Abstract
A phylogenetic analysis of the basic helix-loop-helix (bHLH) gene superfamily was performed using seven different species (human, mouse, rat, worm, fly, yeast, and plant Arabidopsis) and involving over 600 bHLH genes [1]. All bHLH genes were identified in the genomes of the various species, including expressed sequence tags, and the entire coding sequence was used in the analysis. Nearly 15% of the gene family has been updated or added since the original publication. A super-tree involving six clades and all structural relationships was established and is now presented for four of the species. The wealth of functional data available for members of the bHLH gene superfamily provides us with the opportunity to use this exhaustive phylogenetic tree to predict potential functions of uncharacterized members of the family. This phylogenetic and genomic analysis of the bHLH gene family has revealed unique elements of the evolution and functional relationships of the different genes in the bHLH gene family.
Keywords: basic Helix-Loop-Helix transcription factor, bHLH, human, mouse, rat, worm, fly, yeast, plant, nomenclature, phylogenetics
INTRODUCTION
A recent phylogenetic analysis of the basic helix-loop-helix (bHLH) gene superfamily including seven different species re-evaluates the classifications of these genes based on an unbiased sequence comparison across the full length of the proteins [1]. Previous classifications of the bHLH genes were based on a combination of functional characteristics, such as DNA binding activity, dimer partners, and expression patterns [2, 3]. The organization of bHLH genes into clades [1] does not make assumptions about gene function and resulted in the formation of six clades. Clade A contains primarily mammalian genes and no plant genes, while clade F only contains plant genes. The other four clades had a mixture of different species’ genes, and specific ancestral genes are identified [1]. The mouse bHLH gene family is shown in Figure 1. Several members of the bHLH superfamily have been subdivided based on the inclusion of additional functional motifs, including leucine zipper, PAS domain, and Orange domain that provide additional functionality to the transcription factors, as well as alter the DNA binding activity of the bHLH domains [4–7]. These various classes of bHLH genes were categorized appropriately in the new phylogenetic analysis [1].
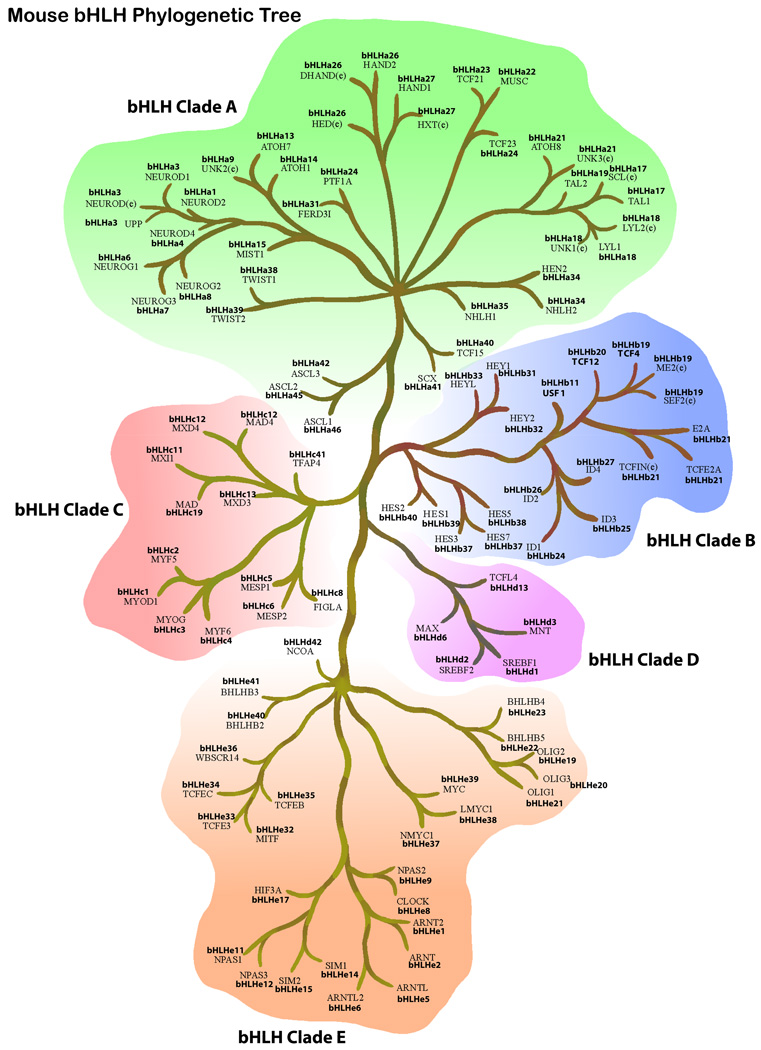
Figure 1.
