Abstract
Rationale: The airflow limitation that defines severity of chronic obstructive pulmonary disease (COPD) is caused by a combination of small airway obstruction and emphysematous lung destruction.
Objectives: To examine the hypothesis that small airway obstructive and emphysematous destructive lesions are produced by differential expression of genes associated with tissue repair.
Methods: The expression of 54 genes associated with repair of repetitively damaged tissue was measured in 136 paired samples of small bronchioles and surrounding lung tissue separated by laser capture microdissection. These samples were collected from 63 patients at different levels of disease severity who required surgery for either lung cancer or lung transplantation for very severe COPD. Gene expression was measured by quantitative polymerase chain reaction in these paired samples and compared with the FEV1 by linear regression analysis.
Measurements and Main Results: After corrections for false discovery rates, only 2 of 10 genes (serpin peptidase inhibitor/plasminogen activator inhibitor member 2 and matrix metalloproteinase [MMP] 10) increased, whereas 8 (MMP2, integrin-α1, vascular endothelial growth factor, a disintegrin and metallopeptidase domain 33, scatter factor/hepatocyte growth factor, tissue inhibitor of matrix metalloproteinase-2, fibronectin, and collagen 3α1) decreased in small airways in association with FEV1. In contrast, 8/12 genes (early growth response factor 1, MMP1, MMP9, MMP10, plasminogen activator urokinase, plasminogen activator urokinase receptor, tumor necrosis factor, and IL13) increased and 4/12 (MMP2, tissue inhibitor of matrix metalloproteinase-1, collagen 1α1, and transforming growth factor-β3) decreased in the surrounding lung tissue in association with progression of COPD.
Conclusions: The progression of COPD is associated with the differential expression of a cluster of genes that favor the degradation of the tissue surrounding the small conducting airways.
Keywords: emphysema; pulmonary disease, chronic obstructive; polymerase chain reaction; laser microdissection; nucleic acid amplification techniques
AT A GLANCE COMMENTARY.
Scientific Knowledge on the Subject
Abnormal tissue repair is a putative mechanism for the development of peripheral lung lesions in chronic obstructive pulmonary disease.
What This Study Adds to the Field
This study provides insight into why small airways thicken in such close proximity to emphysematous destruction by demonstrating differential expression of a cluster of tissue repair genes between small airways and surrounding lung tissue.
The decline in the FEV1 that defines the severity of chronic obstructive pulmonary disease (COPD) is caused by a combination of obstruction of the smaller bronchi and bronchioles less than 2 mm in diameter and a reduction in the force available to drive air out of the lungs due to emphysematous destruction (1). Although both McLean (2) and Leopold and Gough (3) believed that disease in the terminal and preterminal bronchioles preceded emphysematous destruction of respiratory bronchioles to form centrilobular emphysematous lesions in COPD, only Leopold and Gough conducted the three-dimensional reconstructions of the histology required to establish that 60% (54/90) of the 90 centrilobular lesions they examined by this method were supplied by bronchioles that had thickened walls and narrowed lumens (3). Subsequent reports established that the smaller bronchi and bronchioles less than 2 mm in diameter became the major site of airways obstruction in COPD (4–6), and a series of studies reviewed by Hogg and Timens (7) have confirmed and extended the original observations that these airways are chronically inflamed and extensively remodeled in COPD. Moreover, a multivariate analysis performed in a study of lung tissue from patients treated for advanced emphysema by lung volume reduction surgery has shown that thickening of the walls of the smaller bronchi and bronchioles by connective tissue and the accumulation of inflammatory exudates containing mucus explained more of the variance in the decline of FEV1 than the infiltration by any particular inflammatory cell type (8). Just why small airways thicken and narrow in regions of lung affected by emphysematous destruction is an important issue because these two quite different lesions mandate different approaches to therapy.
Several authors have postulated that an aberrant tissue repair process underlies the pathogenesis of COPD because lung fibroblasts from these patients show abnormal responses to cigarette smoke extract (9) and are unable to sustain tissue repair (10). As the deposition of collagen matrix in chronically inflamed tissue undergoing repair is determined by a balance between synthesis and degradation of the matrix (see Figure E1 in the online supplement), it follows that in COPD excess synthesis in bronchiolar tissue would predispose to bronchiolar wall thickening and excess degradation in surrounding lung tissue to emphysematous destruction. The goal of the present study was to test this hypothesis using laser capture microdissection (LCM) to separate bronchioles from the lung tissue immediately surrounding them. RNA isolate from both tissue sources was surveyed for the expression of 3 housekeeping genes that served as comparators for 54 genes previously associated with the accumulation of collagen in chronic wounds (11) (Figure E1). Preliminary results from these studies have previously been reported in abstract form (12, 13).
METHODS
Tissue Acquisition and Processing
Procedures used to obtain human lung tissue for these studies are fully described elsewhere and approved by all participating institutions (14, 15). Briefly, patients requiring lung resection for treatment of either small peripheral tumors (n = 55) at St. Paul's Hospital (Vancouver, Canada) or lung transplantation for advanced COPD (n = 8) at Barnes Jewish Hospital (St. Louis, MO) were divided into a control group with a normal FEV1 and normal ratio of FEV1 to FVC, and into groups with mild (Global Initiative for Chronic Obstructive Lung Disease [GOLD] 1), moderate (GOLD 2), severe (GOLD 3), and very severe (GOLD 4) COPD (1). Forty of the 63 participants smoked up until the time of surgery, 16 stopped smoking for an average of 8 years before resection or transplant, and no data were available on the remaining 7. On completion of the pathological examination of the specimens (14), the lung was inflated with Cryomatrix, (Shandon, Pittsburg, PA) diluted 50% with normal saline, rapidly frozen solid in liquid nitrogen vapor, cut into 2-cm thick transverse slices, and sampled with a power-driven hole-saw to obtain tissue cores 1.5 cm in diameter and 2 cm long that were stored at −80°C.
