Abstract
Proteins play diverse and critical roles in cellular ribonucleoproteins (RNPs) including promoting formation of and stabilizing active RNA conformations. Yet, the conformational changes required to convert large RNAs into an active RNPs have proven difficult to characterize fully. Here we use high-resolution approaches to monitor both local nucleotide flexibility and solvent accessibility for nearly all nucleotides in the bI3 group I intron RNP in four assembly states: the free RNA, maturase-bound RNA, Mrs1-bound RNA, and the complete six-component holocomplex. The free RNA is misfolded relative to the secondary structure required for splicing. The maturase and Mrs1 proteins each stabilized long-range tertiary interactions but neither protein alone induced folding into the functional secondary structure. In contrast, simultaneous binding by both proteins results in large secondary structure rearrangements in the RNA and yielded the catalytically active group I intron structure. Secondary and tertiary folding of the RNA component of the bI3 RNP are thus not independent: RNA folding is strongly non-hierarchical. These results emphasize that protein-mediated stabilization of RNA tertiary interactions functions to pull the secondary structure into an energetically disfavored, but functional, conformation and emphasize a new role for facilitator proteins in RNP assembly.
Large RNAs rarely achieve a functional structure without the aid of a wide variety of protein facilitators (1). Protein facilitators are generally classified into two groups, cofactors and chaperones, which are distinguished by both their mode of action and interactions with RNA (2–4). Cofactors bind tightly to an RNA and promote or enhance the formation of otherwise unfavorable tertiary interactions. Conversely, chaperones make transient interactions with an RNA to facilitate the rearrangement of secondary structure and allow an RNA to more readily achieve a thermodynamically favorable structure. Folding for some RNAs is strongly hierarchal: the secondary structure forms first, followed by tertiary structure (5, 6). However, there are also many examples in which RNA tertiary structure either folds in tandem with the secondary structure or changes the secondary structures sampled during folding (7–15). A protein cofactor that stabilizes an RNA tertiary structure element might thus have a dramatic impact on secondary structure. A protein that made this sort of “non-hierarchical” contribution to RNA folding would exemplify another type of facilitator, distinct from roles thus far associated with cofactors and chaperones.
Group I introns are ideal model systems for study of the impact of protein facilitators on RNA structure. The conserved group I intron catalytic core is composed of two roughly coaxially stacked domains (termed the P5-P4-P6 and P9-P7-P8-P3 domains), which dock with a third domain (the P1-P2 domain) to form a complex active site (19–22). The two-step splicing reaction is initiated by binding of an exogenous guanosine to the folded catalytic core. Distinct group I intron ribozymes interact with both protein cofactors and chaperones to achieve this conserved and catalytically active structure (2–4, 16, 17) and binding of protein cofactors can change the pathway for RNA folding (18).
The bI3 ribonucleoprotein (RNP) is a six-component complex consisting of a group I intron RNA, a monomer of the bI3 maturase protein, and two dimers of the Mrs1 protein (23, 24). The splicing function of the bI3 RNA requires stable binding by both proteins. The maturase monomer binds to a peripheral element in the P5-P4-P6 domain, over 50 Å from the active site (25). Two Mrs1 protein dimers bind cooperatively to the bI3 RNA and promote formation of two GNRA tetraloop-receptor interactions that mediate long-range inter-domain RNA tertiary contacts (26).
We have probed the secondary and tertiary structure of the bI3 RNA in four conformational states: the free RNA, maturase-bound RNA, Mrs1-bound RNA, and the complete holocomplex. We monitored local nucleotide dynamics and solvent accessibility at nearly every nucleotide in the RNA using high-throughput selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) and hydroxyl radical cleavage approaches, respectively. The free RNA is substantially misfolded relative to the known, catalytically active, secondary structure for group I introns. Binding by either the maturase or Mrs1 proteins stabilized long-range RNA tertiary structures, but did not cause the functional secondary structure to form. In the presence of both proteins, however, the RNA underwent large-scale rearrangements in its base pairing pattern to achieve the active group I intron structure. This study thus reveals a new role for protein cofactors. In assembly of the bI3 complex, protein stabilization of long-range tertiary structures induced extensive rearrangements in RNA secondary structure in regions located far from the protein binding sites. The maturase and Mrs1 proteins make “non-hierarchical” contributions to bI3 RNA folding and perform roles distinct from those conventionally assigned to cofactors and chaperones.
Experimental Procedures
bI3 RNA, Maturase and Mrs1
The bI3-ΔL8 RNA was synthesized and purified as described (27). The RNA sequence contained 7 nts of 5'-vector sequence, the entire 77 nt 5'-exon, the 372 nt intron, 30 nts of the 3'-exon, and 60 nts of 3'-vector sequence. The ΔCys-bI3 maturase and Mrs1 proteins were expressed and purified as described (23, 27).
