Abstract
Hepatitis B virus (HBV) genotypes were examined in HIV-infected patients with chronic and occult HBV infection. From a total population of 593 HIV-infected patients, 22 individuals (prevalence 3.7%) were found to be HBsAg while 72 (12.1%) were found to be anti-HBc alone. From them, 20 and 4 were HBV DNA positive, respectively. These last four patients are therefore considered to be HBV infected in an occult form. The genotypes could be determined in all 24 HBV-infected patients. HBV-A was the most common (20/24; 83.3%), followed by HBV-D (2/24; 8.3%) and HBV-F (1/24; 4.2%). The remaining sample exhibited mixed infection involving genotypes A and D as pure ones, thus also forming part of three intergenotypic recombinant forms exhibiting different mosaic S gene patterns. The sexual route of transmission was predominant among HBV genotype A-infected patients. Among the 24 HBV DNA-positive patients, point mutations related to lamivudine resistance were found in four strains. These viral strains showed a methionine-to-valine substitution at codon 204 (rtM204V) in association with an upstream B-domain change at rtL180M. Additionally, two of them exhibited the additional rtV173L mutation. The value of HBV molecular monitoring including both HBV viral genomic characterization and genotypic resistance profile in HIV-HBV-coinfected individuals is discussed.
Hepatitis B virus (HBV) infection is an important cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma worldwide.1 Due to shared modes of transmission, coinfection with HBV and HIV is common.2 Recent data have pointed out that HIV infection accelerates the course of HBV-related liver damage leading more rapidly to cirrhosis and end-stage liver disease.3,4
Eight genotypes of HBV, A to H, are recognized with intergenotypic differences of more than 8% in the entire nucleotide sequence. The HBV genotypes can be further segregated into subgenotypes. Thus, genotypes A and F include two subgenotypes, namely A1–A2 and F1–F2, respectively; genotypes B, C, and D were divided into four subgenotypes.5,6 It is increasingly accepted that in addition to the currently classified genotypes of HBV, recombination between genotypes generates novel variants that contribute to the genetic diversity of HBV.7
HBV genotypes clearly are found to be different in various geographic areas of infection. Data concerning the clinical relevance are less clear and the impact on spontaneous or therapy-induced viral resolution is not yet clearly identified.5 HIV coinfection results in considerable modification of the natural history of HBV infection. Occult HBV infection is diagnosed when HBc antibodies (anti-HBc) and HBV-DNA are detectable in serum while hepatitis B surface antigen (HBsAg) is not.
Between September 2004 and April 2005, serum samples were sequentially collected from a total of 593 HIV-infected patients [age, 39 ± 8.6 (mean ± standard deviation, SD) years; 66% men and 34% women] who attended the Unit of Infectious Diseases at Fernández Hospital, Buenos Aires, Argentina, and who had received previous or current treatment for HIV infection. HIV infection was diagnosed by the presence of anti-HIV antibody by means of enzyme-linked immunosorbent assay (ELISA) and agglutination techniques (Genscreen HIV 1/2 version 2, Bio-Rad; SFD HIV 1/2 PA, Bio-Rad, Fujirebio). Those samples that yielded positive or indeterminate results were subsequently confirmed by Western blot test (HIV: New LAV Blot I, Bio-Rad). The route of transmission is clear for most cases (Table 1) and the sexual route dominates. None of the patients was linked by sexual contact.
Table 1.
Demographic Characteristics and Hepatitis Serological Markers among 593 Argentinean Patients Infected with HIV According to the Mode of Transmission of HIV Infection
| Mode of transmission (n = 593) |
Female (%) |
Male (%) |
Age (median and range, years) |
Percent of the population studied |
Anti-HBc (n = 242) (%) |
HBsAg (n = 22) (%) |
HBeAg (positive/total) |
Anti-HCV (n = 125) (%) |
|---|---|---|---|---|---|---|---|---|
| Heterosexual | 57. 9 | 42. 1 | 37 (23–71) | 39. 3 | 48 (20) | 2 (9) | 2/2 | 35 (28) |
| MSM | 0 | 100 | 39 (24–69) | 31 | 119 (49) | 13 (60) | 10/13 | 14 (11) |
| IDU | 24 | 76 | 37 (25–51) | 16. 2 | 39 (16) | 5 (23) | 2/5 | 61 (49) |
| IDU couple | 92. 8 | 7. 2 | 34 (26–47) | 4. 7 | 7 (3) | 1 (4) | 1/1 | 5 (4) |
| Maternal | 100 | 0 | 19 | 0. 16 | — | — | — | — |
| transmission | ||||||||
| Transfusion | 66. 6 | 33. 3 | 35 (27–47) | 0. 5 | — | — | — | — |
| Occupational exposure |
100 | 0 | 46 | 0. 16 | — | — | — | — |
| Unknown | 31. 6 | 68. 4 | 37 (22–64) | 12 | 28 (12) | 1 (4) | 0/1 | 10 (8) |
Serum samples were tested for HBsAg and anti-HBc by enzyme immunoassay (Abbott, USA) and 41.5% (246/593) had evidence of infection (HBsAg and/or anti-HBc positive). This prevalence was previously investigated by Argentine researchers and was higher8,9 or lower10 depending directly on the contribution of intravenous drug users to the population under study. Those samples that exhibited reactivity for any of the previous HBV serological markers were also tested for HBeAg, anti-HBe, IgM and total anti-HBc, and anti-HBs by ELISA tests (Abbott, USA).