Separation of Small Airways from their Surrounding Parenchyma
Noncartilaginous airways less than 2 mm in internal diameter were isolated using LCM. The surrounding parenchyma was then transferred from the slide with a razor blade. The separated tissues were stored in lysis buffer at −80°C until RNA isolation. Additional detail for this method is provided in the online data supplement.
Isolation and Assessment of RNA
RNA was isolated using the RNeasy Mini kit (Qiagen, Mississauga, Ontario) and an aliquot of this isolate was evaluated using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA) following the manufacturer's instructions (data not shown).
Preparation of Amplified cDNA
Fifty nanograms of RNA from each of the small airway samples and 100 ng from each of the surrounding tissue samples were used as starting material for amplification using the Clontech Super SMART cDNA Amplification Kit (Clontech, Mountain View, CA), which we have previously shown preserves differential expression in the native samples (15).
Taqman Qualitative Polymerase Chain Reaction
Taqman qualitative Polymerase Chain Reaction assays from Applied Biosystems (Foster City, CA) were used for expression profiling the lung cDNA samples following the manufacturer's protocol for 54 genes (Table E1) chosen by a review of the literature on wound healing. Additional detail for this method is provided in the online data supplement.
Gelatin Zymography
Matrix metalloproteinase (MMP) 2 (gelatinase A) activity in whole lung tissue from 11 of the 63 COPD cases that had either high (6 cases) or low (5 cases) MMP2 gene expression was measured using 10% zymogram gels with 0.1% gelatin (Biorad, Hercules, CA) and standard zymography procedures. Additional detail for this method is provided in the online data supplement.
Immunohistochemistry
Sections for immunostaining with primary antibodies to MMP2 (EMD, Gibbstown, NJ), MMP9 (Santa Cruz Biotechnology, Santa Cruz, CA), IL13 (Santa Cruz), and early growth response factor 1 (EGR1; ProSci Inc., Poway, CA) from frozen cores of lung tissue from patients with COPD were visualized using Vulcan Fast Red and counterstained with hematoxylin. Additional detail for this method is provided in the online data supplement.
Data Presentation and Statistical Analysis
All gene and protein expression data were transformed using a cubic-root function to achieve normality. The nonparametric Kruskal-Wallis test was used to compare gene expression and zymography levels across different GOLD stages and the multiple Behrens-Fisher test was used for post hoc pairwise comparisons (16). Linear regression modeling was used to assess the relationship between FEV1 and gene expression. A false discovery rate was used to adjust for multiple testing for the relationship between FEV1 and 54 genes and for the comparison of 54 genes across different GOLD stages separately (17). All statistical analyses were performed using R 2.6.0 software and all statistical tests were two-sided. Statistical significance was defined as P < 0.05 after adjustment for multiple testing. The median value was used because there were variable numbers of samples per case.
RESULTS
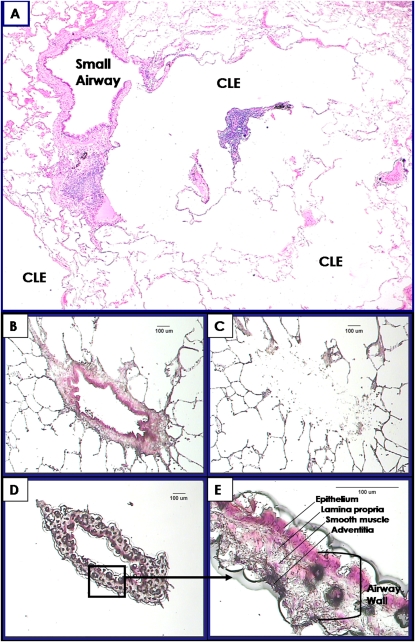
Table 1 shows the data concerning the GOLD stage, origin of lung tissue, the location of the samples within the lung, lung function, smoking history, steroid treatment, and age of the 63 participants providing the lung tissue and the total number of airways and surrounding tissue examined per GOLD stage. Figure 1A shows a thickened small airway and early centrilobular emphysematous destruction in lung tissue from a patient with GOLD 1 COPD, and Figures 1B–1E illustrate the LCM process described in the figure legend. Table 2 shows that 10 genes in small airways and 12 in the surrounding lung tissue maintained statistically significant linear relationships with FEV1 after correction for false discovery rates. Two genes (MMP2 and MMP10) account for 4/22 of these relationships because they were present in both airways and surrounding tissue, 8/22 were found only in airways and 10/22 only in the surrounding tissue. Two of the 10 genes expressed in the airways (serpin peptidase inhibitor/plasminogen activator inhibitor member 2 [SERPINE2] and MMP10) increased as FEV1 declined, whereas 8/10 genes (MMP2, integrin α1 [ITGA1], vascular endothelial growth factor, a disintegrin and metallopeptidase domain 33 [ADAM33], scatter factor/hepatocyte growth factor, fibronectin, tissue inhibitor of matrix metalloproteinase [TIMP]-2 and collagen 3α1 [COL3A1]) all declined in association with FEV1. On the other hand, 8/12 genes (EGR1, MMP9, MMP10, plasminogen activator urokinase [PLAU] and its receptor [PLAUR], tumor necrosis factor [TNF], IL13, and MMP1) increased their expression in the tissue surrounding the airways in association with FEV1, and only 4/12 genes (MMP2, collagen 1α1 [COL1A1], transforming growth factor [TGF]-β3 [B3], and TIMP1) decreased their expression with FEV1 decline.