High-Throughput SHAPE
The bI3 RNA (3 pmol) was heated at 95 °C in water for 1 min, placed on ice for 1 min, and re-folded at 37 °C for 10 min in reaction buffer [40 mM Mops (pH 8.0), 80 mM potassium acetate (KOAc) (pH 8.0), 20 mM MgCl2] in a final volume of 25 μl. Experiments with the free RNA contained 2 μl protein buffer [40 mM Mops (pH 7.5), 400 mM NaCl, 5 mM DTT, 10 % (v/v) glycerol]; if present, 2 μl of 6 μM maturase and Mrs1 were added. Proteins were allowed to bind at 37 °C for 30 min. Reaction mixtures were added to 3 μl of 5 mM 1M7 (1-methyl-7-nitroisatoic anhydride, dissolved in anhydrous DMSO) at 37 °C and allowed to react for 70 sec (equal to 5 hydrolysis half lives) (28). No-reagent control reactions were performed by omitting 1M7. Complexes were subsequently incubated with proteinase K (60 μg, 37 °C, 30 min) and proteins were removed by phenol:cholorform:isoamyl alcohol (25:24:1) extraction. Modified RNA was precipitated with ethanol and resuspended in water. Sites of RNA modification were identified by reverse transcriptase-mediated primer extension using fluorescently labeled primers; products were resolved by capillary electrophoresis (27).
High-Throughput Hydroxyl Radical Cleavage
RNA-protein complexes were prepared as described for SHAPE experiments in a volume of 21 μl. Reactions were initiated by sequential addition of 3 μl each of 2.5 mM (NH4)Fe(SO4)2/3.75 mM EDTA (pH 8.0), 0.1% H2O2, and 50 mM sodium ascorbate. Reactions were quenched after 5 min at 37 °C with 10.5 μl 75% glycerol (v/v). The no-reaction control was performed in the absence of Fe(II)-EDTA. Reactions were treated with proteinase K and visualized by primer extension as described above.
Data Analysis
SHAPE and hydroxyl radical data from the Applied Biosystems 3130 were analyzed using ShapeFinder (29) as described (27). After integration of all peaks in the (+) and (−) reagent traces, reactivities were normalized to a scale in which 1.0 was defined as the mean intensity of highly reactive positions (26). Hydroxyl radical cleavage data were smoothed over a window of three nucleotides. Analysis of RNAs of known structure (26) indicates that hydroxyl radical cleavage intensities of less than one-half the mean correlate with solvent inaccessible positions. For secondary structure predictions, SHAPE data were converted to pseudo-free energy change terms and used to constrain a thermodynamics-based algorithm (slope and intercept were 2.5 and −0.6, respectively) as previously described (30).
Free RNA Structure Model
RNA sequence and base pairing constraints determined from SHAPE experiments were used to prepare an input file for a discrete molecular dynamics (DMD) refinement (31, 32). The RNA was folded from an extended linear structure by reducing the simulation temperature until all base pairing constraints were satisfied. The resulting structural models were clustered (33) and the centroid of the most populated cluster was used as the RNA model. Structure images were composed using PyMOL (Delano Scientific). The model for the final complex was described previously (26).
Results
Strategy
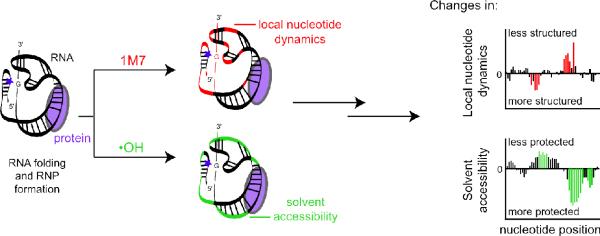
We used a combination of two high-throughput and high resolution techniques to comprehensively evaluate RNA structure during protein-assisted folding (Figure 1). We interrogated local nucleotide dynamics using SHAPE (28, 34–36). In a high-throughput SHAPE experiment, RNA structure is evaluated by monitoring the reactivity of hydroxyl-selective electrophiles towards the ribose 2′-hydroxyl. The resulting bulky 2′-O-ester adducts are detected as stops to primer extension from color-coded fluorescent primers. After separation by capillary electrophoresis and integration of peak intensities, we obtained absolute SHAPE reactivities for almost every nucleotide in the bI3 RNA. Data were normalized to a scale spanning 0 to ~2, where 1.0 is defined as the average intensity of highly reactive nucleotides. Nucleotides with moderate to high reactivities correlate strongly with single-stranded regions. Weakly reactive nucleotides are conformationally constrained and are therefore likely either to be base paired or to participate in highly structured tertiary interactions.
