the disk-addition and the double-diffusion tests, while 13 (10.6%) were positive only by the disk-addition, and two (1.6%), only by the double-diffusion method. The disk-addition and the double-diffusion tests were negative for all isolates not selected by the screening test.
Table 2.
ESBL production by Enterobacteriaceae isolates included in the study
| Bacterial species (number) | Number (%) of positive isolates |
||||
|---|---|---|---|---|---|
| ESBL screening (CLSI) | ESBL confirmatory test |
||||
| Disk-addition (CLSI) | Double-diffusiona | Final positive resultb | Final negative resultc | ||
| E. coli (42) | 17 (40.5) | 9 (21.4) | 6 (14.3) | 10 (23.8) | 32 (76.2) |
| K. pneumoniae (17) | 10 (58.8) | 8 (47.1) | 5 (29.4) | 8 (47.1) | 9 (52.9) |
| P. mirabilis (12) | 3 (25.0) | 3 (25.0) | 3 (25.0) | 3 (25.0) | 9 (75.0) |
| Other species (52) | 44 (84.6) | 13 (25.0) | 8 (15.4) | 14 (26.9)d | 38 (73.1) |
| Total (123) | 74 (60.2) | 33 (26.8) | 22 (17.9) | 35 (28.5) | 88 (71.5) |
At least one confirmatory test positive.
Negative result in both tests.
Two E. aerogenes, two E. cloacae, one M. morganii and one C. freundii isolate were positive only by the disk-addition test. One E. cloacae isolate was positive only by double-diffusion.
3.3. Tests for detection of AmpC
All E. coli (42), K. pneumoniae (17) and P. mirabilis (12) isolates were selected for AmpC testing. One M. morganii isolate (Fer056) was selected as a positive control: the amplicon obtained from this isolate in the ampC multiplex PCR was sequenced and confirmed as blaDHA-1, a M. morganii chromosomal origin AmpC gene fragment (accession # AJ237702). The strain was resistant to cefoxitin and gave a clear positive Hodge and tridimensional test result. No plasmid-mediated ampC genes were detected in any isolate by multiplex PCR. Amplicon products of the respective expected sizes for chromosomal ampC genes were observed when DNA from C. freundii, E. cloacae and M. morganii were used as templates. A clear positive tridimensional test (distortion ≥ 6mm) and Hodge test (distortion = 2) was observed for five (12%) E. coli isolates (Table 3). All these five isolates were resistant to cefoxitin. The other 66 isolates gave indeterminate or negative results.
Table 3.
AmpC test results for 71 Enterobacteriaceae isolates with no chromosomal ampC gene or with constitutive and minimally expressed chromosomal gene
| Distortion (in mm) |
Organism (number of isolates) | Number of isolates within cefoxitin susceptibility category |
Final AmpC result | |||
|---|---|---|---|---|---|---|
| THm | T3Dma | Resistant | Intermediate | Susceptible | ||
| E. coli (37) | 4 | 1 | 32 | 0 | ||
| 0–1 | 0–3 | K. pneumoniae (17) | 3 | 0 | 14 | 0 |
| P. mirabilis (12) | 0 | 1 | 11 | 0 | ||
| 2 | 6–7 | E. coli (5) | 5 | 0 | 0 | 5b |
THm: modified Hodge test; T3Dm: modified tridimentional test
No isolates showed 4–5 mm distortions
Chromosomal AmpC-hyper-producing isolates
For most isolates, the growth patterns of indicator organisms were similar and relatively easy to interpret. The extract of one E. coli isolate and one P. mirabilis isolate, however, inhibited the growth of one of the indicator organisms uniformly along the entire length of the slit (Figure 1E) or growth (Figure 1D), respectively, and interfered with the interpretation of the test result. In contrast, no inhibition was observed along the slit with the second surface organism (Figure 1F). MacConkey agar markedly suppressed growth from unlysed Proteus cells.
3.4. Strain typing
The clonal composition of isolates was evaluated by ERIC2-PCR, including all MDR (65) and a sample (40, 69%) of the susceptible isolates. A variety of ERIC2-strain types were observed: five cluster groups of E. coli isolates (2–5 isolates each, shown in Figure 2); three of P. mirabilis isolates (2–3 and four isolates each); two of S. marcescens isolates (2 isolates each); and one of M. morganii (5 isolates), E. aerogenes (3 isolates), E. cloacae (2 isolates), and K. pneumoniae isolates (2 isolates). Six non-MDR isolates were included in four E. coli clonal groups and six in two P. mirabilis clonal groups. Therefore, the numbers of isolates within a clonal group among MDR isolates (24, 37%), compared to non-MDR isolates (12, 29%), were not significantly different (p 0.5).