TABLE 1.
PATIENT DATA FROM 63 PARTICIPANTS PROVIDING LUNG TISSUE
| GOLD Stage | 0 (Control) | 1 (Mild) | 2 (Moderate) | 3–4 (Severe) |
|---|---|---|---|---|
| Origin of lung tissue | Van | Van | Van | 4 Van, 8 SL |
| Location of lung tissues examined | 3 RUL | 4 RUL | 4 RUL | 2 RUL |
| 4 RLL | 4 RLL | 2 RLL | 2 RLL | |
| 6 LUL | 4 LUL | 7 LUL | 4 LUL | |
| 3 LLL | 6 LLL | 4 LLL | 4 LLL | |
| No. of cases | 16 | 18 | 17 | 12 |
| No. of airways and surrounding tissues examined | 35 | 37 | 36 | 28 |
| FEV1/FVC, mean ± SD | 78.2 ± 4.1 | 65.5 ± 3.6 | 58.8 ± 6.2 | 46.0 ± 12.8 |
| FEV1, mean ± SD | 96.2 ± 8.5 | 92.1 ± 8.9 | 65.6 ± 6.7 | 27.7 ± 12.0 |
| Pack-years, mean ± SD | 36.8 ± 17.2 | 50.9 ± 19.5 | 50.7 ± 30.6 | 49.9 ± 14.4 |
| Former smokers | 6 | 5 (2 n/a) | 2 | 8 (2 n/a) |
| Years stopped smoking, mean ± SD | 15.0 ± 10.3 | 7.4 ± 5.8 | 4.0 ± 4.2 | 5.4 ± 4.7 |
| Steroid treatment, no. of patients | 2 (2 n/a) | 1 (3 n/a) | 0 | 7 (1 n/a) |
| Age, mean ± SD | 64.7 ± 8.5 | 63.6 ± 10.4 | 60.8 ± 9.0 | 60.4 ± 7.8 |
Definition of abbreviations: GOLD = Global Initiative for Chronic Obstructive Lung Disease; LLL = left lower lobe; LUL = left upper lobe; n/a = data not available; RLL = right lower lobe; RUL = right upper lobe; SL = Barnes Jewish Hospital (St. Louis, MO); Van = St. Paul's Hospital (Vancouver, Canada).
Figure 1.
(A) Photomicrograph showing a thickened small airway in close proximity to early centrilobular emphysematous destruction (CLE). (B–E) Process of laser capture microdissection (LCM). (B) A single small airway and surrounding tissue stained with hematoxylin and eosin before LCM. (C) The tissue surrounding the airway after the airway was removed by LCM. (D) The isolated small airway. (E) A higher magnification of boxed area in (D) showing the individual compartments of the isolated small airway wall (Bar = 100 μm).
TABLE 2.
EXPRESSION RESULTS OF 20/54 GENES THAT RETAINED A STATISTICALLY SIGNIFICANT RELATIONSHIP TO FEV1 USING LINEAR REGRESSION MODELING
| Small Airways |
Parenchyma Surrounding Small Airways |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | P Value | Adjusted P Value* | Slope† | R2 | Gene | P Value | Adjusted P Value* | Slope† | R2 |
| MMP2 | <0.001 | <0.001 | 0.037 | 0.34 | EGR1 | <0.001 | <0.001 | −0.005 | 0.22 |
| SERPINE2 | <0.001 | <0.001 | −0.040 | 0.23 | MMP2 | <0.001 | <0.001 | 0.005 | 0.33 |
| ITGA1 | 0.001 | 0.013 | 0.035 | 0.16 | MMP9 | <0.001 | <0.001 | −0.007 | 0.21 |
| VEGF | 0.001 | 0.013 | 0.071 | 0.16 | MMP10 | <0.001 | <0.001 | −0.019 | 0.18 |
| ADAM33 | 0.004 | 0.033 | 0.025 | 0.13 | PLAU | <0.001 | <0.001 | −0.004 | 0.27 |
| MMP10 | 0.005 | 0.033 | −0.090 | 0.13 | TNF | <0.001 | <0.001 | −0.008 | 0.28 |
| SF/HGF | 0.005 | 0.033 | 0.015 | 0.12 | IL13 | 0.001 | 0.006 | −0.005 | 0.17 |
| TIMP2 | 0.005 | 0.033 | 0.010 | 0.13 | PLAUR | 0.001 | 0.006 | −0.003 | 0.17 |
| FN1 | 0.007 | 0.041 | 0.011 | 0.12 | MMP1 | 0.002 | 0.011 | −0.011 | 0.15 |
| COL3A1 | 0.008 | 0.042 | 0.046 | 0.11 | COL1A1 | 0.009 | 0.041 | 0.003 | 0.11 |
| TGFB3 | 0.01 | 0.041 | 0.004 | 0.10 | |||||
| TIMP1 | 0.01 | 0.041 | 0.002 | 0.10 | |||||
Definition of abbreviations: ADAM33 = a disintegrin and metallopeptidase domain 33; COL1A1 = collagen 1α1; COL3A1 = collagen 3α1; EGR1 = early growth response factor-1; FN1 = fibronectin 1; ITGA1 = integrin α1; MMP = matrix metalloproteinase; PLAU = plasminogen activator urokinase; PLAUR = plasminogen activator urokinase receptor; SERPINE2 = serpin peptidase inhibitor/plasminogen activator inhibitor member 2; SF/HGF = scatter factor/hepatocyte growth factor; TGFB3 = transforming growth factor-β3; TIMP2 = tissue inhibitor of matrix metalloproteinase 2; TNF = tumor necrosis factor; VEGF = vascular endothelial growth factor;
The data are segregated between small airways and the surrounding lung, showing the strength of the relationship with declining FEV1 in terms of R2 and the slope of the regression line.