Mouse bHLH super-tree for the 5 different clades and all 107 genes and relatedness presented. Clade B contains the inhibitory bHLH/Orange genes, as well as the Id genes. Clade E contains bHLH/PAS family genes. bHLH/bZip genes are spread throughout. Both the current and new nomenclature is presented. For some genes multiple older alternate names are listed with nearby branches (e.g. bHLHa 17, 18, 26, 27, 34) using the same new nomenclature, which indicates all are a single gene. Some new genes listed in Table S1 are not listed in the tree.
The bHLH motif was first identified in 1989 and initial classification schemes were based on ubiquitous (Class A) and tissue-specific (Class B) expression [8]. The classification system was expanded with the first large scale phylogenetic analysis by Atchley and Fitch (1997). This system used a comparison of the bHLH domains to a Class A set of bHLH genes that included tissue-specific proteins [9], a Class B Group of functionally unrelated proteins [10], a Class C that contained the PAS domain proteins [4], and Class D that are inhibitory (e.g. lack basic domain, Ids). In the most recent phylogenetic analysis, the complete amino acid sequence was used to identify new clades that do not correlate with this previous classification [1], Figure 1. Due to the inclusion of uncharacterized (e.g. EST) bHLH genes and the development of a more accurate relationship of all the genes between species, a new nomenclature is suggested. The previously proposed nomenclature is not unified and provides multiple names for the same gene between species and within the same species. Currently, no functional or structural relationships are considered. The new nomenclature uses the name “bHLH” for the entire gene family and then a letter representing the clade distribution for the gene (e.g. bHLHa for Clade A genes), followed by a number within the clade to help cluster related genes (e.g. bHLHa1-bHLHa46). Homologous genes from multiple species are given the same name and the variants of a single gene have letters [e.g. E12 (bHLHb21a) and E47 (bHLHb21b)]. The old and new nomenclature for all seven species are provided in Supplemental Table S1 and Supplemental Table S2, including recent additions and corrections. The old and new names for the mouse bHLH proteins are shown in the phylogenetic tree in Figure 1.
Several large and small scale phylogenetic analyses of the bHLH transcription factor family have been performed that only use the bHLH domain sequence [2, 11, 12]. A recent analysis in plants used the entire coding sequence [13]. Due to the number of additional domains being associated with the bHLH proteins, utilizing the entire coding region in a large-scale phylogenetic analysis for classification is needed [1]. The unified bHLH gene family nomenclature suggested allows for structural and functional gene relationships within and across species to be considered, Supplemental Table S1. Recently the Human Genome Organization (HuGO) has adopted this nomenclature and integrated it into the GeneBank information. The Arabidopsis (http://www.arabidopsis.org/portals/nomenclature/guidelines.jsp) (Table S5), mouse (http://www.informatics.jax.org/mgihome/nomen/), C elegans (http://www.wormbase.org/) (Table S4), drosophila (http://flybase.bio.indiana.edu/static_pages/docs/nomenclature/nomenclature3.html) (Table S3) and human (http://www.bioscience.org/services/genenome.htm) nomenclature committees have adopted the proposed nomenclature. In the event an EST or un-named gene was identified the nomenclature was adopted as the official name. In the event a previous name existed the new nomenclature was adopted as an alternative name. If the new nomenclature is adopted by the research community in the future the official name assignment will be considered by the appropriate nomenclature committees. The updated nomenclature is proposed to assist in future analysis of cellular differentiation and developmental processes influenced by bHLH proteins.
SPECIES PHYLOGENETIC TREES
The various bHLH proteins for each of the species is shown in Supplemental Table S1 and provides a comparison between the species using the new nomenclature, Supplemental Table 2. Species-specific phylogenetic trees for mouse (Figure 1), human (Figure 2), drosophila (Figure 3) and C. elegans (Figure 4) were constructed to show the clade distribution of the bHLH genes in the specific species. As expected, the C. elegans have the fewest number of bHLH genes, while a larger diversity of gene types are found in drosophila, and the most found in humans (~130), although the number found in mouse is nearly identical (~117). This pattern is suggestive of gene family duplication and diversification as the metazoans diversified. Interestingly, the similar level of diversity among all mammals studied (human, mouse, rat), suggest that most of the functional types of bHLH transcription factors likely originated early in or prior to mammalian diversification. In contrast, Arabidopsis, a flowering plant, has even more (~141) copies of bHLH genes than the mammals (~130 in humans), but many of these plant genes are found in clades completely separate from the metazoan genes [1]. This suggests that while a few copies were present in the shared ancestor of plants and animals, most of the gene diversity was derived later in eukaryotic diversification. This diversification was likely associated with the origins of multicellularity and different lineages of multicellular organisms. The duplication of different copies of bHLH leads to only marginal overlap in gene family membership between these disparate lineages.