Figure 2.
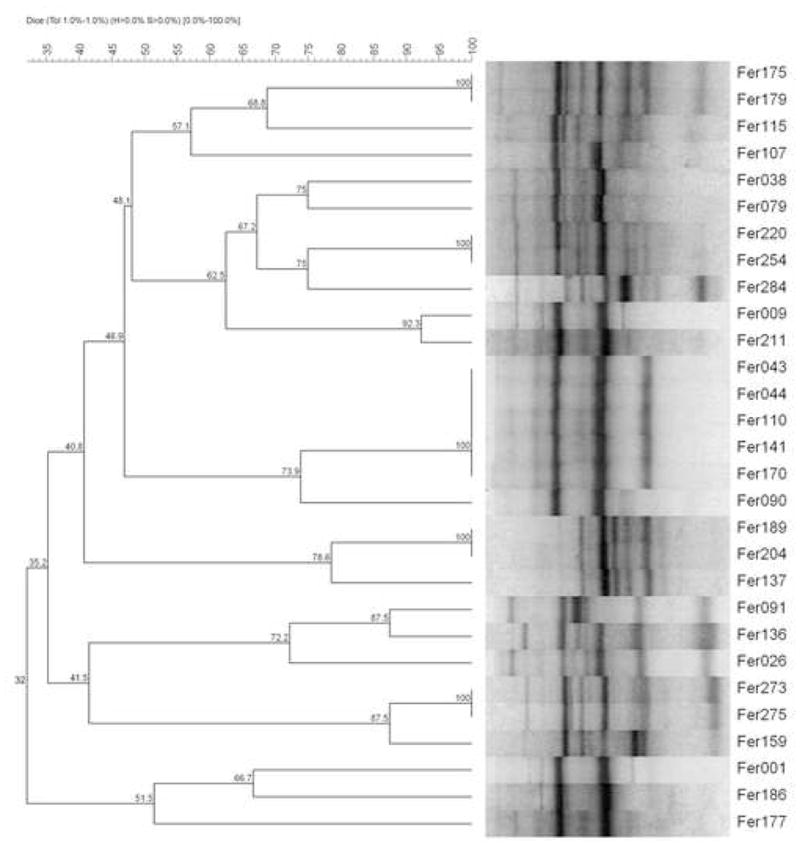
Dendrogram generated from computer-assisted analysis of the ERIC2-PCR fingerprints of 29 E. coli isolates: all 12 MDR and 17 (57%) of the non-MDR isolates. Five clonal E. coli isolates (Fer043, Fer044, Fer110, Fer141 and Fer170) showing similar electrophoretic patterns exhibited high levels of chromosomal AmpC and ESBL production.
4. Discussion
The prevalence of ESBL-producing enterobacterial isolates evaluated in the present study (29%) is much higher than that described in several other parts of the world. In the USA, the prevalence of ESBL-producing enterobacterial isolates is usually less than 9% for K. pneumoniae and less than 4% for E. coli (Jones and Pfaller, 1998; Dandekar et al., 2004). In Europe, the prevalence is around 15%–20% (Babini et al., 2000; Fluit et al., 2000; Luzzaro et al., 2006).
The screening test was able to detect all ESBL-producing isolates in the present collection. A comparison of results for screening with aztreonam, ceftazidime and cefotaxime, revealed that cefotaxime was the most sensitive drug to predict ESBL production. Before 1985, resistance to cefotaxime among enterobacterial clinical isolates involved species possessing inducible AmpC β-lactamase (Jarlier et al., 1988). However, starting in 1985–1987, E. coli and K. pneumoniae isolates showing reduced susceptibility to cefotaxime due to transferable ESBL production became increasingly reported. In the present study, 94% (33 of 35) of ESBL producing isolates showed resistance to cefotaxime. The resistance rates to other β-lactams (ceftazidime and aztreonam) were 77% and 74%, respectively. The double-diffusion test allowed for the detection of 22 (63%) of the 35 ESBL producing isolates, indicating its low sensitivity, while the disk-addition test was able to detect 33 (94%) of the ESBL isolates. The disk-addition test was the most sensitive confirmatory test in the present study.