* Corrected for multiple comparisons and false discovery rates.
Positive slope indicates that gene expression decreased in association with declining FEV1.
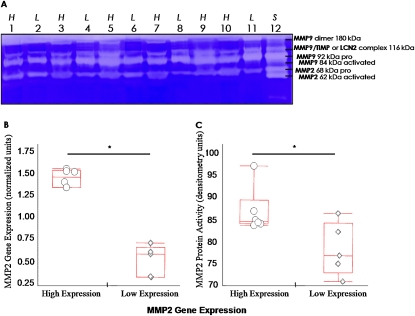
The gelatinase activity of MMP2 (Figure 2A) was measured because the association between MMP2 expression with decreasing FEV1 was ranked first in the small airways and second in the surrounding lung. Figures 2B–2C show that the level of functional gelatinase activity of the MMP2 protein measured was high when gene expression was high (n = 6) and low when gene expression was low (n = 5). Immunohistochemical staining identified MMP2 protein in small airway walls, alveolar walls, and, in particular, alveolar macrophages (Figure E2A–E2C). IL13, EGR1, and MMP9 proteins were also localized in lung tissue (Figures E2D–E2F) using immunohistochemistry. Moderate IL13 staining was typically observed in inflammatory cells located in peripheral airway and alveolar wall tissue. Strong EGR1 and MMP9 staining were found in alveolar and airway walls, stromal cells, arterioles, and inflammatory cells. Immunohistochemical negative controls showed no specific staining (Figures E3A–E3D).
Figure 2.
Matrix metalloproteinase (MMP)-2 activity compared with gene expression in whole lung tissue from patients with chronic obstructive pulmonary disease (COPD). (A) Gelatin zymogram showing MMP activity in lung tissue from five patients with COPD with low (L) and six with high (H) levels of MMP2 gene expression (lanes 1–11) and MMP2/MMP9 zymography standard including various forms and complexes (18–20) (S, lane 12) as identified at the right of the zymogram. (B) Box plots of MMP2 gene expression (in normalized relative gene expression units) in the lungs of these two groups of patients with COPD. (C) The corresponding box plots of MMP2 activity (in relative densitometry units) in these two groups from the gelatin zymogram in (A). In the box plots, the middle horizontal line represent the median, boxes denote lower (Q1) and higher (Q3) quartiles, and whiskers denote range of the data; circles and diamonds represent the values from each sample tested. *Significantly different, P < 0.05.
DISCUSSION
The results show clear differences in the pattern of expression of many of the 54 tissue repair genes examined in bronchiolar tissue compared with the immediately surrounding lung tissue. Only 2 of the 54 genes examined were statistically significantly related to a decline in FEV1 in both small airway and surrounding lung tissue after corrections for multiple comparisons. One of these two genes (MMP10) increased and the other (MMP2) decreased expression in both locations as FEV1 declined. However, an additional 8 genes expressed in the small airways tissue and 10 quite different genes expressed in the surrounding lung tissue were associated with a decline in FEV1. The following discussion concerns the relevance of these findings to the observation that small airways thicken in close proximity to surrounding lung tissue undergoing emphysematous destruction in COPD (3, 4, 7).
The results obtained from the tissue surrounding the small airways provides clear evidence that the pattern of expression of the tissue repair genes examined favors emphysematous lung destruction. More specifically, all eight genes (EGR1, MMP9, MMP10, PLAU, PLAUR, TNF, IL13, and MMP1) that increased their expression in relation to a decline in FEV1 have previously been associated with the pathogenesis of COPD. For example Ning and colleagues (21) found that the transcription factor EGR1 is associated with COPD in studies of human lung tissue and the Yale group led by Jack Elias established that EGR1 drives the production of MMP9 that causes the alveolar enlargement, lung enlargement, lung compliance alterations, and respiratory failure observed in this animal model (22–24). Therefore, the increased expression of IL13, EGR1, and MMP9 in tissue surrounding the bronchioles as FEV1 declined provides strong circumstantial evidence that several genes influence the pathogenesis of emphysematous destruction in COPD. Further support for this conclusion is provided by immunohistochemistry (Figure E2) showing that all these proteins were present in the tissue where gene expression was measured. Moreover, the immunohistochemistry of EGR1 confirms the earlier report by Ning and colleagues (21) showing that it was located in epithelial cells of both alveoli and small airways tissue as well as in stromal and inflammatory cells in human lungs. In addition, the MMP9 was localized to cells in the alveolar wall and IL13 was expressed in the interstitial cells in the same location. The increased expression of MMP1 in association with decline in FEV1 is consistent with the report by D'Armiento and colleagues and Foronjy and coworkers (25, 26), who implicated MMP1 in the pathogenesis of emphysema in both animal and human studies. Furthermore, PLAU and PLAUR, two genes that assist in the conversion of latent MMPs into their active forms by converting plasminogen to plasmin (Figure E1), also increased expression in association with a decline in FEV1. These data on PLAU and PLAUR are consistent with a recent report by Wang and colleagues (27), who used genome-wide expression and pathway analysis tools to identify PLAU and PLAUR as important members of a signature set of 203 differentially regulated genes in human lung tissue from persons with COPD. Moreover the immunohistochemistry performed in their study showing that these two proteins were located in lung epithelial cells, alveolar macrophages, and PMNs compares favorably with the present data showing that PLAU and PLAUR increase their expression in the lung tissue surrounding the small airways. The MMP10 gene, which increased expression in both airways and surrounding lung in this study, was identified by Pinto-Plata and colleagues (28) as one of a number of possible biomarkers for COPD. Understandably they dropped it from further investigation because there was no biologically plausible mechanism linking MMP10 expression to the pathogenesis of COPD. Since then Kassim and colleagues (29) have shown that tracheal epithelial cells from MMP10(−/−) mice express more than 1,600 genes differently than wild-type tracheal epithelial cells, raising the possibility that future studies will identify changes in MMP10 function that are relevant to the pathogenesis of COPD. Finally, the observation that increased TNF expression is associated with decline in FEV1 came as no surprise, in that the phrase “TNF and the pathogenesis of COPD” currently identifies more than 212 articles in PubMed (30). Although a clinical trial showing that the anti-TNF antibody infliximab (Johnson and Johnson, New Brunswick, NJ) had no effect on either symptoms, lung function, or systemic manifestations of COPD has dampened enthusiasm for anti-TNF therapy in this condition (31), genetic studies continue to implicate single nucleotide polymorphisms of TNF as markers of COPD (32) and the analysis of a large database has recently suggested that TNF receptor blockers may be of value in preventing hospital admissions in COPD (33).
