To the editor,
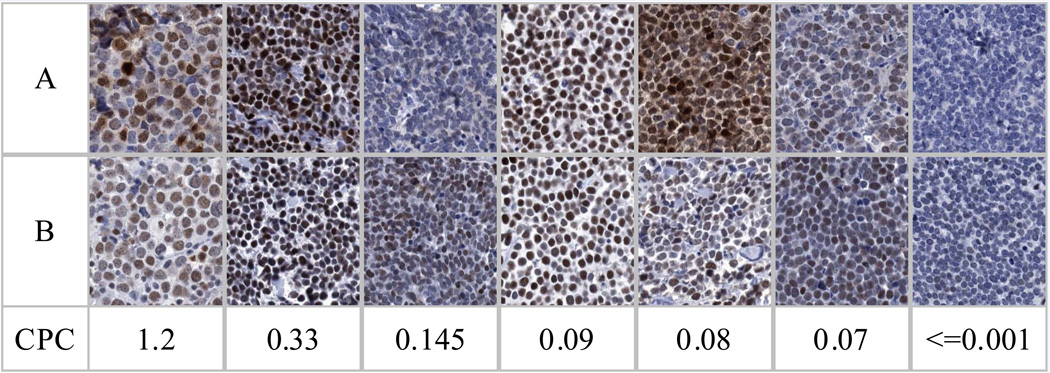
MCPyV, a new human oncogenic polyoma virus, appears etiologic for the classic, rare, aggressive Merkel Cell Carcinomas (MCC) of the skin characterized by MCPyV DNA integration, high virus copy numbers,1 expression of the potentially transforming MCPyV large T antigen (LTA) evidenced by reactivity to monoclonal antibody CM2B4,2–4 signature mutations in LTA,5 expression of retinoblastoma protein3,6 and a vigorous MCPyV specific neutralizing antibody response.7,8 Prevalence of this MCC phenotype has been reported as upward of 77–84% of MCC tested.1,3,9,10 Thus 16–23% of MCC may have a different etiology. MCPyV detection in MCC tissue includes MCCs with high and low virus copy numbers, detection ranges from 24% to 100% (Table 1a).1–4,6,9–19 Differences in relative proportions of MCC with low and high viral copies may also be significantly influenced by geographic region (Table 1b).13 All MCC tumors in our study population6 with viral abundance of 0.06–1.2 DNA copies/cell conform to the reported MCC MCPyV positive phenotype with demonstrable MCPyV LTA (IHC CM2B4, kindly provided by Dr. Moore) and retinoblastoma protein. (Figure 1A) Our tumor study population additionally contained variant MCC with low viral DNA (.0005–.0035 copies/cell) or no detectable virus; these later two subgroups were negative for LTA (CM2B4) and retinoblastoma protein. These data are similar to the more recent data from Busam et al.4 who found 5/15 MCPyV positive MCC non-reactive to CM2B4 and are consistent with the recent comprehensive characterization of MCC cell lines.20
Table 1.