The presence of antibody to HCV (anti-HCV) was determined by two different technical approaches (Abbott, USA) resulting in 125 patients (20.9%) being reactive.
HBsAg was detected in 22 (3.7%) of the 593 patients infected with HIV, showing a tendency to be more frequent among patients aged ≤35 years than among those aged >35 years [6.2% (14/220) versus 2.1% (8/373)]. However, 72 patients exhibited anti-HBc as the only serological marker of HBV infection, characterized as “anti-HBc alone.” This serological pattern was distributed with a similar age tendency among those patients ≤35 years and those >35 years [8.6% (19/220) versus 2.3% (53/373), respectively].
In these two groups of patients, HBsAg reactive (n = 22) and anti-HBc alone (n = 72), the presence of HBV DNA and genotyping were carried out by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) using an amplicon of the HBV surface (S) gene, as described by Zeng et al.11 Twenty out of 22 HBsAg reactive patients had detectable HBV DNA; the remaining two were undetectable by our PCR assay and their HBV viral load was <1000 DNA copies/ml assayed by AMPLICOR HBV MONITOR Test, v2.0, Roche Molecular Diagnostics. Twenty-seven percent (6/22) of them were negative for HBeAg. Regarding the 72 patients with an “anti-HBc alone” profile, four of them (5.6%) evidenced HBV DNA presence exhibiting a viral load lower than 1000 DNA copies/ml (Table 2).
Table 2.
Main Features of the Study Populationa
| Patient | Gender | Risk group |
Anti-HCV | HBeAg | Anti-HBe | Serum ALTb |
CV HBV (log)c |
HBV genotype |
3TC resistance therapy (months) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | IDU | Pos | Neg | Pos | N | <1000 (<3) |
ND | ND (18) |
| 2 | M | HMX | Neg | Pos | Neg | 1–2X | >40, 000, 000 (>7. 6) |
A | wt (3) |
| 3 | M | IDU | Pos | Pos | Neg | 1–2X | 4400 (3. 643) |
F | rtL180M; M204V (>60) |
| 4 | F | HTSX | Neg | Pos | Neg | N | >40, 000, 000 (>7. 6) |
A | wt (5) |
| 5 | M | HTSX | Neg | Pos | Neg | 1–2X | >40, 000, 000 (>7. 6) |
A | wt (0)d |
| 6 | M | HTSX | Neg | Pos | Neg | 1–2X | 28, 000 (4. 45) | A+D + -recA/D |
wt (36) |
| 7 | M | HTSX | Neg | Pos | Neg | N | >40, 000, 000 (>7. 6) |
A | rtV173L/L180M; M204V (94) |
| 8 | M | HTSX | Neg | Neg | Pos | N | <1000 (<3) |
A | wt (NA) |
| 9 | F | IDU | Pos | Neg | Neg | N | <1000 (<3) |
A | wt (>60) |
| 10 | M | HTSX | Neg | Pos | Neg | 1–2X | 3200 (3. 505) | A | wt (30) |
| 11 | M | HTSX | Neg | Pos | Neg | N | >40, 000, 000 (>7. 6) |
A | rtV173L/L180M; M204V (60) |
| 12 | M | HTSX | Neg | Neg | Pos | N | <1000 (<3) |
A | wt (0) |
| 13 | M | HTSX | Neg | Pos | Neg | N | 20, 000 (7. 3) |
A | wt (12) |
| 14 | M | NA | Neg | Neg | Pos | N | <1000 (<3) |
A | wt (60) |
| 15 | M | HTSX | Neg | Pos | Neg | N | >40, 000, 000 (>7. 6) |
A | wt (6) |
| 16 | F | NA | Neg | Pos | Neg | 1–2X | >40, 000, 000 (>7. 6) |
D | wt (0) |
| 17 | M | IDU | Pos | Pos | Neg | 1–2X | 5, 200, 000 (6.716) |
A | wt (0)d |
| 18 | M | HTSX | Neg | Pos | Neg | N | >40, 000, 000 (>7. 6) |
A | wt (0)d |
| 19 | M | HTSX | Neg | Pos | Neg | N | >40, 000, 000 (>7. 6) |
A | rtL180M; M204V (94) |
| 20 | M | IDU | Pos | Neg | Neg | 1–2X | <1000 (<3) |
ND | ND (60) |
| 21 | M | HTSX | Neg | Pos | Neg | 1–2X | >40, 000, 000 (>7. 6) |
A | wt (0) |
| 22 | M | HTSX | Neg | Neg | Neg | N | 2400 (3. 380) | A | wt (0) |
| 23 | M | HTSX | Neg | Neg | Neg | N | <1000 (<3) |
A | wt (0) |
| 24 | M | HTSX | Neg | Neg | Neg | N | <1000 (<3) |
A | wt (0)d |
| 25 | M | HTSX | Neg | Neg | Neg | N | <1000 (<3) |
A | wt (0) |
| 26 | M | IDU | Pos | Neg | Neg | 1–2X | <1000 (<3) |
D | wt (5) |
ND, not done; NA, not available; NC, does not correspond; Wt, wild type; HMX, homosexual; HTSX, heterosexual; IDU, intravenous drug user.