The lung tissue surrounding the bronchioles also showed decreased expression of four genes (MMP2, TIMP1, COL1A1, and TGFB3) as FEV1 declined. MMP2, which is the other gene expressed in both small airways and surrounding lung tissue, ranked second among the 12 genes expressed in surrounding lung tissue and first among the 10 genes expressed in small airway tissue with respect to the strength of its correlation with FEV1. This finding, that MMP2 mRNA decreases its expression and activity, conflicts with the detailed immunohistochemical analyses of MMP2 staining in human lungs by Baraldo and coworkers (34), who reported that MMP2 staining in alveolar macrophages, alveolar walls, peripheral airways, and pulmonary arterioles was greater in severe and in mild COPD than in control smokers and nonsmokers. In the case of alveolar macrophages, more staining was present in severe than mild COPD. Furthermore, MMP2 staining was inversely related to FEV1/FVC and PaO2 and positively related to emphysema and residual volume percent predicted. In a second study based on induced sputum, Chen and colleagues (35) demonstrated that MMP2 concentrations measured by ELISA were higher in both smoking control subjects and patients with COPD than in nonsmokers, and, in patients with COPD, negatively correlated with FEV1 % predicted. A third study by Segura-Valdez and colleagues (36) that examined MMPs 1, 8, 9, and 13, as well as MMP2, claimed that MMP2 was increased in patients with COPD compared with control subjects without providing quantitative data from the representative micrographs containing MMP2-stained cells. Although their gelatin zymographic results from supernatants of bronchoalveolar lavage fluid were quantified, a statistical difference was not presented, nor was activity related to disease progression. Taken together these three studies suggest that MMP2 increases with progression of COPD when quantified by antibody binding but are inconsistent with our limited results showing decreased mRNA expression that is associated with decreased enzymatic activity. Baraldo and coworkers (34) provide a possible reason for this by stating that “anti-MMP2 antibodies identify not only free MMP2, but also MMP2/tissue inhibitor of metalloproteinase complexes.” However, in our opinion it is impossible to determine the precise role of MMP2 in the pathogenesis of COPD based on currently available data because the functions of MMP2 have now expanded far beyond matrix degradation (37). For example, McQuibban and colleagues (38) have shown that MMP2 cleaves monocyte chemotactic protein 3 in ways that allow it to continue to bind to chemokine receptors 1, 2, and 3 without activating them. This suggests that reduced MMP2 activity could contribute to the persistence of the inflammatory response in COPD by allowing unmodified monocyte chemotactic protein 3 to activate chemokine receptors 1, 2, and 3. Furthermore, the demonstration by Corry and colleagues (39) that MMP2(−/−) mice develop an exaggerated lung inflammatory response when sensitized and challenged with inhaled antigen is consistent with the concept that a decline in MMP2 function results in persistent inflammation. To date no one to our knowledge has exposed MMP2(−/−) mice to tobacco smoke to determine if they develop increased lung inflammation and emphysematous destruction. Therefore, further studies are required to reach a decision concerning the precise role of MMP2 in the pathogenesis of COPD.
In the tissue surrounding the airways all 8 of the 54 tissue repair genes that increased their expression as FEV1 declined have previously been implicated in the pathogenesis of emphysema. In contrast, the analysis of the bronchiolar tissue provided minimal evidence to support the hypothesis that increased expression of genes examined in this study is related to bronchiolar wall thickening. In the latter only 2 of the 10 genes increased and the remaining 8 decreased their level of expression as FEV1 declined. SERPINE2, one of the two that increased their expression, has been previously associated with COPD by genetic linkage studies and some, but not all, genetic association studies (40, 41). Moreover, the data from Demeo and colleagues showing that SERPINE2 was primarily located in small airway epithelial cells and in cells in the peribronchial extracellular matrix (40) is consistent with the present results showing increased expression in small airway tissue. Theoretically, SERPINE2 might contribute to airway wall thickening by slowing the conversion of latent MMPs to active enzymes (Figure E1). However, the failure to identify any association between a decline in FEV1 and increased expression of TGF-β1 or any other growth factors, such as connective tissue growth factor, platelet-derived growth factor, and fibroblast growth factor believed to function downstream from TGF-β1 implies that these growth factors do not stimulate collagen synthesis in bronchiolar tissue in late-stage COPD. The possibility that the linear analysis used missed these associations because expression increased at an early stage of COPD and then declined was examined retrospectively by repeating the analysis based on GOLD categories of COPD. These results showed (Figures E4 and E5) that growth factors believed to stimulate fibrosis downstream from TGF-β1, but not TGF-β1 itself, increased in the early stages (GOLD 1–2) of COPD and then declined. The possibility of this biphasic expression with disease progression is support by results from guinea pigs exposed to cigarette smoke in which, in the parenchyma, a 1-month exposure resulted in an initial increase in expression of selected genes involved in tissue repair but this was followed by a decline after 6 months (42). However, the retrospective nature of our current analysis does not allow conclusions to be drawn.