| Table 1a: Summary of reports indicating range of MCPyV prevalence and viral load in Merkel cell carcinomas (MCC). | ||||
|---|---|---|---|---|
| N | Study | Number of MCC samples tested |
Prevalence in MCC (%) |
Range of relative viral DNA levels |
| 1 | Andres11 | 33 | 64 | not available (NA) |
| 2 | Becker10 | 75 | 84.9 | NA |
| 3 | Bhatia6 | 23 | 74 | 0.0005 to 1.2 copies per cell |
| 4 | Busam4 | 43 | 88 | NA |
| 5 | Duncavage12 | 41 | 78 | NA |
| 6 | Feng1 | 10 | 80 | > 1 in 6/8 samples |
| 7 | Garneski13 | 37 | 43 | 0.0001 to 10 viral DNA per cell |
| 8 | Houben3 | 50 | 86 | 0.001 to > 100 copies per cell |
| 9 | Kassem9 | 39 | 77 | NA |
| 10 | Loyo14 | 7 | 86 | 0.05 to 175 copies per genome |
| 11 | Sastre-Garau15 | 10 | 100 | 0.6 to 62.2 genome-equivalent per carcinoma cell |
| 12 | Sihto16 | 114 | 79.8 | 0.0003 to 4224 viral DNA to control DNA ratio |
| 13 | Shuda2 | 36 | 70 | NA |
| 14 | Touzé17 | 32 | 66 | NA |
| 15 | Varga18 | 9 | 78 | NA |
| 16 | Wetzels19 | 5 | 40 | NA |
| Table 1b: Summary by geographic region of samples from three studies reporting three subgroups of MCPyV viral loads in Merkel cell carcinomas (MCC). Percentages of high, low and no viral detection appear to correlate with sun exposure as suggested by Garneski et al.13 | |||||
|---|---|---|---|---|---|
| Region | Studies (subset) | N | % High | % Low | % No |
| Finland | Sihto16 | 114 | 77.2 | 2.6 | 20.2 |
| N. America | Garneski13 (N. America) and Bhatia6 combined | 39 | 41.0 | 30.8 | 28.2 |
| Australia | Garneski13 (Australia) | 21 | 4.8 | 19.0 | 76.2 |
Figure 1.
Figure 1A: Immunohistochemical staining of MCC was performed for the detection of expression of retinoblastoma protein (pRb) using the commercially available antibody from Novocastra (clone 13A10) and for the detection of the LTA encoded by MCPyV using the CM2B4 mAb obtained as a gift from Dr. Patrick Moore and Yuan Chang (University of Pittsburgh). These antibodies were applied to formalin-fixed paraffin-embedded Merkel cell carcinoma tumor core samples in an MCC tissue microarray. MCPyV copies per cell (CPC) were determined by real time PCR using Taqman technology. Details of the PCR assay have been described previously.6 MCPyV CPC associates with (row A) expression of MCPyV LTA using CM2B4 mAb and (row B) expression of pRb.
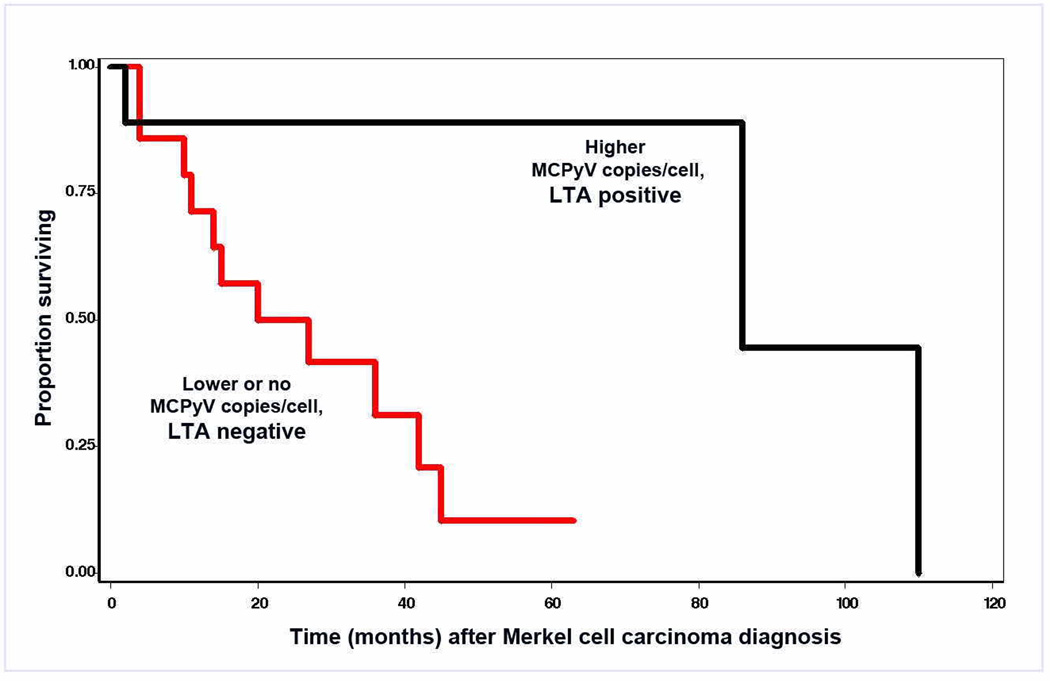
Figure 1B: Correlation between overall survival (Kaplan Meier) and the presence of MCPyV LTA by CM2B4 mAb staining demonstrates that patients with LTA positive tumors survived longer following a diagnosis of MCC than patients with LTA negative tumors. By Cox relative hazard (RH) analysis, likelihood of survival was lower per level (HR=0.35, CI=0.16–0.77, P=0.009). The number of cases in each category is: LTA positive, viral positive, 9; LTA negative, viral positive, 8; and LTA negative, viral negative, 6. Patients with viral positive tumors tended to be younger (P=0.07) but did not differ by sex. Additional characteristics of these patients have been described previously.6