Figure 1.
Schematic of comprehensive, high-throughput, and high resolution structural analysis of the bI3 RNP. Local nucleotide flexibility and dynamics were probed using SHAPE chemistry (top, red) in which the hydroxyl-selective electrophile, 1M7, reacts preferentially at conformationally flexible nucleotides. Solvent accessibility at the RNA backbone was approximated by extent of hydroxyl radical (•OH) mediated cleavage (bottom, green). Changes in local nucleotide dynamics and solvent accessibility were detected at single nucleotide resolution in quantitative difference plots.
In the complementary approach, we measured the solvent accessibility of each nucleotide using high-throughput hydroxyl radical footprinting experiments (Figure 1). Hydroxyl radicals are generated in situ from H2O2 in a reaction catalyzed by Fe(II)-EDTA (37). Backbone cleavage is roughly correlated with ribose solvent accessibility (38–40). Hydroxyl radical-induced cleavages at accessible nucleotides are, again, identified as stops to primer extension (41, 42). Weakly cleaved nucleotides, taken to be cleavage intensities of less than one-half the mean, are solvent inaccessible and buried within the RNA or RNP structure.
Both SHAPE and hydroxyl radical cleavage patterns can be analyzed quantitatively by capillary electrophoresis. Differences between states are identified by subtracting any two reactivity patterns from each other (Figure 1). The nucleotide flexbility of the bI3 RNA monitored by SHAPE and difference plot histograms for maturase-bound, Mrs1-bound, and the holocomplex are shown in Figure 2. Protein-mediated changes in backbone solvent accessibility in the free bI3 RNA and difference plot histograms for RNA bound by maturase alone, Mrs1 alone, and both proteins are shown in Figure 3.
Figure 2.
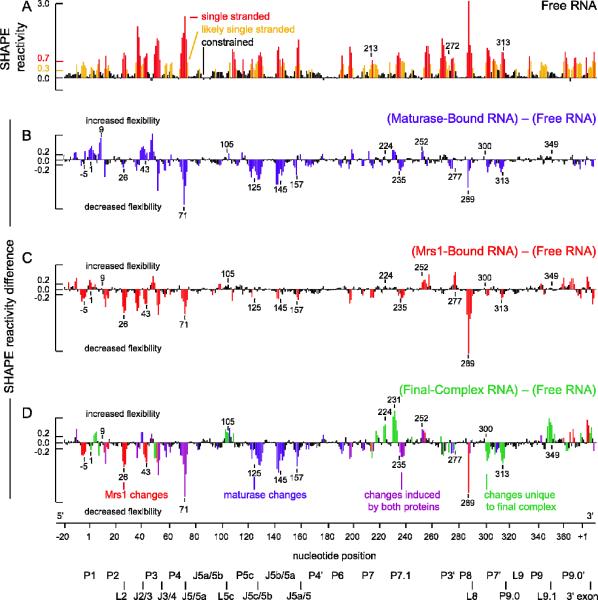
Protein-assisted folding of the bI3 RNA monitored by SHAPE. (A) Reactivity as a function of nucleotide position for the free RNA. Bars are colored by reactivity. Difference plot histograms for RNA bound to (B) maturase, (C) Mrs1, and (D) both maturase and Mrs1. Nucleotides with absolute changes in reactivity greater than 0.2 (two-fold above background) are colored by the protein that induces a given effect: blue for maturase, red for Mrs1, purple if affected by both proteins and green for unique changes seen only in the holocomplex. Structural landmarks are highlighted below the axis.
Figure 3.
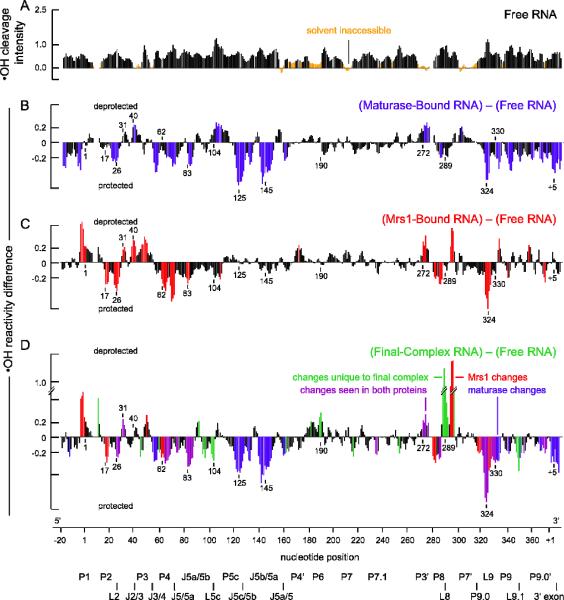
Protein-mediated changes in backbone solvent accessibility in the bI3 RNA. (A) Cleavage intensity versus nucleotide position for the free RNA. Nucleotides with reactivities less than one-half the mean are indicated in orange. Difference plot histograms for RNAs bound by (B) maturase alone, (C) Mrs1 alone, and (D) both proteins. Nucleotides with large protein-induced changes in reactivity are colored as in Figure 2. Structural landmarks are indicated below the axis.
