Summary
Exposure to IR has been shown to induce the formation of senescence markers, a phenotype that coincides with life-long delayed repair and regeneration of irradiated tissues. We hypothesised that IR-induced senescence markers could persist long-term in vivo, possibly contributing to the permanent reduction in tissue functionality. Here we show that mouse tissues exposed to a sublethal dose of IR display persistent (up to 45 weeks, the maximum time analysed) DNA damage foci and increased p16INK4a expression, two hallmarks of cellular senescence and aging. BrdU labelling experiments revealed that IR-induced damaged cells are preferentially eliminated, at least partially, in a tissue dependent manner. Unexpectedly, the accumulation of damaged cells was found to occur independent from the DNA damage response modulator p53, and from an intact immune system, as their levels were similar in wild-type and Rag2−/−γC−/− mice, the latter being deficient in T, B and NK cells. Together, our results provide compelling evidence that exposure to IR induces long-term expression of senescence markers in vivo, an effect that may contribute to the reduced tissue functionality observed in cancer survivors.
Keywords: cancer, cellular senescence, DNA damage, immune system, ionizing radiation, p16, p53
Introduction
Ionizing radiation (IR) and other anticancer drugs generate a high level of DNA damage which often leads to the induction of apoptosis and cellular senescence in vitro and in vivo (Christophorou et al. 2006; Probin et al. 2006; Wang et al. 2006). Induction of apoptosis in rapidly regenerating tissues is believed to be responsible for short-term side effects such as myelosuppression. Intriguingly, up to 70% of cancer survivors also experience, many years after treatment, long-term complications that manifest as a wide array of diseases (Oeffinger et al. 2006; Geenen et al. 2007). We believe the accumulation of dysfunctional senescent cells in tissues combined with a reduction in the proliferative potential of progenitor/stem cells (as a result of their apoptosis/senescence) may account for the development of long-term complications. To support such a hypothesis, one must first determine whether IR-induced senescence markers can persist long-term in tissues. This question is of great interest in light of the recent observation that oncogene-induced senescent cells can be eliminated by immune cells (Xue et al. 2007).
An increase in cells expressing p16INK4a and containing DNA damage foci are arguably the most reliable molecular characteristics of senescent/aged tissues. p16INK4a is a cell cycle inhibitor and tumor suppressor that is in part responsible for cellular senescence, the process that permanently arrests the proliferation of cells at risk for neoplastic transformation (Campisi & d’Adda di Fagagna 2007). Marked age-dependent increases in p16INK4a expression have been described in mouse (Zindy et al. 1997; Krishnamurthy et al. 2004) and human (Nielsen et al. 1999; Melk et al. 2004; Ressler et al. 2006; Liu et al. 2009) tissues. Recent studies show that p16INK4a is not only a marker, but also an effector of age-related decrements in tissue regeneration and repair. For example, compared to wild-type mice, genetically modified mice that lack p16INK4a expression have better regenerative and repair potentials during age, an effect associated with a reduced decline in progenitor cells in renewable tissues such as the bone marrow, brain and pancreas (Janzen et al. 2006; Krishnamurthy et al. 2006; Molofsky et al. 2006). Similarly, p16INK4a deficiency delays the onset of age-related phenotypes in a mouse model of accelerated aging, providing a direct link for the involvement of this protein in organismal aging (Baker et al. 2008). Single nucleotide polymorphisms near the p16INK4a locus have also been linked with several human age-associated pathologies (Sharpless & DePinho 2007). Similar to p16INK4a expression, cells containing DNA damage foci have been shown to increase with age in tissues from mice, baboons and humans (Sedelnikova et al. 2004; Herbig et al. 2006; Jeyapalan et al. 2007; Sedelnikova et al. 2008; Wang et al. 2009). These damaged cells may contribute to the age-related decline in tissue function because deficiencies in DNA repair can accelerate aging phenotypes (Hasty et al. 2003), the DNA damage that accumulates with age in hematopoietic stem cells greatly limits their function (Nijnik et al. 2007; Rossi et al. 2007).
In response to severe DNA lesions such as double strand breaks, DNA damage response (DDR) kinases phosphorylate the histone variant H2A.X (γ-H2A.X), which facilitates the focal assembly of checkpoint and DNA repair factors such as 53BP1 (Rogakou et al. 1998; Peterson & Cote 2004). DDR transducer kinases then activate the p53 tumor suppressor protein, which orchestrates either transient cell cycle inhibition, believed to allow time for DNA repair, or removal of the cell from the proliferative pool by triggering either cellular senescence or apoptosis (cell death). The role of p53 in the damage response and aging phenotypes is context-dependent (Rodier et al. 2007). For example, genetic manipulation of p19Arf, which ultimately stabilizes p53, shows that regulated p53 activity can attenuate cellular senescence and aging phenotypes in mouse models of normal and accelerated aging (Matheu et al. 2007; Baker et al. 2008). Likewise, enhanced regulated p53 activity can suppress the accumulation of cells with damaged telomeres-in telomerase-deficient mice (Garcia-Cao et al. 2006). However, in response to acute DNA damage, such as following exposure to IR, or in mouse models of constitutively active p53, p53 can foster the appearance of aging phenotypes by engaging robust senescence and/or apoptotic responses (Varela et al. 2005; Hinkal et al. 2009). Whether p53 inactivation would reduce (by limiting apoptosis and/or senescence) or increase (by limiting DNA repair) the appearance and accumulation of damaged cells and other markers of aging is of high interest (Rodier et al. 2007), especially because transient inactivation of p53 after DNA damage was shown to prevent apoptosis without significantly reducing cancer-free survival (Christophorou et al. 2006; Strom et al. 2006).
Here, using IR as a source of DNA damage and inducer of senescence markers in mice tissues, we asked whether such markers would persist long-term post IR exposure, and if so under which conditions this would most likely occur. We show that mice exposed to IR have persistent DNA damage and an increased expression of p16INK4a in lung, brain and liver tissues. Unexpectedly, we further show that these markers increased independent of p53 and the T, B and natural killer (NK) cells of the immune system.