The alanine transaminase values are expressed as times the upper limit of normal (N, normal value).
Expressed as copies/ml; using AMPLICOR HBV MONITOR Test, v2. 0, Roche Molecular Diagnostics.
With a previous history of lamivudine therapy (for a period of >1 year before sample collection).
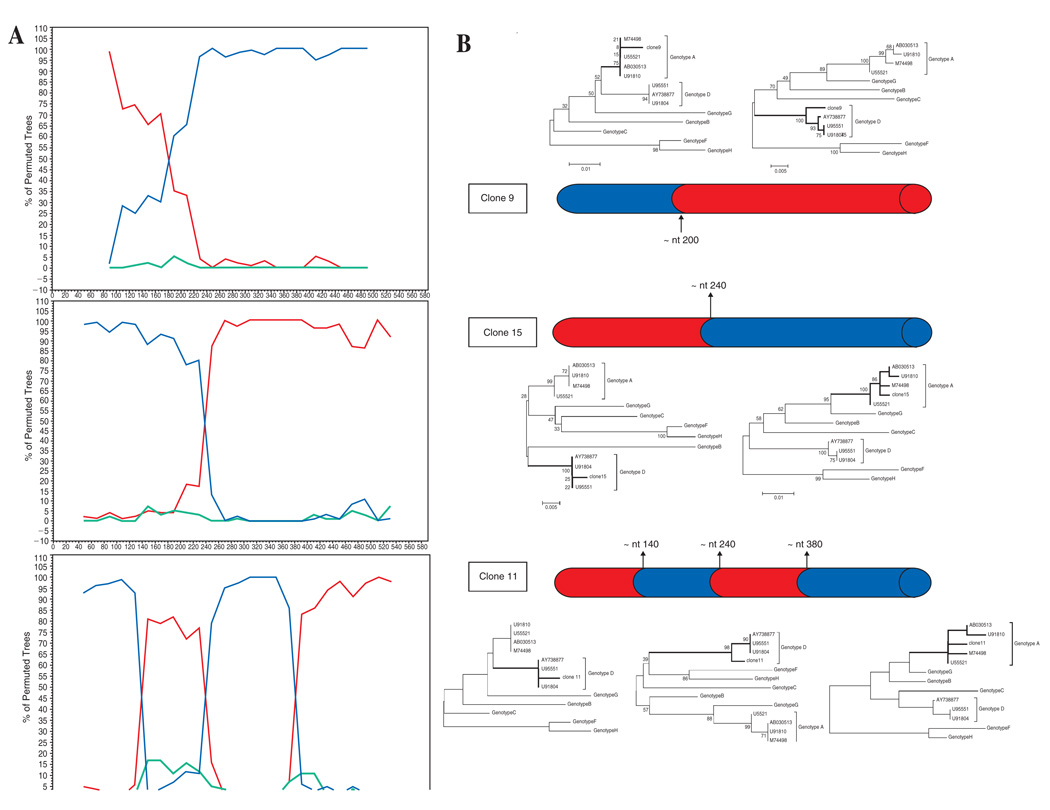
The HBV genotype was further determined in all 24 HBV DNA-positive serum samples by phylogenetic analysis of the above PCR product corresponding to an S gene partial sequence (585 nt, from nucleotide 203 to 787). For this purpose, the amplification products were sequenced directly on both strands, using the BigDye Terminator Cycle Sequencing Ready Reaction Kit on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Only a sample exhibiting unclear phylogenetic clustering (see below) was cloned (pGEM-T Easy Vector, Promega, USA) and 16 clones were further analyzed by sequencing. Sequence analysis was performed by using the programs DNADIST and NEIGHBOR from the Phylip program package version 3.53. Phylogenetic trees were constructed by the neighbor-joining method12 based on the partial nucleotide sequence of the S gene. Bootstrap values were determined on 1000 resamplings of the datasets13 to assess the stability of the nodes. Recombinant analysis for HBV S gene sequences was performed by bootscanning, using the SimPlot program [version 2.5; Stuart Ray, (http://www.med.jhu.edu)] and visual inspection of alignments was used to identify breakpoints in recombinant sequences. After identification of the breakpoints in recombinant viral sequences, subregions of the alignment were reanalyzed by neighbor-joining with bootstrapping to confirm the subtype assignment. Based on both, the S gene RFLP analysis and phylogenetic relatedness, 20 out of the 24 (83.3%) isolates were homologous to genotype A, two (8.3%) isolates to genotype D, and one (4.2%) isolate to genotype F. The remaining sample showed incongruent clustering by phylogenetic analysis suggesting a probable coinfection and/or recombination involving A and D genotypes (Fig. 1). After cloning, 7 out of 16 clones were ascribed to genotype A, while 6/16 were clustered as genotype D. The remaining 3 clones exhibited an A–D intergenotype recombinant pattern with a different gene mosaic for each one. In that sample the genetic heterogeneity of HBV was evident, showing the concomitant presence of A and D genotypes as pure ones and as integrants of three different S gene mosaic recombinant patterns, with one (nt ∼400 for clones 9 and 15) and three (nt ∼340, 400, and 580 for clone 11) breakpoints in the S gene (Fig. 2). For the first time, HBV intergenotype recombinant forms between A and D have been identified in Latin America.7
FIG. 1.