The Free RNA is Misfolded Prior to Protein Binding
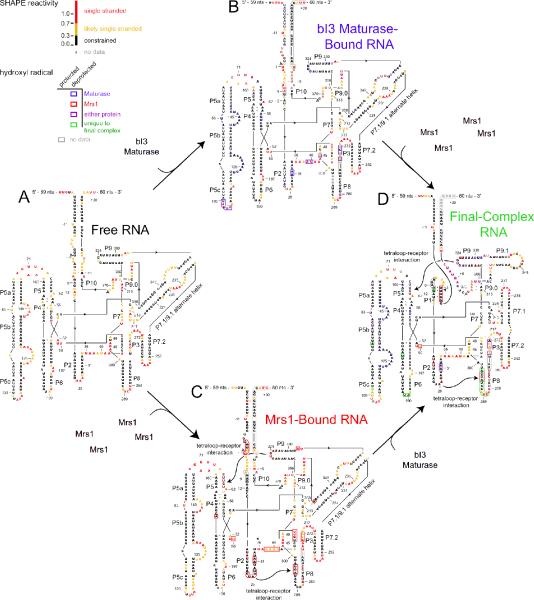
Previous analysis of the free bI3 RNA by SHAPE indicated that about half the RNA does not fold to the catalytically active and conserved group I intron secondary structure (27). Six specific RNA elements are misfolded as compared to the secondary structure expected for the catalytically active structure (Figure 4A). Nucleotides in the conserved P3, P7, and P9.0 helices all had high SHAPE reactivities (nucleotides near positions 272, 213, and 313, respectively; Figures 2A, 4A). High SHAPE reactivities indicate that these nucleotides do not form stable paired elements in the free RNA state. In addition, nucleotides in the P1, P7.1, and P9.1 elements each had SHAPE reactivity patterns consistent with alternate base pairing interactions. Instead of folding as individual helices, the P7.1 and P9.1 sequences base pair with each other to form the P7.1/9.1 alternate helix (Figure 4A). Similarly, sequences in the highly conserved P1 helix base pair with nucleotides in the 3′ exon (27). At a minimum, each of these six misfolded elements must undergo extensive secondary structure rearrangements to form a catalytically active group I intron complex.
Figure 4.
RNA folding in the six-component bI3 RNP. Secondary structural models for the (A) free RNA, (B) maturase-bound RNA, (C) Mrs1-bound RNA, and (D) RNA bound to both proteins. Nucleotides are colored to reflect their SHAPE reactivities in each RNA state. Filled and hollow boxes highlight hydroxyl radical cleavage protections and enhancements, respectively, and are colored according to their protein effector (blue, red, purple, and green for maturase, Mrs1, either protein, or unique to the final complex, respectively). Px labels indicate paired elements in the RNA.
We assessed the degree of solvent inaccessible tertiary structure formed in the free RNA using hydroxyl radical footprinting. Most nucleotides in the free RNA were accessible to cleavage (black nucleotides, Figure 3A), indicating that little tertiary structure was formed in the free RNA prior to protein binding. A few regions were inaccessible to hydroxyl radical cleavage: Those regions included the P1, J2/3, P3, P4, J8/7, P7, and P9.0 elements. Several of these solvent inaccessible regions, including sequences corresponding to P1, P3, and P7, are elements with misfolded base pairs in the protein-free RNA (see Figure 4A). Taken together, SHAPE and hydroxyl radical data indicate that the free RNA forms a catalytically inactive structure that is misfolded in both its secondary and tertiary structure relative to the catalytically active state.
The bI3 RNA Structure in the Functional Complex
When both the bI3 maturase and Mrs1 proteins were stably bound, we observed complex and extensive changes in SHAPE reactivity consistent with large-scale secondary structure rearrangements. We quantified these changes by subtracting the nucleotide-resolution reactivity pattern of the RNP complex from those for the free RNA in a difference plot (Figures 2D and 3D). Regions with the largest changes in reactivity are colored in panels B-D of Figures 3 & 4. In the P7.1 element, for example, nucleotides near positions 224 and 231 increased in SHAPE reactivity, whereas, nucleotides near 235 decreased in reactivity (labeled, Figure 2D). These reactivity changes are consistent with the interpretation that, in the complex, the P7.1/9.1 alternate helix is disrupted and the phylogenetically conserved P7.1 helix has formed (compare structural contexts of P7.1 nucleotides in Figures 4A and 4D). In the functional RNP complex, all six previously identified misfolded elements (P1, P3, P7, P7.1, P9.1, and P9.0) had SHAPE reactivities consistent with formation of the accepted group I intron secondary structure (Figure 4D). In addition, several linker regions and loops had low reactivities in the RNP complex, suggesting that the RNA forms the compact tertiary interactions necessary for group I intron splicing.