Results
Exposure to IR induces persistent DNA damage foci formation in mouse and human tissues
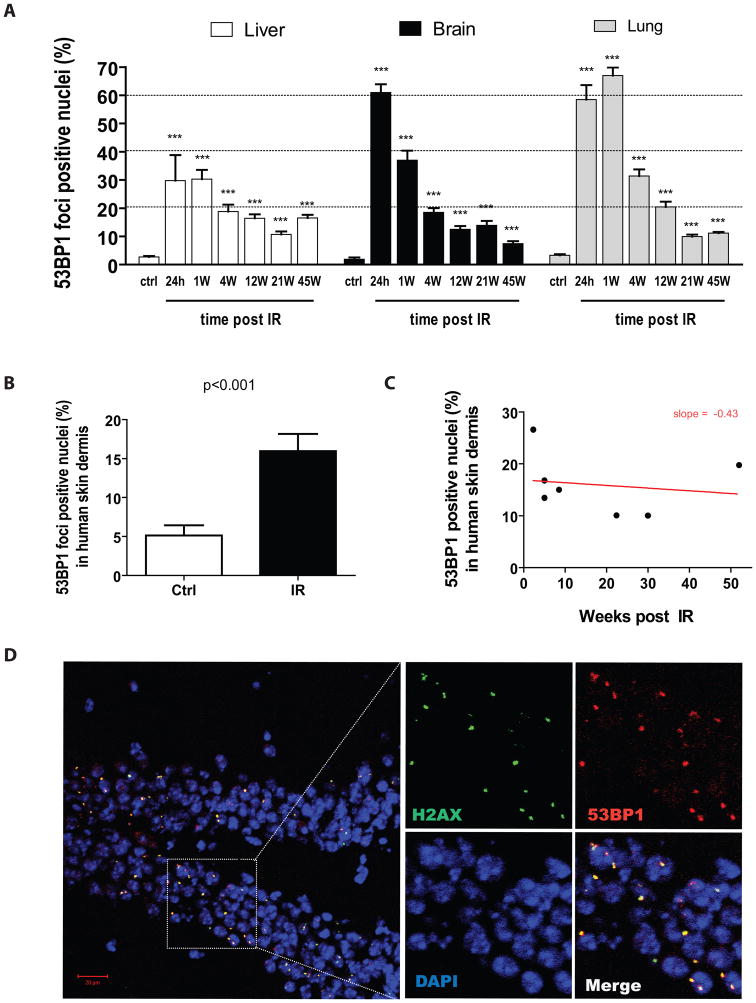
The extent to which exposure to IR can induce the long-term accumulation of putative senescence markers such as DNA damage foci formation is unknown. Therefore, we determined the fraction of cells with distinct 53BP1 and/or γ-H2AX DNA damage foci in the liver, brain and lung tissues of C57BL/6 mice sacrificed in a time dependent manner post their exposure to 8 Gy total body irradiation (TBI). We choose this dose as it represents the maximum tolerated dose at which bone marrow transplantation was not required for 100% of the mice to survive. All three tissues from control unirradiated animals (ctrl) had relatively few cells with 53BP1 foci. These results are in agreement with previous work showing similar or slightly higher background DNA damage levels – differences we believe can be attributed to the methodology used and/or housing conditions of mice (Sedelnikova et al. 2004; Wang et al. 2009). The fraction of cells with 53BP1 foci increased sharply within 24 h after animals were irradiated; thereafter the fraction of positive cells gradually declined, but remained significantly elevated over unirradiated controls for up to 45 weeks, the maximum time analyzed, after IR exposure (Figure 1A). During this time, unirradiated animals did not display any increase in their number of foci-containing cells, at least not before reaching 2 years of age (see figure S1).
Figure 1. Exposure to IR induces persistent DNA damage foci-positive cells in mouse and human tissues.
A) Fraction of cells containing 53BP1 DNA damage foci in liver, brain and lung tissues collected from control (ctrl) and from irradiated (8 Gy TBI) C57BL/6 mice sacrificed at the indicated time in weeks (W) post IR. Data are mean values ± s.e.m (n= 4–7). Statistical significance: *** P < 0.001 was obtained by performing a Student’s t-test relative to control. No increase in the fraction of DNA damage foci-positive cells was observed in control non-irradiated mice until they reached two years of age (see figure S1). B) Proportion of cells containing persistent 53BP1 DNA damage foci in the dermal region of skin collected from cancer patients who, as part of their treatments, received 12 Gy TBI (median of 8 weeks prior from being biopsied) and from age-matched control non-irradiated patients (n = 7–9 per group). p value was obtained by performing a Student’s t-test relative to control. C) Correlation of the amount of 53BP1 DNA damage foci-containing cells to the time to which the biopsies was obtained post exposure to IR. D) Detection by confocal microscopy of 53BP1 (red) and γ-H2AX (green) DNA damage foci from the hippocampus of a mouse sacrificed 12 weeks post its exposure to IR. Nuclei were stained with DAPI.
To determine the relevance of our findings in mice for humans, we obtained skin biopsies from age-matched patients (10±6 versus 13±7 years of age) that had been exposed or not to IR (12 Gy). The group exposed to IR was biopsied over a 2–52 weeks interval following irradiation (median 8 weeks). On average, this group had a significantly higher proportion (15 vs 5%, p<0.001) of cells containing DNA damage foci in the dermis (Figure 1B). Similar results were obtained in the epidermis (data not shown). The negative correlation between the proportion of DNA damaged cells and the interval after exposure to IR was not significant, perhaps because of the limited number of samples (n=7–9) or heterogeneity in DNA repair capacity among patients (Figure 1C).
As expected, we also observed almost complete co-localization between 53BP1 and γ-H2AX foci after IR exposure (Figure 1D), indicating that the 53BP1 foci were indeed DNA damage foci. A higher proportion of cells with persistent damage in the hippocampus compared to other areas of the brain was observed but there were no major differences in the distribution of damaged cells in the other tissues (data not shown). Together, these results are the first to demonstrate that exposure to IR had long lasting effects in that cells with persistent DNA damage foci remained evident for many weeks after the irradiation.
Exposure to IR induces increased p16INK4a expression in mouse tissues
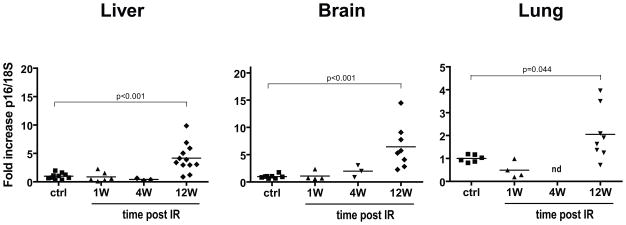
To better understand the relationship between the IR and time-dependent persistence of DNA damaged cells, we assayed tissues from control and irradiated mice for the expression of p16INK4a, another well-defined marker of senescence/aging. Using quantitative real-time PCR, we observed sharp increases in p16INK4a expression (5- to 7-fold on average) in the liver and brain of mice 12 weeks after IR exposure. We observed a more modest increase (~2-fold) in irradiated lung and skin tissues (Figures 2 and S2). Time course analysis revealed that p16INK4a expression increased gradually after irradiation, starting at around 4 weeks after exposure; the reason for this delay is currently unknown (Figure S3). In contrast to mouse tissues, deparaffinated human skin sections failed to show an increase in p16INK4a expression as determined by immunocytochemistry (data not shown). We believe this could reflect the fact that many of the human biopsies were collected relatively soon after IR exposure (median 8 weeks) and by the low sensitivity of the p16INK4a detection method used (histochemistry vs PCR). Nonetheless, these results provide evidence that IR exposure, at least in mice, leads to a tissue wide increase in p16INK4a expression.