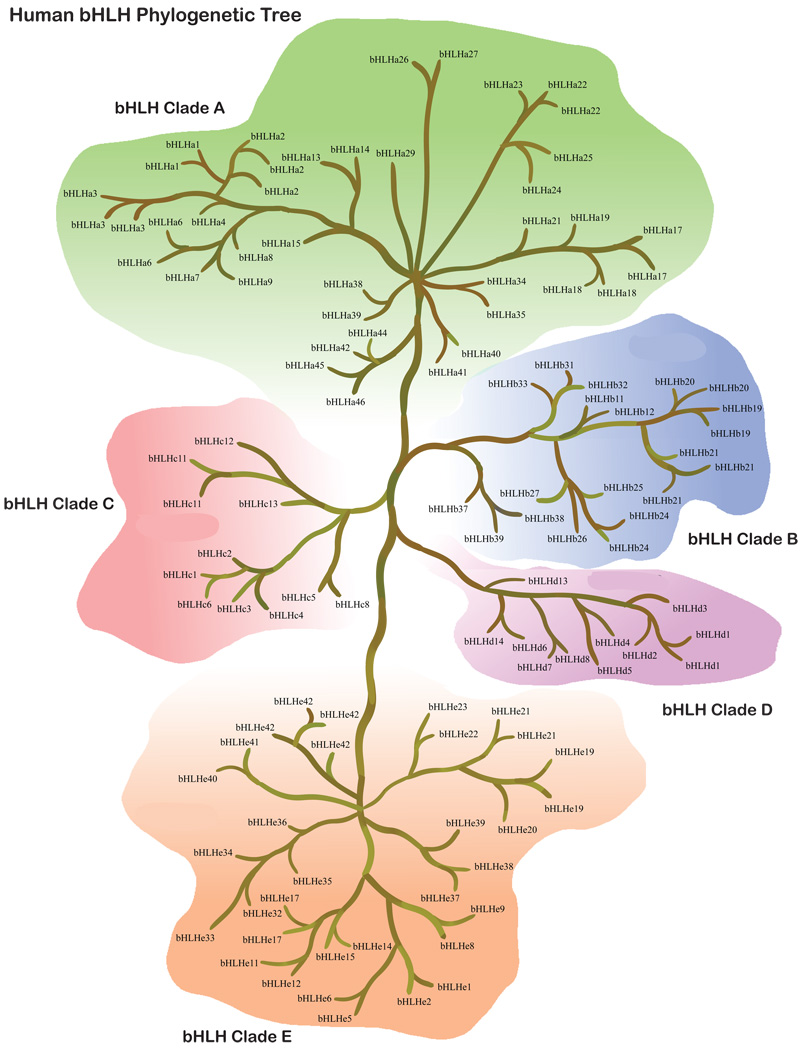
Figure 2.
Human bHLH super-tree for the 5 different clades and all 118 genes and relatedness presented. Multiple listings with the same new nomenclature are the same gene, and some new genes listed in Table S1 are not listed on the tree.