Concerning the ESBL screening test, a high rate of false-positive results was obtained for non-E. coli-K. pneumoniae-P. mirabilis enterobacterial isolates. This observation is in line with the CLSI recommendation for screening E. coli, K. pneumoniae, K. oxytoca and P. mirabilis only. In fact, Schwaber and colleagues (2004) confirmed ESBL production in only 2.2% of 355 isolates that tested positive in the screening test in species other than those recommended by CLSI. However, in the present study, ESLB tests led to the identification of 14 (27%) ESBL-producing isolates in a collection of 52 non-E. coli-K. pneumoniae-P. mirabilis enterobacterial isolates. Most importantly, half of these 14 isolates tested susceptible to cefepime in the antibiogram. We conclude that in a setting with high rates of ESBL production, this CLSI recommendation (to test only E. coli, K. pneumoniae, K. oxytoca and P. mirabilis for ESBL production) is not appropriate and could led to high rates of major susceptibility testing errors.
The critical issue in detecting ESBL production is the potential for simultaneous presence of AmpC β-lactamases (Thomson, 2001), and risk factors for both mechanisms of resistance are similar. Many clinical microbiologists are unaware of plasmid-encoded AmpC because phenotypic detection is difficult, and these β-lactamases can be misidentified as ESBLs (Hanson, 2003). Currently, no guidelines for detection of plasmid-mediated AmpC-producing organisms or organisms harboring multiple β-lactamases are available.
To date, several modifications of the tridimensional test to detect AmpC β-lactamases were tried, but no satisfactory technique has been established. For the tridimensional test, a slit beginning 5 mm from the edge of a cefoxitin disk must be cut in the agar with a sterile scalpel blade in an outward radial direction; cutting this slit is a fastidious and time consuming procedure. In addition, filling the slits homogeneously without overflow onto agar surface is problematic. The Hodge test was proposed as an alternative technique to detect AmpC (Yong et al., 2002). However, in the present study, distinction of positive and negative results depended on cefoxitin inhibition zone distortions of just 1–2 mm. A third method to detect AmpC β-lactamases was proposed, based on disk diffusion using β-lactamase inhibitors (Black et al., 2004), but a high rate of false-negative results precludes its use. Probably, the various inhibitory compounds exhibit different levels of inhibition over heterogeneous groups of AmpC β-lactamases that may be found in clinical isolates (Philippon et al., 2002). An additional method using class-C enzyme inhibitor boronic acid (Crompton et al., 1988) has been proposed. However, boronic acid still fails to detect all AmpC producers (Coudron, 2005; Yagi et al., 2005), probably because the pattern of resistance may be altered by porin loss (Martínez-Martínez et al., 1999; Pangon et al., 1989; Thomson, 2001).
In the present study, we obtained clear positive results for 5 E. coli isolates with the tridimensional test. Positive results gave distortions of 6–7 mm, while negative results gave distortions of 0–3 mm. To facilitate the procedure, we made a metal, autoclavable apparatus to mold the slits into the agar, so they were homogeneous; the technique was much easier to perform. In the present collection of enterobacteria, plasmid AmpC multiplex PCR analysis was negative for all isolates. We found this to be a convenient method to detect plasmid-mediated ampC genes, although suitable to analyze in only certain enterobacterial species. Although reported with increasing frequency, the true rate of occurrence of plasmid AmpC β-lactamases in members of Enterobacteriaceae remains unknown, because only few surveillance studies have examined this resistance mechanism (Coudron et al., 2000; Pérez-Pérez et al., 2002). In Delhi, India, Manchanda and Singh (2003) observed high rates of AmpC production among 9 K. pneumoniae (33.3%), 7 E. coli (14.3%) and 1 P. mirabilis (33.3%) isolates. Gazouli and colleagues (1998), analyzing 2,133 E. coli isolates obtained from 10 hospitals in Greece, observed 55 (2.6%) AmpC-producing isolates. Chromosomal AmpC in P. mirabilis was observed only once in 1998 (Bret et al., 1998); plasmid-mediated AmpC β-lactamase is rarely observed in this species (Bobrowski et al., 1976; Verdet et al., 1998). In the present study, the tridimensional and the Hodge tests revealed that five (41.7%) of 12 E. coli isolates resistant to cefoxitin were AmpC producers. However, these isolates were multiplex PCR-negative, which indicates they were most likely AmpC hyperproducers due to overexpression of the chromosomal ampC gene. Noteworthy was that all these isolates were also ESBL producers, and showed indistinguishable ERIC2-PCR patterns. Four of five patients with AmpC-hyper-producing E. coli acquired these isolates in a surgical unit, which indicates cross-transmission of strains occurred in this ward. We also found nine isolates (five E. coli, three K. pneumoniae, and one P. mirabilis) resistant to cefoxitin with negative results by the tridimensional, the Hodge and multiplex-PCR tests. Therefore, these isolates were considered as non-AmpC-producers. Cefoxitin resistance in AmpC negative isolates could be due to a decreased permeation of porins (Pangon et al., 1989; Thomson, 2001).