The remaining 8 out of 10 genes decreased their expression in bronchiolar tissue as FEV1 declined and included MMP2, which has been discussed earlier. A second gene, ADAM33, a member of a family of membrane-anchored metalloproteinases with integrin-like domains that is known to be expressed in both airway smooth muscle cells and fibroblasts (43), is linked to the pathogenesis of both asthma and COPD in both genetic linkage and association studies (44–46). However, new knowledge concerning the substrates of ADAM33 is required before the details of its role in the pathogenesis of COPD can be determined. The decline in the remaining six (ITGA1, vascular endothelial growth factor, scatter factor/hepatocyte growth factor, TIMP2, fibronectin, and COL3A1) are difficult to relate to the observation that bronchiolar walls thicken in regions of lung undergoing emphysematous destruction.
Of the 20 genes whose expression correlated with the decline in FEV1 in the present study, 6, namely EGR1, COL1A1, COL3A1, TIMP2, PLAU, and PLAUR, have been identified in two high-throughput analyses of gene expression in the lungs of patients with COPD (21, 27) and have been mentioned above. Comparisons with other studies of genome-wide expression in COPD (47, 48), as discussed in the online supplement, did not reveal further overlapping genes.
A recent parallel study from our laboratory demonstrating a previously underappreciated reduction in terminal bronchioles in centrilobular emphysema (49) is consistent with Leopold and Gough's (3) contention that centrilobular emphysematous destruction begins in the terminal and then spreads into the respiratory bronchioles. These observations suggest the alternative hypothesis that terminal bronchioles might be removed by a process similar to that responsible for emphysematous destruction and that this process is shut down in the remaining bronchioles as the destructive process spreads into the gas-exchanging surface.
In summary, the results presented here show a substantially different pattern of expression of many of the 54 tissue-repair genes examined in bronchiolar compared with surrounding lung tissue as FEV1 declines, in that eight genes previously associated with the pathogenesis preferentially increased their expression in the tissue surrounding the bronchioles. However, as no evidence was found to support the hypothesis that increased expression of growth factors might explain why bronchiolar walls thicken in areas of emphysematous destruction, we postulate that the smaller bronchioles may be removed by a process similar to that responsible for centrilobular emphysematous destruction, leaving a reduced number of thickened bronchioles behind.
Supplementary Material
Supported by National Institutes of Health grants NIH-P50HL084922 and P50HL084948, and Canadian Institutes of Health Research grants CIHR 7246 and CIHR MOP-82815.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.200812-1902OC on January 14, 2010
Conflict of Interest Statement: J.V.G. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. S.H. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. W.M.E. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.X. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. B.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. L.Y. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. C.W. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. D.S. received up to $1,000 from Schering Plough in consultancy fees, $1,001–$5,000 from GlaxoSmithKline and $1,001–$5,000 from AstraZeneca in advisory board fees, $10,001–$50,000 from GlaxoSmithKline and $10,001–$50,000 from AstraZeneca in lecture fees, more than $100,001 from AstraZeneca, more than $100,001 from Wyeth Phamaceuticals, $50,001–$100,000 from Merck Frosst, and more than $100,001 from GlaxoSmithKline in industry-sponsored grants. P.D.P. was the principle investigator of a project that was funded by GlaxoSmithKline to develop CT-based algorithms to quantify emphysema and airway disease in COPD. With collaborators he received approximately $300,000 to develop and validate these techniques. P.D.P. was also PI of a Merck Frosst–supported research program to investigate gene expression in the lungs of patients who have COPD. He and collaborators have received approximately $200,000 for this project. P.D.P. has established a new contract with Merck to discover genetic predictors of gene expression in lung tissue. With collaborators he will receive $95,000 over the next year to do this work. P.D.P. sits on an advisory board for Talecris Biotherapeutics, which makes anti-one antitrypsin replacement therapy. J.A.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. R.A.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. A.P. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.C. does not have a financial relationship with a commercial entity that has an interest in the subject of this manuscript. J.C.H. received lecture fees for speaking at conferences that were paid by Boehringer Ingelheim, GlaxoSmithKline, and Merck (total less than $10,000) and serves as the PI on a grant to UBC from the CIFR-FRIC ($100,000) that is sponsored by GlaxoSmithKline. J.C.H. also serves on an advisory board to COPD and organization that is sponsored by Nycomed, for which he receives a fee of $2,000 annually.