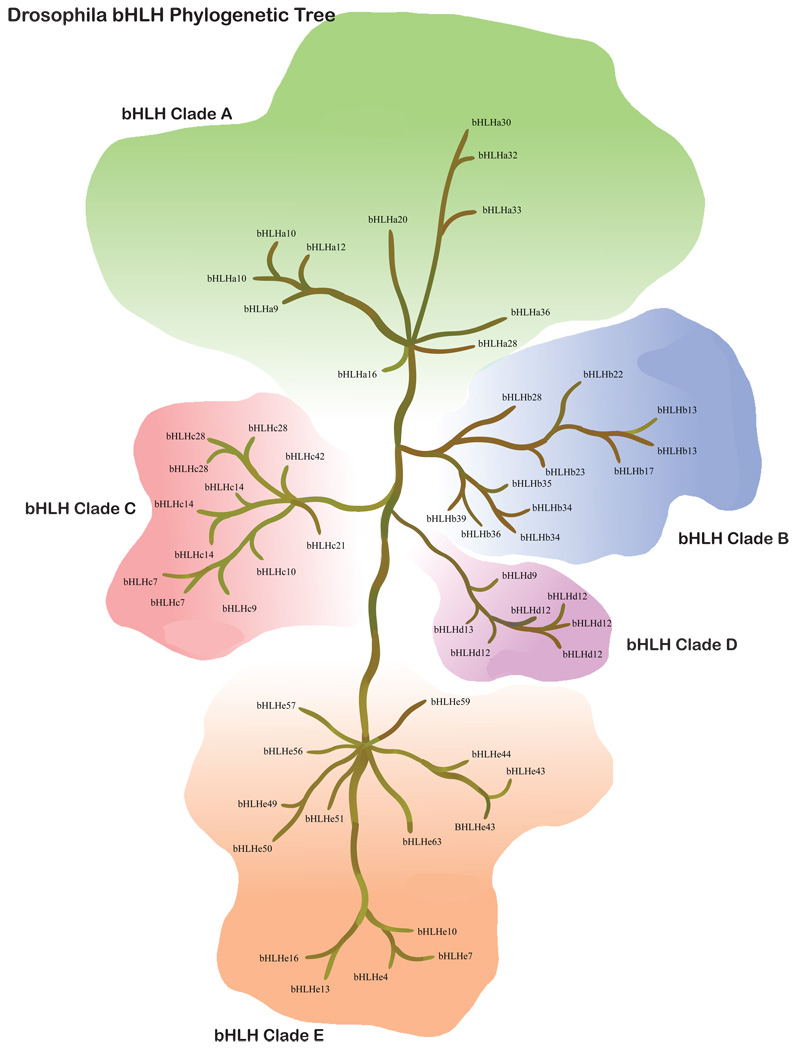
Figure 3.
Drosophila bHLH super-tree for the 5 different clades and all 56 genes and relatedness presented.
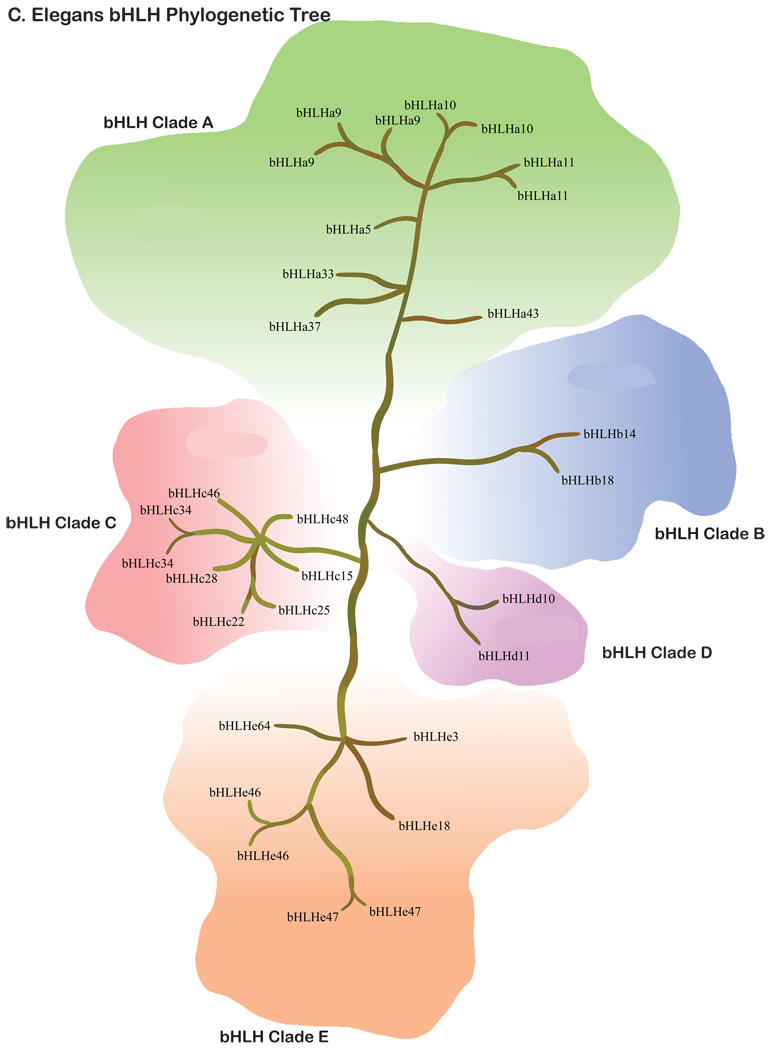
Figure 4.