Analyzing nucleotide solvent accessibility in the complex corroborated these conclusions. Differences in hydroxyl radical cleavage were visualized in a quantitative difference plot (Figure 3D). Upon formation of the holocomplex, one-third of the nucleotides in the bI3 RNA were protected from cleavage. In addition, several sets of nucleotides, such as nucleotides surrounding positions 190 and 289, showed enhanced cleavage indicating that these nucleotides were more exposed to solvent than they were in the RNA alone. We interpret these data as evidence for significant structural rearrangements at these nucleotides. Structural changes were seen in nearly all major group I intron RNA structures when reactivities in the free RNA were compared to those in the complex (colored regions & boxes, Figure 4D).
The bI3 Maturase Stabilizes Tertiary Structures in the P5-P4-P6 Domain
We then used SHAPE to determine which of the structural rearrangements that distinguish the free RNA and the RNP complex were due to maturase binding. The bI3 maturase binds in the P5c helix of the P5-P4-P6 domain and promotes tertiary folding of this domain, including formation of two sets of interaction between adenosine residues and minor groove elements (A-minor interactions) (25). As expected, SHAPE reactivity decreased in the regions of the P5-P4-P6 domain predicted to be flexible in the free RNA, including in the J5/5a, J5c/5b, J5b/5a, and J5a/5 elements (positions 71, 125, 145, and 157 highlighted in blue in Figure 2B). These decreases in reactivity were maintained in the RNP complex (blue/purple bars, Figure 2D). Other changes, outside the P5-P4-P6 domain, were also maintained in the RNP complex, including in the J3/4, P7.1, P3, P7.2, and J8/7 elements (positions 40, 235, 275, 252, and 307 indicated by blue/purple bars in Figure 2D). Maturase binding also induced changes in other regions of the RNA that were not observed in the holocomplex (nucleotides 1, 9, and 43 in Figures 2B, 2D).
Tertiary structure changes induced by maturase binding were examined using hydroxyl radical footprinting. Maturase binding had dramatic effects in the P5-P4-P6 domain and at the 5′ and 3′ ends of the intron (blue bars, Figure 3B). As expected, several elements in the P5-P4-P6 domain became protected upon protein binding, including the two sets of A-minor interactions (nucleotides at positions 125 and 145, Figures 3B, 4B). Additional cleavage differences were seen in the 5′ and 3′ exons and in the P2, P3, P8, P9, and P9.1 helices (positions 1, 26, 272, 289, 324, and +5 indicated in blue in Figures 3B, 4B). Most of these tertiary changes induced by maturase binding were maintained in the holocomplex (blue/purple bars, Figure 3D). However, a subset of changes were not observed in the holocomplex, including differences in regions of the 5′ exon, the 3′ end of the intron, and the P5-P4-P6 domain near nucleotide 104 (compare Figures 3B and 3D). These data suggest, first, that maturase binding promotes tertiary structure folding in the P5-P4-P6 domain and, second, that some regions in the maturase-stabilized structure undergo additional conformational rearrangements in order to form the complete functional RNP.
Mrs1 Stabilizes Two Tetraloop-Receptor Interactions
A difference plot analysis of the SHAPE data for the Mrs1-bound and the free RNA shows decreases in reactivity throughout the RNA (red bars, Figure 2C). Mrs1 binding induced extensive changes in the stability of individual secondary structure elements of the bI3 RNA (Figure 4C). However, as with the maturase, Mrs1 binding had little effect on the P1, P7.1, or P9.1 elements, indicating that these structures were still misfolded relative to the catalytically active conformation. Most Mrs1 effects on the bI3 RNA structure were maintained in the complex (red/purple bars, Figure 2D). Hydroxyl radical footprinting of the Mrs1-bound RNA emphasized that most changes were localized at the two GNRA tetraloop-receptor interactions in the bI3 RNA (red bars, Figure 3C). Both the L2 and L9 tetraloops were strongly protected from cleavage upon Mrs1 binding (at positions 26 and 324, Figure 3C). Residues in receptor helices in P8 and P5 displayed both protections and enhancements from cleavage relative to the free RNA (see nucleotides 286, 295, and 62 in Figure 3C). Elements adjacent to the tetraloop-receptor motifs were also affected by Mrs1 binding, including the P1, P3, and P4 helices and the J2/3 and J5/5a joiner regions. In the SHAPE analysis, single-stranded sequences proximal to the receptor helices and tetraloops, such as the J2/3, J3/4, J5/5a, and L8, showed reduced reactivities upon Mrs1 binding (red nucleotides around 43, 51, 71, and 289, Figure 2C). These data indicate that Mrs1 promotes the formation of the tetraloop tertiary interactions in the bI3 RNA but does not directly modulate other higher-order interactions required to fold to the active structure.