Figure 2. Exposure to IR induces delayed p16INK4a expression in mouse tissues.
RNA was isolated from homogenized liver, brain and lung tissues collected from control (ctrl) and from irradiated (8 Gy TBI) C57BL/6 mice sacrificed at the indicated time in weeks (W) post IR. RNA was then used to determined p16INK4a expression by quantitative real-time PCR (n = 3–12, each symbol representing an individual mouse). p values were obtained by performing a Student’s t-test relative to control. nd = not determined.
Active clearance of IR-induced DNA damaged cells is tissue dependent
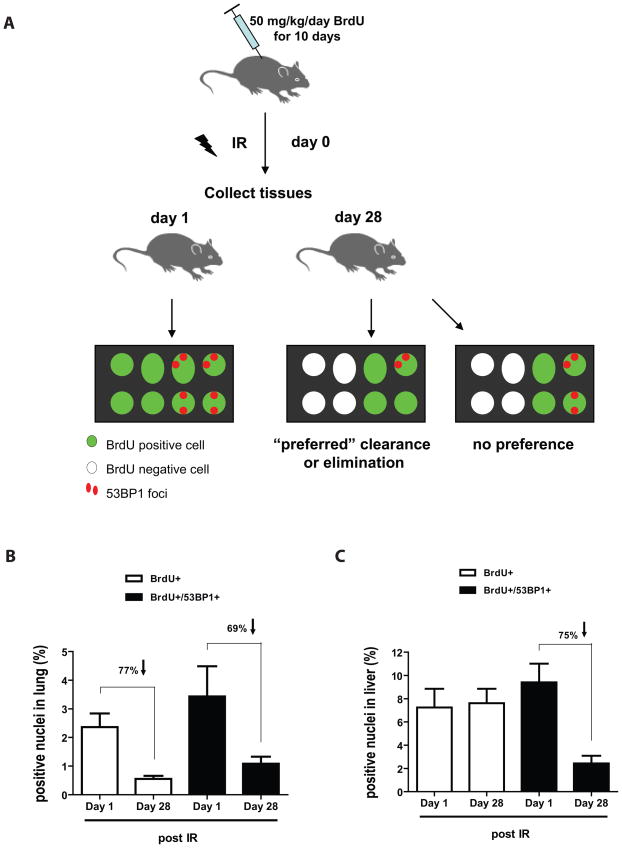
Despite that damaged cells undoubtedly accumulate in tissues long-term post irradiation, our result showed that a significant proportion of these cells are cleared/replaced over time. This prompted us to ask whether a fraction of damaged cells were preferentially eliminated in vivo. To answer this question, we injected mice with BrdU for 10 consecutive days and then immediately subjected these mice to 8 Gy TBI before collecting tissues on 1 and 28 days after irradiation. We then determined whether the proportion of double positive cells (BrdU+ and 53BP1+) declined more rapidly than the proportion of undamaged cells (BrdU+ alone). If damaged cells are preferentially eliminated, one would expect the proportion of BrdU+53BP1+ cells to decline more rapidly than the proportion of BrdU+ cells during the 28 days period post irradiation. Alternatively, if damaged cells are not preferentially eliminated, the decrease in BrdU+53BP1+ cells should be similar to the decline observed in the BrdU+ cell population (or even slower if damaged cells are not dividing, preventing dilution of the BrdU signal, see Figure 3A). Unexpectedly, we found that damaged cells in lung were not preferentially eliminated. That is, the proportion of BrdU+53BP1+ and BrdU+ cells cleared was similar (77% vs 69%) over the same 28 days period (Figure 3B). In contrast, damaged cells in liver were preferentially cleared with the proportion of BrdU+53BP1+ declining by 75% over the 28 days period in sharp contrast to BrdU+ cells, which surprisingly persisted (Figure 3C). These results suggest that the rate at which IR-induced damaged cells are cleared, at least partially, is tissue dependent.
Figure 3. Clearance of IR-induced DNA damaged cells is tissue dependent.
A) Schematic representation of the protocol used and the expected outcomes of BrdU+ cells containing or not 53BP1 DNA damage foci. B and C) Proportion of cells labelled with BrdU (alone) or BrdU and 53BP1 foci at 1 or 28 days post exposure to 8 Gy TBI. Indicated is the proportion of cells that have lost BrdU signal during the 28 days period post exposure to IR. n = 4 in each group.
IR-induced p16INK4a expression and accumulation of DNA damage foci-containing cells is independent of T, B and NK cells
The accumulation of DNA damaged cells and other senescence markers with age coincide with reduced immune function (Sansoni et al. 1993; Min et al. 2006). Immune cells, particularly natural killer (NK), have a role in eliminating damaged cells. Indeed, it was shown that DNA damage stimulates the expression of NKG2D ligands, the cell surface proteins by which NK and a subset of T cells recognize and target cells for killing; thus, supporting the notion that damaged cells might be eliminated by these immune cells (Gasser et al. 2005; Soriani et al. 2009). Based on these observations, we asked whether the partial clearance and long-term accumulation of senescence markers following exposure to IR could be related to the fitness of the immune system.
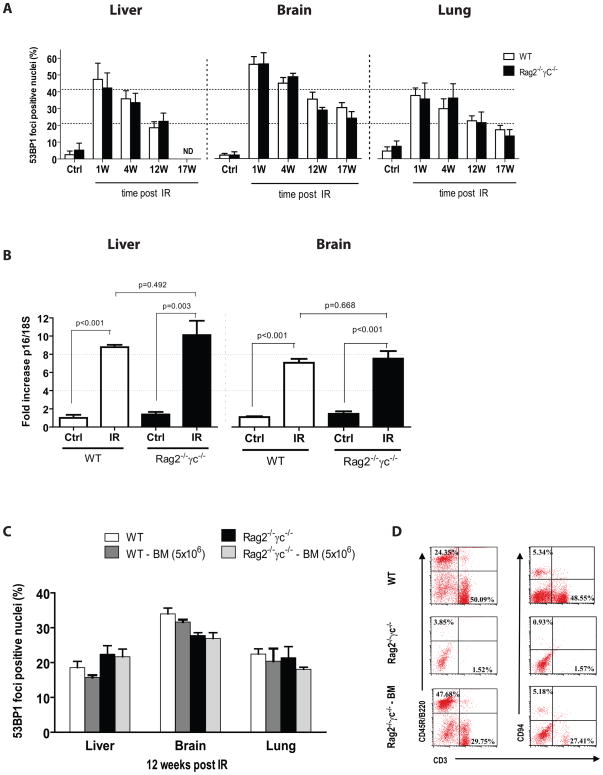
To answer this question, we used Rag2−/−γc−/− mice, which are completely deficient in T, B and NK cell functions (Goldman et al. 1998). In contrast to immune deficient mice on a NOD/SCID background, BALB/c Rag2−/−γc−/− mice have no known defect in DNA repair and their tolerance to irradiation is similar to that of wild-type mice (Chang et al. 1993). We subjected Rag2−/−γc−/− mice and their wild type counterparts to 6 Gy TBI (maximum tolerated dose in absence of bone marrow transplantation on the BALB/c background). We then assessed liver, brain and lung tissues for the accumulation of 53BP1 DNA foci-containing cells. As we observed in C57BL/6 mice, IR initially generated many cells with 53BP1 foci (Figure 4A). The proportion of cells containing DNA damage foci then gradually declined in both groups, but, 12–17 weeks after IR exposure, the proportion of damaged cells was still much higher than the proportion in aged-match unirradiated mice (Figure 4A). Of significance, at all times tested, the fraction of DNA damage foci-containing cells was not significantly different in Rag2−/−γc−/− mice compared to their wild-type counterparts. Likewise, we found that IR-induced p16INK4a expression was similar in wild-type and Rag2−/−γc−/− mice 17–18 weeks after IR exposure (Figure 4B). These findings suggest that damaged cells with senescence-associated markers are not cleared by B, T or NK cells in mice in vivo.