C. elegans bHLH super-tree for the 5 different clades and all 30 genes and relatedness presented.
Gene diversity within clades also varies among species. While the patterns are very similar between human and mouse (Fig. 1–Fig. 2), the diversity in C. elegans is very different (Fig. 4). The largest obvious difference, other than fewer total genes, is the paucity of bHLH-B clade genes. The functional significance of this lack of Clade B genes is unclear. In drosophila, the bHLH gene family is reasonably diverse, but it should be noted that while drosophila has more diversity than C. elegans, and less than mammals, the distribution of gene copies in the phylogenies are quite different between the Ecdysozoa (the clade that includes both arthropods and nematodes) and mammals. For instance, the drosophila bHLHa28 is sister to the clade of mammalian bHLHa26/a27 (HAND) genes, suggesting that the drosophila copy diverged prior to this duplication. However, there does not appear to be an ortholog in C. elegans. Since C. elegans and drosophila share a more recent common ancestor than either does with the mammals, this is suggestive that this copy might have been lost in the C. elegans lineage or has not been recognized as a bHLH gene family member yet. Alternatively, there are some clades with multiple copies in drosophila or C. elegans, or both, but without any known mammalian copies. This suggests duplications and possible functional diversification in the Ecdysozoa or within more recently diverged lineages. In order to address the timing of duplications and deletions, and their possible functional significance, more detailed sampling of the bHLH gene family will be necessary outside of the commonly used model systems. Further phylogenetic analysis will be required to better predict functional diversification and the differences between duplications and deletions in the bHLH gene family.
bHLH FAMILY FUNCTIONAL RELATIONSHIPS
The bHLH transcription factors represent an ancient family found in fungi, plants, and animals that are essential for organisms to respond to a broad set of environmental factors. Individual members of the family are able to direct differential gene expression that allows cells to respond to select nutrients in the environment (glucose, sterols, inositol, choline, iron, and calcium), promote cellular and systemic protection from hypoxic conditions and xenobiotics, and coordinate organismal growth in response to light. Other members are required for diversification of the cell types during embryogenesis through cell type-specific specification, differentiation, proliferation, and apoptosis. A comprehensive analysis of the bHLH gene family in metazoans revealed multiple rounds of expansion [3, 14]. The first round occurred with the split of the metazoans from the choanoflagellates and the second round was associated with the increase in specialized cell types in higher metazoans. The most recent phylogenetic analysis of bHLH genes by [1] using 541 known bHLH genes across both plants and animals divides the genes into new clades based solely on the sequence comparison of the entire protein, instead of the highly conserved bHLH domain. The organization of these clades provides new insight into the functional relationships and potential ancestral origins of the bHLH genes.
The best-described role for bHLH genes during embryogenesis is the development of the neurogenic circuitry [15–17]. The differentiation and specification of individual neurons as well as their patterning in the central and peripheral nervous system are dependent on the Atonal-related [Atonal (bHLHa12, bHLHa13, and bHLHa21), NeuroD (bHLHa1, bHLHa2, bHLHa3, and bHLHa4), Neurogenin (bHLHa6, and bHLHa7), Mist (bHLHa15)], Acheate-scute complex (AS-C) (bHLHa42, bHLHa44, bHLHa45, bHLH46, bHLH47), Hairy/Enhancer of Split (HES) [Hairy and E(spl) (bHLHb37, bHLHb38, bHLHb39, bHLHb40, bHLHb41, bHLHb42, and bHLHb43), Hey (bHLHb31, bHLHb32, bHLHb33)], N-Twist (bHLHa31) and NHLH (bHLHa34, and bHLHa35) genes. Orthologs for these subfamilies have been identified in all cnidarians and bilaterians underscoring the fundamental role of these bHLH proteins in neurogenesis [14]. The Atonal-related AS-C, and NHLH genes are grouped in Clade A, consistent with their linked functions. Interestingly, the HES genes are present in Clade B. The HES genes are downstream targets of the Notch signaling pathway, where they function to inhibit the transcription of proneural genes [18]. This predicts that HES genes are a later addition that increased the complexity of the proneural regulatory pathway.
In addition to the HES genes, Clade B also contains the Id/emc family (bHLHb24, bHLHb25, bHLHb26, bHLHb27) of transcriptional repressors. A previous classification of bHLH genes placed the Id/emc family into a separate class (Class D) based on the absence of the basic residues within the bHLH domain required for DNA binding [2]. Repression by these factors occurs by forming heterodimers with bHLH transcriptional activators and disrupting DNA binding. Inclusion of these factors in the same clade as the HES subfamily predicts a previously unappreciated ancestral link between these two subfamilies of transcriptional repressors. Hes and E(spl) genes direct repression by binding the transcriptional co-repressor, Groucho, through a carboxy-terminal tetrapeptide WRPW motif [19]. However, several members of the HES subfamily (HES1, 3, 5, and 6 and Hey1 and 2) have also been reported to repress transcription by a mechanism analogous to Id/emc proteins [20–22] supporting the functional link between these gene subfamilies.