Cooperative Interactions in the bI3 RNP
Individually, both the maturase and Mrs1 proteins stabilize distinct tertiary structures (blue and red bars; Figure 3D). In general, tertiary structure changes induced by binding of either protein alone occurred in peripheral structural elements distant from the catalytic core; however, effects of protein binding were seen throughout the RNA, indicating that structural changes propagate through connected RNA structures. There also exists a dynamic interplay between the maturase and Mrs1 proteins. For example, SHAPE reactivity around nucleotide 43 increased upon maturase binding, but decreased significantly upon Mrs1 binding (labeled, Figure 2). In the RNP complex, reactivity resembled that of Mrs1 binding alone, suggesting that Mrs1 has the stronger influence on RNA structure in this region. Thus, stabilization of tertiary structures upon protein binding influences the stability of local and distal secondary structures.
Both SHAPE and hydroxyl radical experiments revealed reactivity differences that occur only in the complex and not in either the maturase- or Mrs1-bound RNAs (green bars, Figures 2D and 3D). A major consequence of binding by both proteins is that all six of the RNA elements misfolded in the free RNA showed significant SHAPE reactivity changes in the complex (green nucleotides in P1, P3, P7, P7.1, P9.1, and P9.0 in Figure 2D). These differences in SHAPE reactivity correlated directly with the formation of the active secondary structure for the bI3 RNA (see P1, P3, P7, P7.1, P9.1, and P9.0, Figure 4D). Hydroxyl radical data corroborated this observation and demonstrated clear RNA structure rearrangements unique to the RNP complex RNA (green bars, Figure 3D).
Discussion
The folding of the bI3 RNP is a strikingly cooperative process in which two proteins that stabilize distinct tertiary interactions, work together to induce large-scale RNA secondary structure rearrangements. SHAPE and hydroxyl radical experiments showed that both the maturase and Mrs1 proteins perform traditional RNA cofactor functions by stabilizing specific tertiary interactions. The maturase stabilizes the U-shaped structure of the P5-P4-P6 domain, whereas Mrs1 promoted the formation of two GNRA tetraloop-receptor interactions that link the three RNA domains. Neither protein alone repaired the extensive misfolding seen in the free RNA. Instead, the RNA underwent a large-scale change in secondary structure only when bound by both proteins. Overall, assembly of this RNP was highly cooperative and non-hierarchical: Protein-mediated tertiary structure stabilization strongly influenced the RNA secondary structure adopted in the active complex.
Hybrid Roles for Proteins in the bI3 RNP
Proteins that facilitate RNA folding are typically categorized as cofactors or chaperones (2–4, 16). Both the maturase and Mrs1 stably bind the bI3 RNA (24) and function individually to stabilize the tertiary structure, much like conventional cofactors. In addition, protease treatment causes the RNA to relax back to the inactive secondary structure, which reinforces the idea that the maturase and Mrs1 proteins function as cofactors and must remain stably bound to influence the RNA secondary structure. Yet simultaneous binding of both proteins induces large-scale rearrangement of the secondary structure, a role related to that played by some RNA chaperones. In sum, these results indicate that Mrs1 and bI3 maturase exert their effects via a distinct mechanism.
The binding by both proteins to the bI3 RNA appears to induce tension in the connecting RNA segments that forces secondary structure rearrangement in regions distant from the direct protein interaction sites. In the individually bound states, both maturase and Mrs1 proteins induced long-range structural changes throughout the RNA. A subset of these effects was not maintained in the RNP complex. For instance, local structures in the misfolded P1 and P3 elements in the catalytic core were clearly modulated by the individual maturase or Mrs1 protein binding. These structural changes were not retained in the final complex (for example, positions near 1, 9, 43, and 277 in Figure 2). However, the structural effects of the individual protein bound states are amplified when both proteins bound the RNA. We propose that this accumulation of structural strain propagates through the bI3 RNA, eventually causing large-scale structural rearrangements that were observed in the functional RNA-protein holocomplex.