Classic MCC viral oncogenesis characterized by abundance of MCPyV is clinically relevant as suggested by Sihto et al,16 who demonstrated that viral abundance in MCC correlates with survival. Our result6 supports this conclusion. When our patient MCC population is segregated by high vs. low or no viral copies/cell, survival is best for those with high viral copies/cell and worse for MCC likely initiated through other modes of oncogenesis (Figure 1B). We differ from the report by Becker et al.3,10 where high viral copy numbers were present in most of their samples. Lack of CM2B4 reactivity in a tumor with high viral copy number has been described.2 Some CM2B4 negative MCC might be ascribed to variations in large T antigenic epitopes. However, high correlation among viral load, retinoblastoma protein and CM2B4 reactivity in our data suggests that viral low-abundance or negative MCC tumor cells may have no or insufficient LTA for detection with CM2B4. Several recent reports have demonstrated presence of MCPyV in normal skin and other tumors.6,11,14,21 Our results do not allow us to discriminate between the presence of MCPyV in MCC tumor cells versus the presence of MCPyV in surrounding non-tumor cells; this issue is germane to the relevance of MCPyV mediated tumorigenesis particularly in MCC with low viral abundance. One approach to address this would be to examine whether MCPyV genome is integrated in the host genome, a process that is likely to indicate a direct role for MCPyV mediated oncogenesis. The lack of fresh/frozen tumor samples however, makes such a study technically unfeasible. Integration of MCPyV genome is also often associated with the presence of truncating mutations in the LTA and while it is unknown whether mutations occur prior to integration, the presence of such mutations may provide an alternative surrogate for supporting a pathogenic role of MCPyV. However recent data21 suggest that identical truncating mutations are detected in both tumor and adjacent normal skin tissue, thus the pathogenic relevance of detecting truncating mutations warrant additional investigation. While technically feasible, the sensitivities for detecting mutations particularly from low abundance viral DNA pose several challenges and will still fail to directly address the issue whether the viral DNA is associated with the tumor cells or the non tumor cells in the sample. Nonetheless the rarity of cells in MCC with low amplification of virus would be inconsistent with the presence of the viral DNA prior to clonal expansion in these tumors.
To appreciate the differences in oncogenesis and MCC survival, the MCC study population must be heterogeneous and contain tumors with high, low and no viral detection. While artifact related to specimen quality and differences in detection methods could add to this heterogeneity, tumor factors such as mutations, ultraviolet (UV) DNA damage and patient clinical parameters such as immunosuppression must contribute. Compared to specimens from North America, MCC tissues from Australia might be expected to be virus negative because high UV radiation exposure of melanin-deficient skin probably contributes to MCC development.13 Our data (Figure 1A) imply that MCPyV positive and negative tumors differ in oncoprotein expression and prognosis. This may be important in establishing clinical correlates that will be useful worldwide.