Strain typing analysis revealed that most of the patients in our study had sporadic infections; only 36 (32.1%) patients had infection or colonization by isolates belonging to clonal groups. Thus, a large proportion of hospital infections caused by ESBL-producing enterobacteria identified during the study period were due to sporadic infections rather than undetected outbreaks. This observation thus emphasizes the need to improve our detection methods for ESBL- and AmpC-producing organisms in hospitals where extended-spectrum cephalosporins are in wide use.
Acknowledgments
This study was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ) of Brazil and Fogarty International Program in Global Infectious Diseases (TW006563) of the National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves MS, Dias RCS, Castro ACD, Riley LW, Moreira BM. Identification of Clinical Isolates of Indole-Positive and Indole-Negative Klebsiella spp. J Clin Microbiol. 2006;44:3640–3646. doi: 10.1128/JCM.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa Y, Shibata N, Shibayama K, Kurokawa H, Yagi T, Fujiwara H, Masafumi G. Convenient test for screening metallo-beta-lactamase-producing Gram-negative bacteria by using thiol compounds. J Clin Microbiol. 2000;38:40–43. doi: 10.1128/jcm.38.1.40-43.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babini GS, Livermore DM. Antimicrobial resistance amongst Klebsiella spp. Collected from intensive care unit in Southern and Western Europe in 1997–1998. J Antimicrob Chemother. 2000;42:183–189. doi: 10.1093/jac/45.2.183. [DOI] [PubMed] [Google Scholar]
- Bell JM, Turnidge JD, Gales AC, Pfaller MA, Jones RN. Prevalence of extended spectrum beta-lactamase (ESBL)-producing clinical isolates in the Asia-Pacific region and South Africa: Regional results from SENTRY Antimicrobial Surveillance Program (1998–99) Diagn Microbiol Infect Dis. 2002;42:193–198. doi: 10.1016/s0732-8893(01)00353-4. [DOI] [PubMed] [Google Scholar]
- Ben Redjeb S, Ben Yaghlane H, Boujnah A, Philippon A, Labia R. Synergy between clavulanic acid and newer beta-lactams on nine clinical isolates of Klebsiella pneumoniae, Escherichia coli and Salmonella typhimurium resistant to third generation cephalosporins. J Antimicrob Chemother. 1988;21:263–266. doi: 10.1093/jac/21.2.263. [DOI] [PubMed] [Google Scholar]
- Black JJ, Thomson KS, Pitout JDD. Use of beta-lactamase inhibitors in disk tests to detect plasmid-mediated AmpC beta-lactamases. J Clin Microbiol. 2004;42:2203–2206. doi: 10.1128/JCM.42.5.2203-2206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski MM, Mthew M, Barth PT, Datta N, Grinter NJ, Jacob AE, Kontomichalou P, Dale JW, Smith JT. Plasmid-determined beta-lactamase indistinguishable from the chromosomal beta-lactamase of Escherichia coli. J Bacteriol. 1976;123:149–157. doi: 10.1128/jb.125.1.149-157.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bret L, Chanal-Claris C, Sirot D, Chaibi EB, Labia R, Sirot J. Chromosomally encoded AmpC-type beta-lactamase in a clinical isolate of Proteus mirabilis. Antimicrob Agents Chemother. 1998;42:1110–1114. doi: 10.1128/aac.42.5.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI)/NCCLS. CLSI document M100-S15. Clinical and Laboratory Standards Institute; 940 West Valley Road, Suite 1400, Wayne, Pennsylvania 19087–1898 USA: 2005. Performance standards for antimicrobial susceptibility testing; Fifteenth informational supplement. [Google Scholar]
- Cohen ML. Epidemiology of drug resistance: implications for a post-antimicrobial era. Science. 1992;257:1050–1055. doi: 10.1126/science.257.5073.1050. [DOI] [PubMed] [Google Scholar]