A Strain-Propagation Model for RNP Assembly
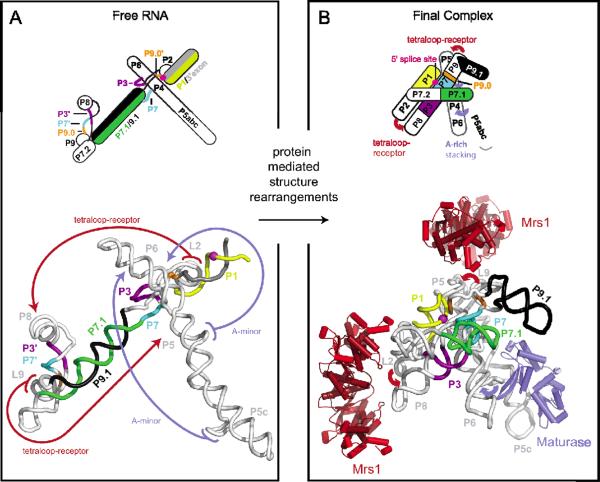
To further understand the proposed “strain-propagation” mechanism, we modeled the three-dimensional architecture of the RNA in both the protein-free and RNP states (Figure 5). Solvent accessibility probing indicated that the free RNA did not contain large solvent inaccessible regions and was therefore likely to be extended in solution (Figure 3A). Only local RNA interactions, base pairing and stacking, were included in the model of the free RNA (Figure 5A). The bI3 holocomplex (Figure 5B), including proteins, was previously shown to have the same tertiary architecture as visualized in high resolution group I intron structures (22, 26).
Figure 5.
Structural models of the (A) free bI3 RNA and (B) the RNA in the six-component RNP complex. RNA structural elements whose base pairing changes between the two models are emphasized in color. Red and blue arrows indicate long-range tertiary interactions that form in the holocomplex. Interactions promoted by the Mrs1 and maturase proteins are colored red and blue, respectively. The scissile phosphate, cleaved in the first step of splicing, is shown as a magenta sphere.
Critically, formation of the Alt P7.1/P9.1 helix has a dramatic effect on the architecture of the RNA (in black and green, Figure 5A). The long, stable Alt P7.1/P9.1 helix prevents the formation of the P3, P7, and P9.0 helices by physically separating base pairing partners (purple, cyan, and orange, respectively in Figure 5). Additionally, formation of the two GNRA tetraloop-receptor interactions (highlighted with red arrows, Figure 5A) is unfavorable due to the distance between interacting partners as enforced by the P7.1/P9.1 helix. The experimental data indicate that these two tetraloop-receptor interactions do form upon Mrs1 binding (Figures 3C, 3D, and 4C). However, binding of Mrs1 did not alter the secondary structure of the alternate helix (Figures 2C, 4C). Therefore, Mrs1 binding and subsequent formation of the two tetraloop-receptor interactions appears to strain the integrity of the long P7.1/P9.1 helix. Similarly, maturase binding induced tertiary folding of the P5-P4-P6 domain (blue arrows, Figure 5A) and induced other structural changes in the RNA, including in regions of the alternate helix (Figures 2B, 3B). We infer that binding by both proteins exerts sufficient combined stress on the alternate helix to induce or stabilize large-scale secondary structural rearrangement.
These structural studies on the cooperative and non-hierarchical folding of the bI3 RNP highlight the importance of obtaining comprehensive and high-resolution information when studying complex RNA-protein interactions. It was possible to generate high-resolution models of the various bI3 complexes because local nucleotide dynamics and solvent accessibility were measured at >96% of all nucleotides in the RNA. Maturase and Mrs1 binding on bI3 RNA structure impacted tertiary folding in regions of the bI3 RNA located far from their binding sites and, in so doing, changed both the secondary and tertiary structure of the RNA. These long-range protein-induced effects indicate that, collectively, the maturase and Mrs1 proteins play a newly identified role for protein facilitators of RNA folding. Given sufficiently comprehensive nucleotide-resolution information, it seems likely that complex contributions will also be identified in other RNA-protein systems.
Acknowledgments
This work was supported by a grant from the US National Institutes of Health (AI068462 to K.M.W.).