Figure 4. IR-induced persistent DNA damage foci and p16INK4a expression is independent of T, B and NK cell functions.
A) Proportion of cells containing 53BP1 DNA damage foci in liver, brain and lung tissues collected from wild type (WT) or Rag2−/−γC−/− immune deficient mice sacrificed at the indicated time in weeks (W) post 6 Gy TBI (n = 3–5 mice per group). nd = not determined. No statistical difference was observed between groups of mice at all time point analyzed post IR. B) Expression of p16INK4a as determined by quantitative real-time PCR using RNA isolated from homogenized liver or brain tissues of wild type and Rag2−/−γC−/− mice 17–18 weeks post 6 Gy TBI, p values were obtained by performing a Student’s t-test. C) As described for panel A, proportion of cells containing 53BP1 DNA damage foci 12 weeks post IR in wild type and Rag2−/−γC−/− mice transplanted or not with 5×106 bone marrow cells (BM) collected from a wild type donor mouse (n = 4 per group). No statistical difference was observed between transplanted and non-transplanted groups. D) Representative flow cytometry profiles showing the proportion of T cells (CD3+), B cells (CD45R/B220) and NK cells (CD94+, CD3−) in blood collected from wild type (WT), and Rag2−/−γC−/− mice transplanted (Rag2−/−γC−/− BM) or not with bone marrow cells.
Leukocytes counts are greatly reduced soon after exposure to IR, and remain low for a few weeks. We therefore considered the possibility that cells with permanent DNA damage and/or p16INK4a expression might escape immune clearance if they were capable of alerting the immune system only transiently. To test this possibility, we transplanted wild-type and Rag2−/− γc−/− mice immediately after irradiation with 5×106 freshly isolated bone marrow cells from an unirradiated donor mouse (or an equivalent number of spleenocytes, data not shown), and then measured the fraction of damaged cells over time. At 12 weeks after transplantation, we observed no significant reduction in the fraction of damaged cells in all tissues tested, despite confirmation by flow cytometry that the transplantation was successful, as judged by the appearance of T (CD3+), B (B220+) and NK (CD94+/CD3−) cells in Rag2−/−γc−/− mice (Figure 4C,D). Similarly, transplantation of immune cells immediately after irradiation did not affect IR-induced p16INK4a expression in liver and brain tissues (data not shown).
We also assessed the role T, B and NK cells in eliminating damaged cells and p16INK4a expression in the context of chronological aging by assaying for these markers in young (3 months) and old (18 months) immune deficient Rag2−/−γc−/− mice. Aged wild-type and Rag2−/−γc−/−mice had similarly increased expression of p16INK4a compared to their young counterparts (Fig. S4). 18 months old wild type and Rag2−/−γc−/− mice did not show a significant increase in the proportion of cells containing DNA damage foci (data not shown), possibly because they were not old enough, but reinforcing the idea that the absence of T, B and NK cells is unlikely to yield premature accumulation of damaged cells with age. Together, these finding suggest that whether induced following exposure to IR or during the course of chronological aging, the clearance/accumulation of senescence markers is independent of T, B or NK cells functions.
IR-induced p16INK4a expression and persistent DNA damage is independent of p53
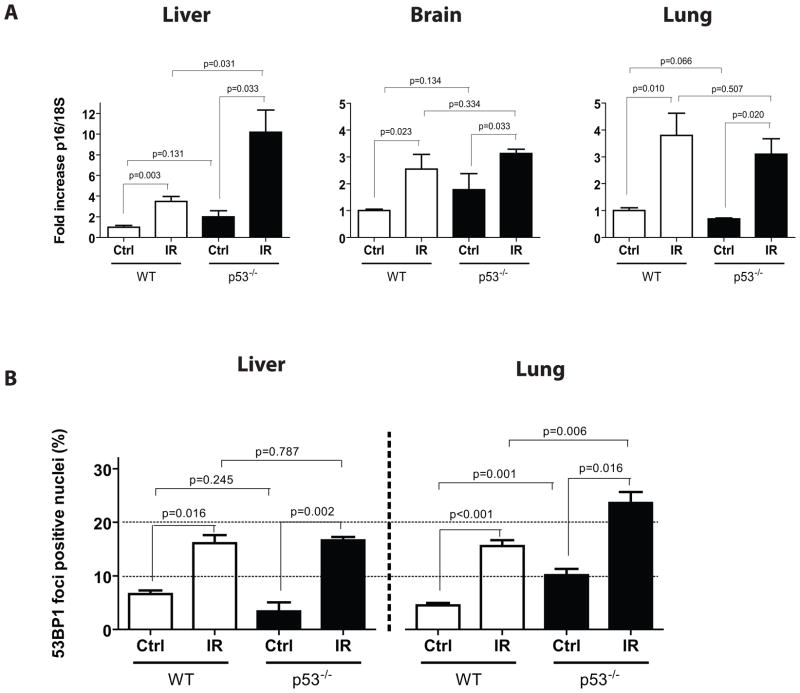
Because of the pivotal role p53 plays in orchestrating DNA damage responses and cell fate, we asked whether p53 inactivation could prevent (by interfering with DNA damage response signalling) or increase (by limiting DNA repair and apoptosis) the long-term accumulation of IR-induced senescence markers. Answering this question is important because inhibitors of apoptosis, many targeting the p53 pathway, are currently being tested for their ability to prevent side-effects of cancer therapies (Green & Kroemer 2005). We subjected p53−/− mice and wild-type littermate controls to 5 Gy TBI and analyzed lung, liver and brain tissues for p16INK4a expression 8 weeks later. Because of the cancer susceptibility of p53-deficient mice, we limited their exposure to 5 Gy TBI in order to allow 8 weeks cancer-free survival. We found that p16INK4a expression increased equally well (lung and brain) or to even higher levels (liver) in p53-deficient mice compared to wild-type controls (Figure 5A). While these results confirmed that IR-induced p16INK4a expression is independent of p53, they also suggest that the p53-dependent apoptosis that occurs after IR exposure (Christophorou et al. 2006) has little impact on the accumulation of p16INK4a. Surprisingly, the absence of p53 had little effect on the proportion of cells with DNA damage foci 8 weeks after exposure to IR (Figure 5B). While it is entirely possible that IR produced more damaged cells in p53-deficient tissues immediately after exposure, our observations suggest that p53 does not play a major role in the clearance or persistence of damaged cells long-term after IR.