References
- 1.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2007;176:532–555. [DOI] [PubMed] [Google Scholar]
- 2.McLean KH. The macroscopic anatomy of pulmonary emphysema. Australas Ann Med 1956;5:73–88. [DOI] [PubMed] [Google Scholar]
- 3.Leopold JG, Gough J. The centrilobular form of hypertrophic emphysema and its relation to chronic bronchitis. Thorax 1957;12:219–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogg JC, Macklem PT, Thurlbeck WM. Site and nature of airway obstruction in chronic obstructive lung disease. N Engl J Med 1968;278:1355–1360. [DOI] [PubMed] [Google Scholar]
- 5.Van Brabandt H, Cauberghs M, Verbeken E, Moerman P, Lauweryns JM, Van de Woestijne KP. Partitioning of pulmonary impedance in excised human and canine lungs. J Appl Physiol 1983;55:1733–1742. [DOI] [PubMed] [Google Scholar]
- 6.Yanai M, Sekizawa K, Ohrui T, Sasaki H, Takishima T. Site of airway obstruction in pulmonary disease: direct measurement of intrabronchial pressure. J Appl Physiol 1992;72:1016–1023. [DOI] [PubMed] [Google Scholar]
- 7.Hogg JC, Timens W. The pathology of chronic obstructive pulmonary disease. Annu Rev Pathol 2009;4:435–459. [DOI] [PubMed] [Google Scholar]
- 8.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med 2004;350:2645–2653. [DOI] [PubMed] [Google Scholar]
- 9.Zandvoort A, Postma DS, Jonker MR, Noordhoek JA, Vos JT, Timens W. Smad gene expression in pulmonary fibroblasts: indications for defective ECM repair in COPD. Respir Res 2008;9:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Togo S, Holz O, Liu X, Sugiura H, Kamio K, Wang X, Kawasaki S, Ahn Y, Fredriksson K, Skold CM, et al. Lung fibroblast repair functions in patients with chronic obstructive pulmonary disease are altered by multiple mechanisms. Am J Respir Crit Care Med 2008;178:248–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar V, Abbas AK, Fausto N, Robbins SL, and Cotran RS. Robins and Cotran pathologic basis of disease. Philidelphia, PA: Elsevier Saunders; 2005.
- 12.Gosselink J, Kim GH, Javardifard A, Ellitott WM, Hayashi S, Pierce J, Cooper J, Hogg JC. The nature of small airway remodeling in COPD [Abstract]. Proc Am Thorac Soc 2006;3:A630. [Google Scholar]
- 13.Gosselink JV, Hayashi S, Elliott WM, Chan B, Sin D, Pare P, Pierce J, Pierce R, Patterson A, Cooper J, et al. Gene expression of peripheral lung remodeling in COPD [abstract]. Am J Respir Crit Care Med 2008;177:A656. [Google Scholar]
- 14.Ding L, Quinlan KB, Elliott WM, Hamodat M, Pare PD, Hogg JC, Hayashi S. A lung tissue bank for gene expression studies in chronic obstructive pulmonary disease. COPD 2004;1:191–204. [DOI] [PubMed] [Google Scholar]
- 15.Gosselink JV, Hayashi S, Chau E, Cooper J, Elliott WM, Hogg JC. Evaluation of small sample cDNA amplification for microdissected airway expression profiling in COPD. COPD 2007;4:91–105. [DOI] [PubMed] [Google Scholar]
- 16.Munzel U, Ludwig LA. A unified approach to simultaneous rank test procedures in the unbalanced one-way layout. Biom J 2001;43:553–569. [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57:289–300. [Google Scholar]
- 18.Kjeldsen L, Johnsen AH, Sengelov H, Borregaard N. Isolation and primary structure of NGAL, a novel protein associated with human neutrophil gelatinase. J Biol Chem 1993;268:10425–10432. [PubMed] [Google Scholar]
- 19.Pucci-Minafra I, Minafra S, La Rocca G, Barranca M, Fontana S, Alaimo G, Okada Y. Zymographic analysis of circulating and tissue forms of colon carcinoma gelatinase A (MMP-2) and B (MMP-9) separated by mono- and two-dimensional electrophoresis. Matrix Biol 2001;20:419–427. [DOI] [PubMed] [Google Scholar]
- 20.Triebel S, Blaser J, Reinke H, Tschesche H. A 25 kDa alpha 2-microglobulin-related protein is a component of the 125 kDa form of human gelatinase. FEBS Lett 1992;314:386–388. [DOI] [PubMed] [Google Scholar]
- 21.Ning W, Li CJ, Kaminski N, Feghali-Bostwick CA, Alber SM, Di YP, Otterbein SL, Song R, Hayashi S, Zhou Z, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci USA 2004;101:14895–14900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho SJ, Kang MJ, Homer RJ, Kang HR, Zhang X, Lee PJ, Elias JA, Lee CG. Role of early growth response-1 (Egr-1) in interleukin-13-induced inflammation and remodeling. J Biol Chem 2006;281:8161–8168. [DOI] [PubMed] [Google Scholar]
- 23.Lanone S, Zheng T, Zhu Z, Liu W, Lee CG, Ma B, Chen Q, Homer RJ, Wang J, Rabach LA, et al. Overlapping and enzyme-specific contributions of matrix metalloproteinases-9 and -12 in IL-13-induced inflammation and remodeling. J Clin Invest 2002;110:463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ Jr, Chapman HA Jr, Shapiro SD, Elias JA. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest 2000;106:1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Armiento J, Dalal SS, Okada Y, Berg RA, Chada K. Collagenase expression in the lungs of transgenic mice causes pulmonary emphysema. Cell 1992;71:955–961. [DOI] [PubMed] [Google Scholar]
- 26.Foronjy RF, Okada Y, Cole R, D'Armiento J. Progressive adult-onset emphysema in transgenic mice expressing human MMP-1 in the lung. Am J Physiol Lung Cell Mol Physiol 2003;284:L727–L737. [DOI] [PubMed] [Google Scholar]
- 27.Wang IM, Stepaniants S, Boie Y, Mortimer JR, Kennedy B, Elliott M, Hayashi S, Loy L, Coulter S, Cervino S, et al. Gene expression profiling in patients with chronic obstructive pulmonary disease and lung cancer. Am J Respir Crit Care Med 2008;177:402–411. [DOI] [PubMed] [Google Scholar]
- 28.Pinto-Plata V, Toso J, Lee K, Park D, Bilello J, Mullerova H, De Souza MM, Vessey R, Celli B. Profiling serum biomarkers in patients with COPD: associations with clinical parameters. Thorax 2007;62:595–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassim SY, Gharib SA, Mecham BH, Birkland TP, Parks WC, McGuire JK. Individual matrix metalloproteinases control distinct transcriptional responses in airway epithelial cells infected with Pseudomonas aeruginosa. Infect Immun 2007;75:5640–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pubmed [Internet]. [Accessed 2008. April 23]. Available from: http://www.pubmed.gov.