References
- 1.Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd ed. Cold Spring Harbor Laboratory Press; New York: 2006. [Google Scholar]
- 2.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 3.Weeks KM. Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 4.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat. Rev. Mol. Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 5.Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 6.Tinoco I, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 7.LeCuyer KA, Crothers DM. The Leptomonas collosoma spliced leader RNA can switch between two alternate structural forms. Biochemistry. 1993;32:5301–5311. doi: 10.1021/bi00071a004. [DOI] [PubMed] [Google Scholar]
- 8.Gluick TC, Draper DE. Thermodynamics of folding a pseudoknotted mRNA fragment. J. Mol. Biol. 1994;241:246–262. doi: 10.1006/jmbi.1994.1493. [DOI] [PubMed] [Google Scholar]
- 9.Wu M, Tinoco I., Jr. RNA folding causes secondary structure rearrangement. Proc. Natl. Acad. Sci. U.S.A. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed non-native RNA folding state. Nat. Struct. Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 11.Zheng M, Wu M, Tinoco I., Jr. Formation of a GNRA tetraloop in P5abc can disrupt an interdomain interaction in the Tetrahymena group I ribozyme. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3695–3700. doi: 10.1073/pnas.051608598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chauhan S, Caliskan G, Briber RM, Perez-Salas U, Rangan P, Thirumalai D, Woodson SA. RNA tertiary interactions mediate native collapse of a bacterial group I ribozyme. J. Mol. Biol. 2005;353:1199–1209. doi: 10.1016/j.jmb.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson KA, Merino EJ, Weeks KM. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- 14.Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J. Am. Chem. Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang B, Wilkinson KA, Weeks KM. Complex ligand-induced conformational changes in tRNA(Asp) revealed by single-nucleotide resolution SHAPE chemistry. Biochemistry. 2008;47:3454–3461. doi: 10.1021/bi702372x. [DOI] [PubMed] [Google Scholar]
- 16.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 17.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webb AE, Rose MA, Westhof E, Weeks KM. Protein-dependent transition states for ribonucleoprotein assembly. J. Mol. Biol. 2001;309:1087–1100. doi: 10.1006/jmbi.2001.4714. [DOI] [PubMed] [Google Scholar]
- 19.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 20.Cech TR. Self-Splicing of Group I Introns. Annu. Rev. Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 21.Woodson SA. Structure and assembly of group I introns. Curr. Opin. Struct. Biol. 2005;15:324–330. doi: 10.1016/j.sbi.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Vicens Q, Cech TR. Atomic level architecture of group I introns revealed. Trends Biochem. Sci. 2006;31:41–51. doi: 10.1016/j.tibs.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Bassi GS, de Oliveira DM, White MF, Weeks KM. Recruitment of intron-encoded and co-opted proteins in splicing of the bI3 group I intron RNA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:128–133. doi: 10.1073/pnas.012579299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bassi GS, Weeks KM. Kinetic and thermodynamic framework for assembly of the six-component bI3 group I intron ribonucleoprotein catalyst. Biochemistry. 2003;42:9980–9988. doi: 10.1021/bi0346906. [DOI] [PubMed] [Google Scholar]
- 25.Longo A, Leonard CW, Bassi GS, Berndt D, Krahn JM, Hall TM, Weeks KM. Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase. Nat. Struct. Mol. Biol. 2005;12:779–787. doi: 10.1038/nsmb976. [DOI] [PubMed] [Google Scholar]
- 26.Duncan CD, Weeks KM. The Mrs1 splicing factor binds the bI3 group I intron at each of two tetraloop-receptor motifs. PLoS One. 2010;5:e8983. doi: 10.1371/journal.pone.0008983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 29.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U.S.A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding F, Sharma S, Chalasani P, Demidov VV, Broude NE, Dokholyan NV. Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA. 2008;14:1164–1173. doi: 10.1261/rna.894608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gherghe CM, Leonard CW, Ding F, Dokholyan NV, Weeks KM. Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J. Am. Chem. Soc. 2009;131:2541–2546. doi: 10.1021/ja805460e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barton GJ. OC - A cluster analysis program. University of Dundee; Scotland, UK: 1993. [Google Scholar]
- 34.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 35.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 36.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tullius TD, Dombroski BA. Hydroxyl radical “footprinting”: high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc. Natl. Acad. Sci. U.S.A. 1986;83:5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 39.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr. Opin. Chem. Biol. 2005;9:127–134. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Balasubramanian B, Pogozelski WK, Tullius TD. DNA strand breaking by the hydroxyl radical is governed by the accessible surface areas of the hydrogen atoms of the DNA backbone. Proc. Natl. Acad. Sci. U.S.A. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shcherbakova I, Brenowitz M. Monitoring structural changes in nucleic acids with single residue spatial and millisecond time resolution by quantitative hydroxyl radical footprinting. Nature Protocols. 2008;3:288–302. doi: 10.1038/nprot.2007.533. [DOI] [PubMed] [Google Scholar]
- 42.McGinnis JL, Duncan CDS, Weeks KM. High-throughput SHAPE and hydroxyl radical analysis of RNA structure and ribonucleoprotein assembly. Meth. Enzymol. 2009;468:67–89. doi: 10.1016/S0076-6879(09)68004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]