- Coudron PE. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J Clin Microbiol. 2005;43:4163–4167. doi: 10.1128/JCM.43.8.4163-4167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudron PE, Moland ES, Thomson KS. Occurrence and detection of AmpC beta-lactamases among Escherichia coli, Klebsiella pneumoniae, and Proteus mirabilis isolates at a Veterans Medical Center. J Clin Microbiol. 2000;38:1791–1796. doi: 10.1128/jcm.38.5.1791-1796.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crompton IE, Cuthbert BK, Lowe G, Waley SG. Beta-lactamase inhibitors. The inhibition of serine beta-lactamases by specific boronic acids. Biochem J. 1988;251:453–459. doi: 10.1042/bj2510453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dandekar PK, Tetreault J, Quinn JP, Nightingale CH, Nicolau DP. Prevalence of extended spectrum beta-lactamase producing Escherichia coli and Klebsiella isolates in a large community teaching hospital in Connecticut. Diagn Microbiol Infect Dis. 2004;49:37–39. doi: 10.1016/j.diagmicrobio.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Davin-Regli A, Bosi C, Charrel RN, Ageron E, Papazian L, Grimont PAD, Cremieux A, Bollet C. A nosocomial outbreak due to Enterobacter cloacae strains with the E. hormaechei genotype in patients treated with fluoroquinolones. J Clin Microbiol. 1997;35:1008–1010. doi: 10.1128/jcm.35.4.1008-1010.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer JJ., III . Enterobacteriacea: Introduction and Identification. In: Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. Manual of Clinical Microbiology. 7. Vol. 41. ASM Press; Washington, D.C: 2003. pp. 636–653. [Google Scholar]
- Fluit AC, Jones ME, Schmitz FJ, Acar J, Gupta R, Verhoef J. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY antimicrobial surveillance program, 1997 and 1998. Clin Infect Dis. 2000;30:454–460. doi: 10.1086/313710. [DOI] [PubMed] [Google Scholar]
- Gazouli M, Tzouvelekis LS, Vatopoulos AC, Tzelepi E. Transferable class C beta-lactamases in Escherichia coli strains isolated in Greek hospitals and characterization of two enzyme variants (LAT-3 and LAT-4) closely related to Citrobacter freundii AmpC beta-lactamase. J Antimicrob Chemother. 1998;42:419–425. doi: 10.1093/jac/42.4.419. [DOI] [PubMed] [Google Scholar]
- Hanson ND. AmpC beta-lactamases: what do we need to know for the future? J Antimicrob Chemother. 2003;52:2–4. doi: 10.1093/jac/dkg284. [DOI] [PubMed] [Google Scholar]
- Hobson RP, MacKenzie FM, Gould IM. An outbreak of multiply-resistant Klebsiella pneumoniae in the Grampian region of Scotland. J Hosp Infect. 1996;33:249–262. doi: 10.1016/s0195-6701(96)90011-0. [DOI] [PubMed] [Google Scholar]
- Holmberg S, Solomon S, Blake P. Health and economic impact of antimicrobial resistance. Rev Infect Dis. 1987;9:1065–1078. doi: 10.1093/clinids/9.6.1065. [DOI] [PubMed] [Google Scholar]
- Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10:867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- Jones RN, Pfaller MA. Bacterial resistance: A worldwide problem. Diagn Microbiol Infect Dis. 1998;31:379–388. doi: 10.1016/s0732-8893(98)00037-6. [DOI] [PubMed] [Google Scholar]
- Luzzaro F, Mezzatesta M, Mugnaioli C, Perilli M, Stefani S, Amicosante G, Rossolini GM, Toniolo A. Trends in Production of Extended-Spectrum β-Lactamases among Enterobacteria of Medical Interest: Report of the Second Italian Nationwide Survey. J Clin Microbiol. 2006;44:1659–1664. doi: 10.1128/JCM.44.5.1659-1664.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchanda V, Singh NP. Occurrence and detection of AmpC beta-lactamases Among Gram-negative clinical isolates using a modified three-dimensional test at Guru Tegh Bahadur Hospital, Delhi, India. J Antimicrob Chemother. 2003;51:415–418. doi: 10.1093/jac/dkg098. [DOI] [PubMed] [Google Scholar]
- Martínez-Martínez L, Pascual A, Hernández-Allés S, Alvarez-Díaz D, Suárez AI, Tran J, Benedí VJ, Jacoby GA. Roles of beta-lactamases and porins in activities of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob Agents Chemother. 1999;43:1669–1673. doi: 10.1128/aac.43.7.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KS, Urban C, Eagan JA, Berger BJ, Rahal JJ. Nosocomial outbreak of Klebsiella infection resistant to late-generation cephalosporins. Annals of Internal Medicine. 1993;119:353–358. doi: 10.7326/0003-4819-119-5-199309010-00001. [DOI] [PubMed] [Google Scholar]