Notably absent from Clade B are BHLHB2/DEC1 (bHLHe40) and BHLHB3/DEC2 (bHLHe41) (1). These transcriptional repressors play a pivotal role in regulating circadian rhythm control, metabolite-sensing, hypoxia response and the development of the vascular, neural, muscle and cartilage tissue [23–25]. Based on the sequence comparison of the bHLH domain alone, BHLHB2 and BHLHB3 have been grouped with the Hes and E(spl) genes [26]. However, when using the entire protein for a phylogenetic analysis, BHLHB2 and BHLHB3 fall in Clade E that includes the bHLH-Pas genes that are essential for circadian oscillation, hypoxia response, and vascular and neuronal development (Figure 1) [27]. This reveals a closer phylogenetic link between bHLH and bHLH-Pas genes participating in environmental adaptation and differentiation than previously appreciated.
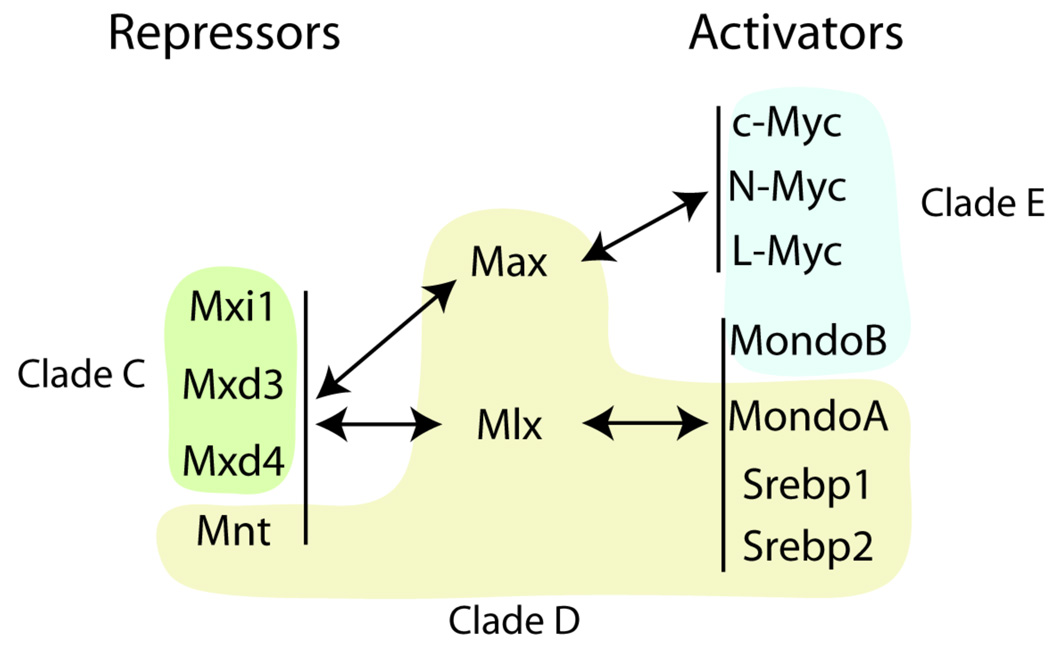
One of the outcomes of the analysis by [1] is the distribution of a family of bHLH genes that contain another functionally important motif known as the Leucine Zipper (Zip), which participates in both hetero- and homodimerization. Though these genes share two functional domains they are distributed among four clades. The best described of the bHLH-Zip genes belong to the Myc/Mad/Max network. Max (bHLHb6) serves as an obligate heterodimer partner for members of both the Myc and Mad subfamilies in the regulation of cell proliferation, differentiation, and death [28]. Myc genes are transcriptional activators, while Mad genes function as repressors through a Sin3a-mediated mechanism. An interconnected network uses a Max-like factor (Mlx/ bHLHd13) as an obligate partner for Mad factors and a separate family of transcriptional activators that regulate lipid and glucose metabolism [SREBF1 (bHLHd1), SREBF2 (bHLHd2), MondoA (bHLHe36), and MondoB (bHLHd14) [29, 30]. Both Max and Mlx fell into clade D, while the Mad and Myc subfamilies were in clades C and E, respectively (Figure 5). Interestingly, the Mad-like factor, Mnt was present in clade D instead of clade C with the Mad factors. This predicts Mnt shares functional similarity with other Mad transcriptional repressors, but a phylogenetic relationship closer to Max. Similarly, SREBF1 and 2, as well as MondoA are found in clade D, predicting that these transcriptional activators have a closer relationship to Max than the Myc genes. A second bHLH- Zip family called Mitf-Tcfe [Mitf (bHLHe32), Tcfe3 (bHLHe33), Tcfeb (bHLHe35), and Tcfec (bHLHe34)] forms heterodimers similar to the Myc/Mad/Max network. Natural and induced mutations predict that this family regulates neural crest and osteoclast differentiation [31–33]. Though these genes do not interact with Myc factors, the phylogenetic analysis places them in the same clade. Twist (bHLHa38) and members of the Twist subfamily are known to alter their function based on binding partner [34–36]. Consistent with other analyses, the Twist subfamily fell into the same clade [1]
Figure 5.