Figure 5. p53 is not required for the induction of persistent DNA damage foci and increased p16INK4a expression post IR.
A) Expression of p16INK4a was determined by quantitative real-time PCR on liver, brain and lung tissues collected from age-match wild type (WT) or p53−/− deficient mice (n = 3–5 mice per group) 8 weeks following their exposure to 5 Gy TBI. B) Proportion of cells containing 53BP1 DNA damage foci in tissues collected from mice as described in panel A. p values were obtained by performing a Student’s t-test.
Discussion
Here, we provide the first evidence that exposure to IR, independent of the immune status and p53 function, induces the long-term accumulation of DNA damaged cells. The persistence of these damaged cells is unlikely to reflect poor DNA repair as cells were able to repair most of the damage formed by IR (>10 foci per nuclei on average), leaving only one or two foci remaining. Several factors can lead to persistent DNA damage, including telomere attrition/dysfunction (d’Adda di Fagagna et al. 2003; Herbig et al. 2006) and telomere independent mechanisms such as oxidative stress or the expression of certain oncogenes (Bartkova et al. 2006; Di Micco et al. 2006; Mallette et al. 2007; Matheu et al. 2007; Wang et al. 2009). Cells might not fully resolve IR-induced DNA damage foci for a variety of reasons. One possibility is that the structure or localization of some genomic sequences, for example telomeres, may be more difficult to repair than others. In support of this idea, an increase in the proportion of DNA damage foci that co-localized at telomeres was observed in brain sections collected long-term after exposure to IR (Fumagalli et al. submitted). Another possibility is that DNA lesions themselves have been repaired, but the foci not disassembled. It is also possible, albeit a rare event, that DNA damage occurs subsequent to the irradiation, for example if IR induced a mutation that activated an oncogene.
The proportion of BrdU+ cells containing 53BP1+ DNA damage foci 1 or 28 days after exposure to IR showed that at significant fraction of IR-induced damaged cells are preferentially eliminated in liver. These results greatly differed from what we observed in lung, and suggest the absence of a central mechanism by which IR-induced damaged cells are removed from tissues. In agreement with these observations, the turnover of the total proportion of BrdU+ cells observed in presence and absence of irradiation was comparable in lung but slightly faster in irradiated liver (Figure S5A–B). Notably, clearance of damaged cells in brain was not determined given the very low proportion of cells that incorporated BrdU in this tissue. It was not possible to reliably identify the type of BrdU and 53BP1 double positive cells (e.g., epithelial vs infiltrating leukocyte), and thus our results reflect the clearance of multiple cell types. Nonetheless, co-localization of BrdU and CD45 (a pan-leukocyte marker) signals revealed that more than 90% of CD45+ infiltrating cells had lost BrdU+ signal 28 days after IR, in agreement with the expected fast turnover of hematopoietic cells. In comparison, less then 40% of CD45− cells had lost BrdU+ signal during the same period, compatible with a much longer half-life for non hematopoietic cells (Figure S5C). Hence, we believe the relatively rapid decline in the fraction of damaged cells observed in the first month post-irradiation can be attributed to a high proportion of damaged cells either represented by infiltrating hematopoietic cells or differentiated epithelial/stromal cells having an intrinsic fast turnover.
Experiments using immune deficient Rag2−/−γc−/− mice revealed the dispensability of B, T and NK cells for the clearance of damaged cells. This was somewhat surprising because work by others suggested an active role for the immune system in the clearance of senescent/damaged cells. Indeed, cultured mouse and human cells, whether normal or cancerous, increase expression of NKG2D ligands when exposed to IR or other DNA damaging agents (Gasser et al. 2005; Soriani et al. 2009). Moreover, senescent cells induced in mouse models were shown to be eliminated by NK and other immune cells (Xue et al. 2007; Krizhanovsky et al. 2008). As a possible explanation for our results, we were unable to detect increased expression of mouse NKG2D ligands (Rae-1 and Mult-1) in irradiated cells or tissues, in contrast to expectations (Gasser et al. 2005). Alternatively, as suggested (Xue et al. 2007), neutrophils and macrophages, two cell types present in Rag2−/−γc−/− mice, may play an important role in the clearance of IR-induced damaged cells. However, it should be noted that IR induces transient leucopoenia (including loss of neutrophils and macrophages) and that bone marrow transplantation immediately after IR did not prevent the persistence of damaged cells. Assuming that transplantation of 5×106 whole bone marrow cells was quantitatively and/or qualitatively sufficient to reconstitute a fully functional immune system, our results suggest that neither innate nor adaptive immune cells are entirely effective at preventing IR-induced age marker formation.
Unexpectedly, and in contrast with the previously known role of p53 in inducing apoptosis shortly after irradiation (Christophorou et al. 2006), here we observed that deletion of p53 had little consequences in the establishment and long-term expression of senescence markers following exposure to IR. That is, elimination of DNA damaged cells was not inhibited in absence of p53, suggesting that damaged cells, at least in liver, must have p53 independent mechanisms by which they undergo delayed cell death/clearance in response to IR. This is surprising because deletion of p53 was recently found to impair clearance of damaged cells in aging telomere-dysfonctional mice (Begus-Nahrmann et al. 2009). Differences in the mouse models (irradiated vs telomerase knockout) and tissues analysed may account for this apparent disparity in the role of p53.
We also found that IR-induced p16INK4a expression is independent of p53 and parallel to p53/p21Cip1/Waf1, the latter increasing only transiently shortly after exposure to IR (Fig. S6). Since p16INK4a expression increases only several weeks after exposure to IR, p16INK4a may respond secondarily to cellular and tissue environmental changes induced by IR. For example, the senescence-associated secretory phenotype (SASP) has been shown to alter the tissue microenvironment and has been proposed to fuel the development of secondary cancers (Krtolica et al. 2001; Coppe et al. 2006; Coppe et al. 2008; Kuilman et al. 2008). Indeed, IR-induced p16INK4a expression in p53−/− mice is compatible with the previous observation that p53 is not required for the formation of the SASP (Coppe et al. 2008; Rodier et al. 2009). Because a reliable antibody for the detection of murine p16INK4a by immunofluorescence is lacking, it was not possible to determine whether cells containing DNA damage foci also had high p16INK4a expression. However, our observations that 18 months old tissues from wild type and Rag2−/−γc−/−mice have high p16INK4a expression and yet no detectable increase in DNA damage foci suggest these two markers might not necessarily be linked (Figure S4). Intriguingly, p16INK4a expression inceasing only several weeks post exposure to IR suggests that p16INK4a rises as a consequence of IR on the tissue rather than as a cell cycle check-point in response to IR itself. Notably, IR-induced p16INK4a expression and accumulation of DNA damage did not coincide with the expression of senescence associated β-galactosidase (data not shown). Reasons for this are currently unknown.