- NCCLS. NCCLS document M2-A7. NCCLS; 940 West Valley Road, Suíte 1400, Wayne, Pennsylvania 19087–1898, USA: 2000. Performance Standards for Antimicrobial Disk Susceptibility Tests; Approved Standard Seventh Edition. [Google Scholar]
- Pangon B, Bizet C, Bure A, Pichon F, Philippon A, Ragnier B, Gutmann L. In vivo selection of a cephamycin-resistant, porin-deficient mutant of K. pneumoniae producing a TEM-3 beta-lactamase. J Infect Dis. 1989;159:1005–1006. doi: 10.1093/infdis/159.5.1005. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez FJ, Hanson ND. Detection of plasmid-mediated AmpC beta-lactamase gene in clinical isolates by using multiplex PCR. J Clin Microbiol. 2002;40:2153–2162. doi: 10.1128/JCM.40.6.2153-2162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A, Arlet G, Jacoby GA. Plasmid-determined AmpC-type beta-lactamases. Antimicrob Agents Chemoth. 2002;46:1–11. doi: 10.1128/AAC.46.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippon A, Ben Redjeb S, Fournier G, Ben Hassen A. Epidemiology of extended spectrum beta-lactamase. Infection. 1989;17:347–354. doi: 10.1007/BF01650727. [DOI] [PubMed] [Google Scholar]
- Ramchandani M, Manges AR, DebRoy C, Smith SP, Johnson JR, Riley LW. Possible animal origin of human-associated, multidrug-resistant, uropathogenic Escherichia coli. Clin Infect Dis. 2005;40:251–257. doi: 10.1086/426819. [DOI] [PubMed] [Google Scholar]
- Rice LB, Willey SH, Papanicolaou GA, Medeiros AA, Eliopoulos GM, Moellering RC, Jacoby GA. Outbreak of ceftazidime resistance caused by extended-spectrum beta-lactamases at a Massachusetts chronic-care facility. Antimicrob Agents Chemoth. 1990;34:2193–2199. doi: 10.1128/aac.34.11.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaber MJ, Raney PM, Rasheed JK, Biddle JW, Williams P, McGowan JE, Tenover FC. Utility of NCCLS guidelines for identifying extended-spectrum beta-lactamases in non-Escherichia coli and non-Klebsiella spp. Of Enterobacteriaceae. J Clin Microbiol. 2004;42:294–298. doi: 10.1128/JCM.42.1.294-298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson KS. Controversies about extended-spectrum and AmpC beta-lactamases. Emerg Infect Dis. 2001;7:333–336. doi: 10.3201/eid0702.010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdet C, Arlet G, Ben Redjeb S, Ben Hassen A, Lagrange PH, Philippon A. Characterization of CMY-4, an AmpC-type plasmid-mediated beta-lactamase, in a Tunisian clinical isolate os Proteus mirabilis. FEMS Microbiol Lett. 1998;169:235–240. doi: 10.1111/j.1574-6968.1998.tb13323.x. [DOI] [PubMed] [Google Scholar]
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in Eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn WC Jr, Allen SD, Janda WM, Koneman EW, Procop GW, Schreckenberg PC, Woods GL, editors. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 6. Vol. 6. Lippincott Williams & Wilkins; Philadelphia, EUA: 2006. The Enterobacteriacea; pp. 213–308. [Google Scholar]
- Winokur PL, Canton R, Casellas JM, Legakis N. Variations in the prevalence of strains expressing an extended-spectrum beta-lactamase phenotype and characterization of isolates from Europe, the Americas and the Western Pacific region. Clin Infect Dis. 2001;32:S94–S103. doi: 10.1086/320182. [DOI] [PubMed] [Google Scholar]
- Yagi T, Wachino J-i, Kurokawa H, Suzuki S, Yamane K, Doi Y, Shibata N, Kato H, Shibayama K, Yoshichika Arakawa. Practical methods using boronic acid compounds for identification of class C beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli. J Clin Microbiol. 2005;43:2151–2558. doi: 10.1128/JCM.43.6.2551-2558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong D, Park R, Yum JH, Lee K, Choi EC, Chong Y. Further modification of the Hodge test to screen AmpC beta-lactamase (CMY-1)-producing strains of Escherichia coli and Klebsiella pneumoniae. J Microbiol Methods. 2002;51:407–410. doi: 10.1016/s0167-7012(02)00053-2. [DOI] [PubMed] [Google Scholar]