Clade distribution of bHLH-Zip genes that heterodimerize with Max or Mlx. Max and Mlx are obligate dimer partners for both transcriptional activators and transcriptional repressors.
CONCLUSION
The ability of bHLH factors to alter their DNA binding specificity based on their dimmer partners has long been recognized as a mechanism to maximize the diversity in target gene transcription that can be controlled by a relatively small number of genes. The new organization of the bHLH genes suggests novel linkages between subfamilies that were not previously appreciated, but now uncovered by their clade association. This represents a potentially fertile area of study for further investigation into the interactions and functions of bHLH genes. The unified nomenclature proposed now assists comparative studies between species and provides a more accurate functional relationship of the bHLH genes. Clearly as research in the bHLH area advances the gene associations and nomenclature will need to be corrected and evolve. Consideration of this phylogenetic analysis provides insights into the evolutionary biology of this gene family and potential functional relationships of specific bHLH genes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ms. Heather Johnson for assistance in preparation of the manuscript. None of the authors have any financial interests or disclosures. This work was supported by NIH grants to Michael K. Skinner.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Stevens JD, Roalson EH, Skinner MK. Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation. 2008;76:1006–1022. doi: 10.1111/j.1432-0436.2008.00285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ledent V, Paquet O, Vervoort M. Phylogenetic analysis of the human basic helix-loop-helix proteins. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-6-research0030. RESEARCH0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews ST. Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev. 1998;12:607–620. doi: 10.1101/gad.12.5.607. [DOI] [PubMed] [Google Scholar]
- 5.Taelman V, Van Wayenbergh R, Solter M, Pichon B, Pieler T, Christophe D, Bellefroid EJ. Sequences downstream of the bHLH domain of the Xenopus hairy-related transcription factor-1 act as an extended dimerization domain that contributes to the selection of the partners. Dev Biol. 2004;276:47–63. doi: 10.1016/j.ydbio.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 6.Massari ME, Murre C. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 8.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 9.Hassan BA, Bellen HJ. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- 10.Facchini LM, Penn LZ. The molecular role of Myc in growth and transformation: recent discoveries lead to new insights. Faseb J. 1998;12:633–651. [PubMed] [Google Scholar]
- 11.Buck MJ, Atchley WR. Phylogenetic analysis of plant basic helix-loop-helix proteins. J Mol Evol. 2003;56:742–750. doi: 10.1007/s00239-002-2449-3. [DOI] [PubMed] [Google Scholar]
- 12.Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20:735–747. doi: 10.1093/molbev/msg088. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Duan X, Jiang H, Sun Y, Tang Y, Yuan Z, Guo J, Liang W, Chen L, Yin J, Ma H, Wang J, Zhang D. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simionato E, Ledent V, Richards G, Thomas-Chollier M, Kerner P, Coornaert D, Degnan BM, Vervoort M. Origin and diversification of the basic helix-loop-helix gene family in metazoans: insights from comparative genomics. BMC Evol Biol. 2007;7:33. doi: 10.1186/1471-2148-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunet JF, Ghysen A. Deconstructing cell determination: proneural genes and neuronal identity. Bioessays. 1999;21:313–318. doi: 10.1002/(SICI)1521-1878(199904)21:4<313::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 16.Verzi MP, Anderson JP, Dodou E, Kelly KK, Greene SB, North BJ, Cripps RM, Black BL. N-twist, an evolutionarily conserved bHLH protein expressed in the developing CNS, functions as a transcriptional inhibitor. Dev Biol. 2002;249:174–190. doi: 10.1006/dbio.2002.0753. [DOI] [PubMed] [Google Scholar]