Finally, permanent growth arrest in the form of cellular senescence is a recognized tumour suppressive response (Collado et al. 2005; Michaloglou et al. 2005; Cosme-Blanco et al. 2007; Feldser & Greider 2007) that can be induced by p16INK4a expression (Baker et al. 2008) and in many cases DNA damage (d’Adda di Fagagna et al. 2003). Exposure to DNA damaging treatments such as IR and busulfan selectively induces premature senescence in murine hematopoietic progenitor cells (Meng et al. 2003; Wang et al. 2006). Similarly, neurons, much more than astrocytes, have been reported to accumulate DNA damage foci with age (Enokido et al. 2008). Hence, we hypothesize that IR-induced p16INK4a expression could have long-term consequences by preventing tissues stem and progenitor cells renewal. Alternatively, IR-induced persistent DNA damage signalling could be deleterious to cancer survivors by altering the tissue microenvironment by inducing the SASP (Rodier et al. 2009). In future work, it will be interesting to determine if a sub-population of cells (e.g., neural progenitors in the hippocampus) is more likely to accumulate DNA damage foci following exposure to IR. We believe increased p16INK4a expression and/or the proportion of cells containing DNA damage foci could be used as biomarkers for the identification of less damaging drugs and perhaps for the screening of cancer survivors at greater risk of developing late-effects.
Methods
Mice
8–12 week old female C57BL/6 or BALB/c mice were purchased from Charles River Laboratories (Saint-Constant, QC). Rag2−/−γc−/− mice (Goldman et al. 1998) were generated by in-house breedings of BALB/c mice carrying a null mutation of the cytokine receptor common gamma chain (γC) with RAG2 null mice purchased respectively from the Jackson Laboratories (Bar Harbor, ME) and Taconic (Albany, NY) under Material Transfer Agreement. BALB/c and Rag2−/−γc−/− mice were aged on site (18 months) while tissues from aged C57BL/6 mice (24 months) were purchased individually from the National Institute of Aging. p53 null mice on a BALB/c background were purchased from Jackson Laboratories (Jacks et al. 1994). All in vivo manipulations were approved by the Comité Institutionnel des Bonnes Pratiques Animales en Recherche (CIBPAR) of CHU-Ste-Justine and/or University of Colorado.
Irradiation
Mice were exposed to a single sublethal dose of total body irradiation (TBI) using a Gammacell 220 and cobalt 60 as a source. C57BL/6 mice received 8 Gy TBI while mice on a BALB/c background were exposed to 6 Gy TBI, to take into account their higher susceptibility to irradiation compared to C57BL/6 mice (Yu et al. 2001). p53 null mice were exposed to 5 Gy TBI (to minimize their probability of developing lymphoma) using a RadSource RS2000 irradiator as the X-ray source (Marusyk et al., submitted). Unless indicated otherwise, mice were not transplanted post-irradiation.
Bone marrow transplantation and flow cytometry
Where indicated, immediately after their exposure to 6 Gy TBI, BALB/c and Rag2−/−γc−/− mice received a tail vein injection of 5×106 whole bone marrow cells or an equivalent number of spleenocytes freshly isolated from a non-irradiated donor BALB/c mouse. Bone marrow reconstitution of Rag2−/−γc−/− mice was monitored weekly by flow cytometry on blood. Erythrocytes were first lysed (Lysing Buffer, BD Biosciences) and leukocytes stained with the following conjugated antibodies or their isotype controls: FITC anti-mouse CD3 (Clone 145-2C11), PE anti-mouse CD45R/B220 (Clone RA3-6B2), APC anti-mouse CD11b (Clone M1/70) all from BioLegend (San Diego, CA) and a PE anti-mouse CD94 (Clone 18d3) from eBioscience (San Diego, CA) Flow cytometry was performed by a FACSCalibur apparatus (BD).
RNA isolation and quantitative real-time PCR
Excised tissues sliced into ~2 mm2 pieces and preserved in RNA Later (Qiagen) were mechanically dissociated in 500ul Qiazol lysis reagent (Qiagen) using a power-driven homogenizer (OMNI International), and total RNA was extracted using the RNeasy lipid tissue Mini Kit (Qiagen). One μg of RNA was then reverse-transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative differences in gene expression were determined by real-time quantitative PCR using SYBR-Green PCR master mix (BioRad) and a spectrofluorometric thermal cycler (Mx3000P from Stratagene). Values are presented as the ratio of target mRNA to 18S rRNA obtained using the relative standard curve method of calculation. Primers sequences used were: p16INK4a (Gene Bank accession number AF332190.1) forward primer: AACTCTTTCGGTCGTACCCC, reverse primer: GCGTGCTTGAGCTGAAGCTA, 18S (Gene Bank accession number X00686) forward primer: CGCCGCTAGAGGTGAAATTCT, reverse primer: CGAACCTCCGACTTTCGTTCT: p21Cip1/Waf1 (Gene Bank accession number NM_007669.3) forward primer: TTGTCGCTGTCTTGCACTCTGGT, reverse primer: AGACCAATCTGCGCTTGGAGTGAT, p19ARF (Gene Bank accession number L76092.1) forward primer: GCCGCACCGGAATCCT, reverse primer: TTGAGCAGAAGAGCTGCTACGT.
Immunological examinations
Immediately after sacrificed, mouse tissues were frozen on dry ice and embedded in OCT compound (Sakura Finetek USA). Cryosections were mounted on glass slides previously treated with 1% gelatin and 0.05% chromo alum, dried at ambient temperature and then stored at −80 °C until usage. For DNA damage foci detection by immunofluorescence, thawed sections were fixed for 20 minutes in 3.7% formaldehyde, permeabilized with 0.5% Triton in PBS for 5 minutes and soaked for 60 minutes in blocking solution (5% Goat serum and 1% BSA in PBS). Sections were then incubated overnight at 4 °C in PBS containing 1% BSA and antibodies against 53BP1 (rabbit polyclonal, Novus – Littleton, CO; #NB100–304; 1:1500) and/or H2A.X (mouse monoclonal, Millipore – Temencula, CA; clone #JBW301; 1:250). Alexa Fluor goat anti Mouse 488 and/or goat anti-rabbit 594 (Invitrogen, CA; 1:750) were used as secondary antibodies along with DAPI for nuclei staining. Sections were mounted with mounting-medium (Vectashield H-1000) and analyzed using a Nikon Eclipse E800 epifluorescent microscope or a Zeiss LSM 510 confocal microscope. Paraffin embedded human skin biopsies obtained from patients previously exposed or not to IR, and from which we obtained informed consent, were deparaffinised with xylene and antigens retrieved by steaming slides in sodium citrate buffer (pH.6.0) using an Oster food steamer (model 5715) for 45 minutes. Sections were then processed as described above for detection of 53BP1 foci. Alternatively, immunohistochemical detection of p16INK4a was performed using a mouse anti human p16INK4a antibody (Santa Cruz, CA – clone JC8; 1: 50) and an HRP-conjugated goat anti-mouse secondary antibody was detected with a peroxidase kit (SK-4100) from Vector Laboratories. In all situations, DNA damage foci were counted from at least 200 cells from a minimum of 3 random fields per section.