Clarifying the role of MCPyV in MCC will require accurate quantification of viral abundance. Houben et al.3 contend that most MCCs carry at least one viral copy per cell and do not find MCC with low viral abundance. In contrast, we and others6,13,16 have found very low viral loads (.0001–.0035 copies/cell) in 19 of 174 MCC tumors tested (Table 1). Both we and Garneski et al.6,13 observe what appear to be discrete categories of high- and low-abundance MCC tumors. Estimation of viral abundance is susceptible to differences in tissue quality, proportion of tumor and non tumor cells in the sample, primers selected for PCR amplification, target cellular genomic sequences used as a reference, and other technical differences, but it appears that at least a proportion of MCCs carry viral DNA that equates to less than 1 in 300 cells infected with MCPyV. Notably, similar low abundance of viral association has been described in MCC cell lines, suggesting that mere viral association may not provide growth advantage, at least in vitro, over non viral infected MCC.20
Finally, there is the question of how expression of oncoproteins in MCC differs among the classic MCPyV DNA and variant phenotypes. We find heterogeneity in MCC as to abundance or absence of detectable MCPyV and the presence of oncoproteins as well as geographic, clinical, or patient factors. All may be critically important for prognosis and therapy. To this end, the development of a multi-investigator, multi-institutional consensus study using an international, geographically diverse MCC tumor population with a pre-determined common set of reagents and methods to estimate MCPyV abundance and oncoproteins in primary, recurrent and metastatic lesions, geographic distributions of MCC types and correlations with clinical features of MCC would be an important step.
Acknowledgments
We are grateful to Drs. Patrick Moore and Yuan Chang (University of Pittsburgh) for sharing the antibody CM2B4 prior to its publication. We thank Dr Allan Hildesheim (IIB, DCEG, NCI) for useful discussions and for critical reading of the manuscript. The work described in this letter was supported by NCI Intramural Research Funding and OSU Comprehensive Cancer Center Research Enhancement and Assistance Program (REAP 200925).
References
- 1.Feng H, Shuda M, Chang Y, Moore PS. Clonal Integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernández-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, et al. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int J Cancer. 2009;125:1243–1249. doi: 10.1002/ijc.24510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houben R, Schrama D, Alb M, Pföhler C, Trefzer U, Ugurel S, Becker JC. Comparable expression and phosphorylation of the retinoblastoma protein in merkel cell polyoma-virus positive and negative merkel cell carcinoma. Int J Cancer. 2009 Jul 27; doi: 10.1002/ijc.24790. [Epub ahead of print] PMID 19637243. [DOI] [PubMed] [Google Scholar]
- 4.Busam KJ, Jungbluth AA, Rekthman N, Coit D, Pulitzer M, Bini J, Arora R, Hanson NC, Tassello JA, Frosina D, Moore P, Chang Y. Merkel Cell Polyomavirus Expression in Merkel Cell Carcinomas and Its Absence in Combined Tumors and Pulmonary Neuroendocrine Carcinomas. Am J Surg Pathol. 2009 Jul 15; doi: 10.1097/PAS.0b013e3181aa30a5. [Epub ahead of print] PMID 19609295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Nat. Acad. Sci (USA) 105:16272–16277. doi: 10.1073/pnas.0806526105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatia K, Goedert JJ, Modali R, Preiss L, Ayers LW. Merkel cell carcinoma subgroups by merkel cell polyomavirus DNA relative abundance and oncogene expression. Int J Cancer. 2009 Jun 23; doi: 10.1002/ijc.24676. [Epub ahead of print] PMID 19551862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pastrana DV, Tolstov YL, Becker JC, Moore PS, Chang Y, Buck CB. Quantitation of human seroresponsiveness to Merkel cell polyomavirus. PLoS Pathog. 