- 17.Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Curr Opin Genet Dev. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- 19.Paroush Z, Finley RL, Jr., Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 20.Chin MT, Maemura K, Fukumoto S, Jain MK, Layne MD, Watanabe M, Hsieh CM, Lee ME. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 21.Hirata H, Ohtsuka T, Bessho Y, Kageyama R. Generation of structurally and functionally distinct factors from the basic helix-loop-helix gene Hes3 by alternative first exons. J Biol Chem. 2000;275:19083–19089. doi: 10.1074/jbc.M001075200. [DOI] [PubMed] [Google Scholar]
- 22.Gao X, Chandra T, Gratton MO, Quelo I, Prud'homme J, Stifani S, St-Arnaud R. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J Cell Biol. 2001;154:1161–1171. doi: 10.1083/jcb.200104058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St-Pierre B, Flock G, Zacksenhaus E, Egan SE. Stra13 Homodimers Repress Transcription through Class B E-box Elements. Biol Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- 24.Yamada K, Miyamoto K. Basic helix-loop-helix transcription factors, BHLHB2 and BHLHB3; their gene expressions are regulated by multiple extracellular stimuli. Front Biosci. 2005;10:3151–3171. doi: 10.2741/1772. [DOI] [PubMed] [Google Scholar]
- 25.Sato F, Bhawal UK, Kawamoto T, Fujimoto K, Imaizumi T, Imanaka T, Kondo J, Koyanagi S, Noshiro M, Yoshida H, Kusumi T, Kato Y, Kijima H. Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively regulates vascular and endothelial growth factor expression. Genes Cells. 2008;13:131–144. doi: 10.1111/j.1365-2443.2007.01153.x. [DOI] [PubMed] [Google Scholar]
- 26.Zheng X, Wang Y, Yao Q, Yang Z, Chen K. A genome-wide survey on basic helix-loop-helix transcription factors in rat and mouse. Mamm Genome. 2009;20:236–246. doi: 10.1007/s00335-009-9176-7. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh BE, Hogenesch JB, Bradfield CA. Mammalian per-arnt-sim proteins in environmental adaptation. Annu Rev Physiol. 2010;72:625–645. doi: 10.1146/annurev-physiol-021909-135922. [DOI] [PubMed] [Google Scholar]
- 28.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 29.Billin AN, Ayer DE. The Mlx network: evidence for a parallel Max-like transcriptional network that regulates energy metabolism. Curr Top Microbiol Immunol. 2006;302:255–278. doi: 10.1007/3-540-32952-8_10. [DOI] [PubMed] [Google Scholar]
- 30.Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, Ayer DE. MondoA-Mlx heterodimers are candidate sensors of cellular energy status: mitochondrial localization and direct regulation of glycolysis. Mol Cell Biol. 2006;26:4863–4871. doi: 10.1128/MCB.00657-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weilbaecher KN, Hershey CL, Takemoto CM, Horstmann MA, Hemesath TJ, Tashjian AH, Fisher DE. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J Exp Med. 1998;187:775–785. doi: 10.1084/jem.187.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verastegui C, Bertolotto C, Bille K, Abbe P, Ortonne JP, Ballotti R. TFE3, a transcription factor homologous to microphthalmia, is a potential transcriptional activator of tyrosinase and TyrpI genes. Mol Endocrinol. 2000;14:449–456. doi: 10.1210/mend.14.3.0428. [DOI] [PubMed] [Google Scholar]
- 33.Steingrimsson E, Tessarollo L, Pathak B, Hou L, Arnheiter H, Copeland NG, Jenkins NA. Mitf and Tfe3, two members of the Mitf-Tfe family of bHLH-Zip transcription factors, have important but functionally redundant roles in osteoclast development. Proc Natl Acad Sci U S A. 2002;99:4477–4482. doi: 10.1073/pnas.072071099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castanon I, Von Stetina S, Kass J, Baylies MK. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development. 2001;128:3145–3159. doi: 10.1242/dev.128.16.3145. [DOI] [PubMed] [Google Scholar]
- 35.Firulli BA, Krawchuk D, Centonze VE, Vargesson N, Virshup DM, Conway SJ, Cserjesi P, Laufer E, Firulli AB. Altered Twist1 and Hand2 dimerization is associated with Saethre-Chotzen syndrome and limb abnormalities. Nat Genet. 2005;37:373–381. doi: 10.1038/ng1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int J Dev Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.