BrdU injection and detection
10–12 week old C57BL/6 mice were injected intraperitoneally with BrdU (Roche, Montréal, QC) at a dose of 50mg/kg (0.8mg/mL in PBS) once a day for 10 consecutive days. 24 hours after the last injection, mice were exposed to 8 Gy TBI and sacrificed 1 or 28 days later, a time at which tissues were collected and stored frozen until ready for sectioning. In brief, cryosections were fixed with 3.7% formaldehyde (for lung) or methanol (for liver), denatured in 1N HCl for 30 min, neutralized with 0.1 M borate buffer pH 8.5, and then immune detection was processed as described above using antibodies against BrdU (mouse monoconal conjugated to FITC, Roche, clone BMC 9318 ; 1:50), 53BP1 (rabbit anti-human from Novus #NB100–304: 1:1500) or the pan-leukocyte CD45 marker (rat anti-mouse from Biolegend clone 30-F11BD, 1: 200). Sections were stained with DAPI and co-localizations of BrdU with 53BP1 and/or CD45 were determined by counting cells as described above.
Statistical analysis
Statisticaldifferences were assessed using unpaired Student’s t-tests performed with the Graph Pad software (version 5.0).
Supplementary Material
Acknowledgments
We would like to thanks Dr Utz Herbig for its help with DNA damage foci detection of paraffin-embedded human skin tissues and Vadym Zaberezhny for providing irradiated tissues from p53 deficient mice.
Financial support: This work was supported by grants from: The Canadian Institute of Health Research #IAO-79317 and la Fondation des Gouverneurs de l’espoir to C.M.B., NIH CA109657 to J.D., NIH AG017242, CA12654 and AG09909 to J.C., and NIH AG025708 to the Buck Institute. C.M.B. holds a Health Research Foundation Rx&D/CIHR young investigator award.
Footnotes
Authors contributions: Oanh Le: Collection of data, data analysis and interpretation
Francis Rodier: Collection of data, data analysis and interpretation, manuscript editing
Francois Fontaine: Collection of data
Jean-Philippe Coppe: Data analysis and interpretation
Judy Campisi: Provision of study material, manuscript editing
Viktor Kokta: Provision of study material
Caroline Laverdiére: Provision of study material
James Degregori: Provision of study material, manuscript editing
Elie Haddad: Provision of study material, data analysis and interpretation
Christian Beauséjour: Conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- Baker DJ, Perez-Terzic C, Jin F, Pitel K, Niederlander NJ, Jeganathan K, Yamada S, Reyes S, Rowe L, Hiddinga HJ, Eberhardt NL, Terzic A, van Deursen JM. Opposing roles for p16Ink4a and p19Arf in senescence and ageing caused by BubR1 insufficiency. Nat Cell Biol. 2008;10:825–836. doi: 10.1038/ncb1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, Takaoka M, Nakagawa H, Tort F, Fugger K, Johansson F, Sehested M, Andersen CL, Dyrskjot L, Orntoft T, Lukas J, Kittas C, Helleday T, Halazonetis TD, Bartek J, Gorgoulis VG. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Begus-Nahrmann Y, Lechel A, Obenauf AC, Nalapareddy K, Peit E, Hoffmann E, Schlaudraff F, Liss B, Schirmacher P, Kestler H, Danenberg E, Barker N, Clevers H, Speicher MR, Rudolph KL. p53 deletion impairs clearance of chromosomal-instable stem cells in aging telomere-dysfunctional mice. Nat Genet. 2009;41:1138–1143. doi: 10.1038/ng.426. [DOI] [PubMed] [Google Scholar]
- Campisi J, d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- Chang C, Biedermann KA, Mezzina M, Brown JM. Characterization of the DNA double strand break repair defect in scid mice. Cancer Res. 1993;53:1244–1248. [PubMed] [Google Scholar]
- Christophorou MA, Ringshausen I, Finch AJ, Swigart LB, Evan GI. The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature. 2006;443:214–217. doi: 10.1038/nature05077. [DOI] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguria A, Zaballos A, Flores JM, Barbacid M, Beach D, Serrano M. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Kauser K, Campisi J, Beausejour CM. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J Biol Chem. 2006;281:29568–29574. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosme-Blanco W, Shen MF, Lazar AJ, Pathak S, Lozano G, Multani AS, Chang S. Telomere dysfunction suppresses spontaneous tumorigenesis in vivo by initiating p53-dependent cellular senescence. EMBO Rep. 2007;8:497–503. doi: 10.1038/sj.embor.7400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, Maestro R, Pelicci PG, d’Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Enokido Y, Yoshitake A, Ito H, Okazawa H. Age-dependent change of HMGB1 and DNA double-strand break accumulation in mouse brain. Biochem Biophys Res Commun. 2008;376:128–133. doi: 10.1016/j.bbrc.2008.08.108. [DOI] [PubMed] [Google Scholar]
- Feldser DM, Greider CW. Short telomeres limit tumor progression in vivo by inducing senescence. Cancer Cell. 2007;11:461–469. doi: 10.1016/j.ccr.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Tomas-Loba A, Martin-Caballero J, Flores JM, Klatt P, Blasco MA, Serrano M. Increased p53 activity does not accelerate telomere-driven ageing. EMBO Rep. 2006;7:546–552. doi: 10.1038/sj.embor.7400667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geenen MM, Cardous-Ubbink MC, Kremer LC, van den Bos C, van der Pal HJ, Heinen RC, Jaspers MW, Koning CC, Oldenburger F, Langeveld NE, Hart AA, Bakker PJ, Caron HN, van Leeuwen FE. Medical assessment of adverse health outcomes in long-term survivors of childhood cancer. Jama. 2007;297:2705–2715. doi: 10.1001/jama.297.24.2705. [DOI] [PubMed] [Google Scholar]
- Goldman JP, Blundell MP, Lopes L, Kinnon C, Di Santo JP, Thrasher AJ. Enhanced human cell engraftment in mice deficient in RAG2 and the common cytokine receptor gamma chain. Br J Haematol. 1998;103:335–342. doi: 10.1046/j.1365-2141.1998.00980.x. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. Pharmacological manipulation of cell death: clinical applications in sight? J Clin Invest. 2005;115:2610–2617. doi: 10.1172/JCI26321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- Herbig U, Ferreira M, Condel L, Carey D, Sedivy JM. Cellular senescence in aging primates. Science. 2006;311:1257. doi: 10.1126/science.1122446. [DOI] [PubMed] [Google Scholar]