2009 Sep;5(9):e1000578. doi: 10.1371/journal.ppat.1000578. Epub 2009 Sep 11. PMID: 19750217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carter JJ, Paulson KG, Wipf GC, Miranda D, Madeleine MM, Johnson LG, Lemos BD, Lee S, Warcola AH, Iyer JG, Nghiem P, Galloway DA. Association of Merkel cell polyoma virus specific antibodies with Merkel Cell Carcinoma. Jour Natl Can Inst. 2009 Sep 23; doi: 10.1093/jnci/djp332. [Epub ahead of print]PMID: 19776382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kassem A, Schöpflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- 10.Becker JC, Houben R, Ugurel S, Trefzer U, Pföhler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- 11.Andres C, Belloni B, Puchta U, Sander CA, Flaig MJ. Prevalence of MCPyV in Merkel cell carcinoma and non-MCC tumors. J Cutan Pathol. 2009 Jul 14; doi: 10.1111/j.1600-0560.2009.01352.x. [Epub ahead of print] PMID 19615033. [DOI] [PubMed] [Google Scholar]
- 12.Duncavage EJ, Zehnbauer BA, Pfeifer JD. Prevalence of Merkel cell polyomavirus in Merkel cell carcinoma. Mod Pathol. 2009;22:516–521. doi: 10.1038/modpathol.2009.3. [DOI] [PubMed] [Google Scholar]
- 13.Garneski KM, Warcola AH, Feng Q, Kiviat NB, Leonard JH, Nghiem P. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J Invest Dermatol. 2009;129:246–248. doi: 10.1038/jid.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loyo M, Guerrero-Preston R, Brait M, Hoque M, Chuang A, Kim M, Sharma R, Liégeois N, Koch W, Califano J, Westra W, Sidransky D. Quantitative detection of merkel cell virus in human tissues and possible mode of transmission. Int J Cancer. 2009 Jul 8; doi: 10.1002/ijc.24737. [Epub ahead of print] PMID 19588496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sastre-Garau X, Peter M, Avril MF, Laude H, Couturier J, Rozenberg F, Almeida A, Boitier F, Carlotti A, Couturaud B, Dupin N. Merkel cell carcinoma of the skin: pathological and molecular evidence for a causative role of MCV in oncogenesis. J Pathol. 2009;218:48–56. doi: 10.1002/path.2532. [DOI] [PubMed] [Google Scholar]
- 16.Sihto H, Kukko H, Koljonen V, Sankila R, Böhling T, Joensuu H. Clinical factors associated with Merkel cell polyomavirus infection in Merkel cell carcinoma. J Natl Cancer Inst. 2009;101:938–945. doi: 10.1093/jnci/djp139. [DOI] [PubMed] [Google Scholar]
- 17.Touzé A, Gaitan J, Maruani A, Le Bidre E, Doussinaud A, Clavel C, Durlach A, Aubin F, Guyétant S, Lorette G, Coursaget P. Merkel cell polyomavirus strains in patients with merkel cell carcinoma. Emerg Infect Dis. 2009;15:960–962. doi: 10.3201/eid1506.081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varga E, Kiss M, Szabó K, Kemény L. Detection of Merkel cell polyomavirus DNA in Merkel cell carcinomas. Br J Dermatol. 2009 May 12; doi: 10.1111/j.1365-2133.2009.09221.x. [Epub ahead of print] PMID 19438857. [DOI] [PubMed] [Google Scholar]
- 19.Wetzels CT, Hoefnagel JG, Bakkers JM, Dijkman HB, Blokx WA, Melchers WJ. Ultrastructural proof of polyomavirus in Merkel cell carcinoma tumour cells and its absence in small cell carcinoma of the lung. PLoS One. 2009;4:e4958. doi: 10.1371/journal.pone.0004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischer N, Brandner J, Fuchs F, Moll I, Grundhoff A. Detection of Merkel cell polyomavirus (MCPyV) in Merkel cell carcinoma cell lines: Cell morphology and growth phenotype do not reflect presence of the virus. Int J Cancer. 2009 Sep 8; doi: 10.1002/ijc.24877. [Epub ahead of print] PMID: 19739110. [DOI] [PubMed] [Google Scholar]
- 21.Dworkin AM, Tseng SY, Allain DC, Iwenofu OH, Peters SB, Toland AE. Merkel Cell Polyomavirus in Cutaneous Squamous Cell Carcinoma of Immunocompetent Individuals. J Invest Dermatol. 2009 Jun 25; doi: 10.1038/jid.2009.183. [Epub ahead of print] PMID: 19554019. [DOI] [PubMed] [Google Scholar]