- 31.Rennard SI, Fogarty C, Kelsen S, Long W, Ramsdell J, Allison J, Mahler D, Saadeh C, Siler T, Snell P, et al. COPD Investigators. The safety and efficacy of infliximab in moderate to severe chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;175:926–934. [DOI] [PubMed] [Google Scholar]
- 32.Gingo MR, Silveira LJ, Miller YE, Friedlander AL, Cosgrove GP, Chan ED, Maier LA, Bowler RP. Tumour necrosis factor gene polymorphisms are associated with chronic obstructive pulmonary disease. Eur Respir J 2008;31:1005–1012. [DOI] [PubMed] [Google Scholar]
- 33.Suissa S, Ernst P, Hudson M. TNF-alpha antagonists and the prevention of hospitalisation for chronic obstructive pulmonary disease. Pulm Pharmacol Ther 2008;21:234–238. [DOI] [PubMed] [Google Scholar]
- 34.Baraldo S, Bazzan E, Zanin ME, Turato G, Garbisa S, Maestrelli P, Papi A, Miniati M, Fabbri LM, Zuin R, et al. Matrix metalloproteinase-2 protein in lung periphery is related to COPD progression. Chest 2007;132:1733–1740. [DOI] [PubMed] [Google Scholar]
- 35.Chen Y, Chen P, Hanaoka M, Droma Y, Kubo K. Enhanced levels of prostaglandin E2 and matrix metalloproteinase-2 correlate with the severity of airflow limitation in stable COPD. Respirology 2008;13:1014–1021. [DOI] [PubMed] [Google Scholar]
- 36.Segura-Valdez L, Pardo A, Gaxiola M, Uhal BD, Becerril C, Selman M. Upregulation of gelatinases A and B, collagenases 1 and 2, and increased parenchymal cell death in COPD. Chest 2000;117:684–694. [DOI] [PubMed] [Google Scholar]
- 37.auf dem Keller U, Doucet A, Overall CM. Protease research in the era of systems biology. Biol Chem 2007;388:1159–1162. [DOI] [PubMed] [Google Scholar]
- 38.McQuibban GA, Gong JH, Tam EM, McCulloch CA, Clark-Lewis I, Overall CM. Inflammation dampened by gelatinase a cleavage of monocyte chemoattractant protein-3. Science 2000;289:1202–1206. [DOI] [PubMed] [Google Scholar]
- 39.Corry DB, Rishi K, Kanellis J, Kiss A, Song Lz LZ, Xu J, Feng L, Werb Z, Kheradmand F. Decreased allergic lung inflammatory cell egression and increased susceptibility to asphyxiation in MMP2-deficiency. Nat Immunol 2002;3:347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet 2006;78:253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chappell S, Daly L, Morgan K, Baranes TG, Roca J, Rabinovich R, Millar A, Donnelly SC, Keatings V, MacNee W, et al. The SERPINE2 gene and chronic obstructive pulmonary disease. Am J Hum Genet 2006;79:184–186, author reply 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Churg A, Zhou S, Preobrazhenska O, Tai H, Wang R, Wright JL. Expression of profibrotic mediators in small airways versus parenchyma after cigarette smoke exposure. Am J Respir Cell Mol Biol 2009;40:268–276. [DOI] [PubMed] [Google Scholar]
- 43.Umland SP, Garlisi CG, Shah H, Wan Y, Zou J, Devito KE, Huang WM, Gustafson EL, Ralston R. Human ADAM33 messenger RNA expression profile and post-transcriptional regulation. Am J Respir Cell Mol Biol 2003;29:571–582. [DOI] [PubMed] [Google Scholar]
- 44.Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 2002;418:426–430. [DOI] [PubMed] [Google Scholar]
- 45.van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Schouten JP, Boezen HM. A disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general population. Am J Respir Crit Care Med 2005;172:329–333. [DOI] [PubMed] [Google Scholar]
- 46.Gosman MM, Boezen HM, van Diemen CC, Snoeck-Stroband JB, Lapperre TS, Hiemstra PS, Ten Hacken NH, Stolk J, Postma DS. A disintegrin and metalloprotease 33 and chronic obstructive pulmonary disease pathophysiology. Thorax 2007;62:242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Golpon HA, Coldren CD, Zamora MR, Cosgrove GP, Moore MD, Tuder RM, Geraci MW, Voelkel NF. Emphysema lung tissue gene expression profiling. Am J Respir Cell Mol Biol 2004;31:595–600. [DOI] [PubMed] [Google Scholar]
- 48.Spira A, Beane J, Pinto-Plata V, Kadar A, Liu G, Shah V, Celli B, Brody JS. Gene expression profiling of human lung tissue from smokers with severe emphysema. Am J Respir Cell Mol Biol 2004;31:601–610. [DOI] [PubMed] [Google Scholar]
- 49.Hogg JC, McDonough JE, Sanchez PG, Cooper JD, Coxson HO, Elliott WM, Naiman D, Pochettino M, Horng D, Gefter WB, et al. Micro-computed tomography measurements of peripheral lung pathology in chronic obstructive pulmonary disease. Proc Am Thorac Soc 2009;6:546–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.