- Hinkal GW, Gatza CE, Parikh N, Donehower LA. Altered senescence, apoptosis, and DNA damage response in a mutant p53 model of accelerated aging. Mech Ageing Dev. 2009;130:262–271. doi: 10.1016/j.mad.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Dombkowski DM, Cheng T, DePinho RA, Sharpless NE, Scadden DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Jeyapalan JC, Ferreira M, Sedivy JM, Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V, Yon M, Dickins RA, Hearn S, Simon J, Miething C, Yee H, Zender L, Lowe SW. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A, Parrinello S, Lockett S, Desprez PY, Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc Natl Acad Sci U S A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Vredeveld LC, Douma S, van Doorn R, Desmet CJ, Aarden LA, Mooi WJ, Peeper DS. Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell. 2008;133:1019–1031. doi: 10.1016/j.cell.2008.03.039. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Ibrahim JG, Thomas NE, Sharpless NE. Expression of p16 in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009 doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette FA, Gaumont-Leclerc MF, Ferbeyre G. The DNA damage signaling pathway is a critical mediator of oncogene-induced senescence. Genes Dev. 2007;21:43–48. doi: 10.1101/gad.1487307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, Serrano M. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- Melk A, Schmidt BM, Takeuchi O, Sawitzki B, Rayner DC, Halloran PF. Expression of p16INK4a and other cell cycle regulator and senescence associated genes in aging human kidney. Kidney Int. 2004;65:510–520. doi: 10.1111/j.1523-1755.2004.00438.x. [DOI] [PubMed] [Google Scholar]
- Meng A, Wang Y, Van Zant G, Zhou D. Ionizing radiation and busulfan induce premature senescence in murine bone marrow hematopoietic cells. Cancer Res. 2003;63:5414–5419. [PubMed] [Google Scholar]
- Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, Majoor DM, Shay JW, Mooi WJ, Peeper DS. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Min H, Montecino-Rodriguez E, Dorshkind K. Effects of aging on the common lymphoid progenitor to pro-B cell transition. J Immunol. 2006;176:1007–1012. doi: 10.4049/jimmunol.176.2.1007. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen GP, Stemmer-Rachamimov AO, Shaw J, Roy JE, Koh J, Louis DN. Immunohistochemical survey of p16INK4A expression in normal human adult and infant tissues. Lab Invest. 1999;79:1137–1143. [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, Enver T, Bell JI, Slijepcevic P, Goodnow CC, Jeggo PA, Cornall RJ. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Oeffinger KC, Mertens AC, Sklar CA, Kawashima T, Hudson MM, Meadows AT, Friedman DL, Marina N, Hobbie W, Kadan-Lottick NS, Schwartz CL, Leisenring W, Robison LL. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355:1572–1582. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. doi: 10.1101/gad.1182704. [DOI] [PubMed] [Google Scholar]
- Probin V, Wang Y, Bai A, Zhou D. Busulfan selectively induces cellular senescence but not apoptosis in WI38 fibroblasts via a p53-independent but extracellular signal-regulated kinase-p38 mitogen-activated protein kinase-dependent mechanism. J Pharmacol Exp Ther. 2006;319:551–560. doi: 10.1124/jpet.106.107771. [DOI] [PubMed] [Google Scholar]
- Ressler S, Bartkova J, Niederegger H, Bartek J, Scharffetter-Kochanek K, Jansen-Durr P, Wlaschek M. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5:379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- Rodier F, Campisi J, Bhaumik D. Two faces of p53: aging and tumor suppression. Nucleic Acids Res. 2007;35:7475–7484. doi: 10.1093/nar/gkm744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Coppe JP, Patil CK, Hoeijmakers WA, Munoz DP, Raza SR, Freund A, Campeau E, Davalos AR, Campisi J. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11:973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Bryder D, Seita J, Nussenzweig A, Hoeijmakers J, Weissman IL. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Sansoni P, Cossarizza A, Brianti V, Fagnoni F, Snelli G, Monti D, Marcato A, Passeri G, Ortolani C, Forti E, et al. Lymphocyte subsets and natural killer cell activity in healthy old people and centenarians. Blood. 1993;82:2767–2773. [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Redon C, Nakamura A, Zimonjic DB, Popescu NC, Bonner WM. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell. 2008;7:89–100. doi: 10.1111/j.1474-9726.2007.00354.x. [DOI] [PubMed] [Google Scholar]
- Sedelnikova OA, Horikawa I, Zimonjic DB, Popescu NC, Bonner WM, Barrett JC. Senescing human cells and ageing mice accumulate DNA lesions with unrepairable double-strand breaks. Nat Cell Biol. 2004;6:168–170. doi: 10.1038/ncb1095. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- Soriani A, Zingoni A, Cerboni C, Iannitto ML, Ricciardi MR, Di Gialleonardo V, Cippitelli M, Fionda C, Petrucci MT, Guarini A, Foa R, Santoni A. ATM-ATR-dependent up-regulation of DNAM-1 and NKG2D ligands on multiple myeloma cells by therapeutic agents results in enhanced NK-cell susceptibility and is associated with a senescent phenotype. Blood. 2009;113:3503–3511. doi: 10.1182/blood-2008-08-173914. [DOI] [PubMed] [Google Scholar]
- Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- Varela I, Cadinanos J, Pendas AM, Gutierrez-Fernandez A, Folgueras AR, Sanchez LM, Zhou Z, Rodriguez FJ, Stewart CL, Vega JA, Tryggvason K, Freije JM, Lopez-Otin C. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Wang C, Jurk D, Maddick M, Nelson G, Martin-Ruiz C, von Zglinicki T. DNA damage response and cellular senescence in tissues of aging mice. Aging Cell. 2009;8:311–323. doi: 10.1111/j.1474-9726.2009.00481.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Schulte BA, LaRue AC, Ogawa M, Zhou D. Total body irradiation selectively induces murine hematopoietic stem cell senescence. Blood. 2006;107:358–366. doi: 10.1182/blood-2005-04-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Okayasu R, Weil MM, Silver A, McCarthy M, Zabriskie R, Long S, Cox R, Ullrich RL. Elevated breast cancer risk in irradiated BALB/c mice associates with unique functional polymorphism of the Prkdc (DNA-dependent protein kinase catalytic subunit) gene. Cancer Res. 2001;61:1820–1824. [PubMed] [Google Scholar]
- Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.