Phylogenetic tree constructed by the neighbor-joining method based on the partial nucleotide sequence of the S gene (585 nt) of 27 hepatitis B virus (HBV) isolates, using a woolly monkey HBV isolate (WMHBV: AF046996) as an out-group. In addition to the Argentinean isolates found in the present study, several representative HBV isolates of genotypes A to H were included (indicated with the GenBank accession number). HBV viral isolates from HBsAg-positive and “anti-HBc alone” patients are numbered according to Table 2; clones obtained from sample 6 are also shown by number. Bootstrap values are indicated for the major nodes as a percentage of the data obtained from 1000 resamplings.
FIG. 2.
(A) SIMPLOT analysis for clones 9 (upper), 15 (middle), and 11 (lower) belonging to each mosaic pattern. Reference sequences of genotype A (U55521) in red, D (U91804) in blue, and F, as outgroup (AY179794), in green were used. Plots represent bootstrap values, based on 100 resamplings, supporting branching with the reference sequences within a 100 nt window moving in steps of 20 bases. (B) Mosaic structure and phylogenetic confirmation of three recombination patterns in the S gene (nt: 203–787). Neighbor-joining trees with bootstrap values were built for each segment. Red, genotype A; blue, genotype D. Vertical arrows show potential breakpoints for recombination.
Previous studies on HBV isolated from blood donors from blood banks distributed nationwide in Argentina,14 as well as in chronically infected adults15 and children16 without concurrent infection with HIV, reported the occurrence of genotypes A (17%), B (1%), C (1%), D (17%), and mainly F (64%). A significant difference (p = 0.0009) was found when comparing HBV genotype distribution regarding HIV coexistence. The precise reason why genotype A HBV has become largely prevalent among Argentine patients coinfected with HIV is unknown.
The highest associated risk was unprotected sexual intercourse between men (17/24); all of them were ascribed to genotype A, in agreement with a previous report,17 providing additional support to this genotype being more efficiently transmitted sexually than genotype D.
Our findings suggest that chronic hepatitis B infection has a low prevalence (<5%) among HIV-infected individuals in Argentina. It is more frequent among homosexual men (Table 2), and it is associated with a high rate of HBeAg positivity. Although chronic hepatitis B with detectable HBV DNA but negative HBeAg reflects in most cases infection by precore mutant HBV strains with slow replicative capacity1, 2 (five HBsAg-positive/HBeAg-negative genotype A-infected patients, with an HBV viral load level <2400 copies/ml), our data suggest that in Argentina, genotype A may include the selection of these variants.
The presence of genotypes A and D could be a consequence of the immigrant origins of the Buenos Aires population, which has received immigration from the Mediterranean area. Magnius and Norder18 suggested that genotype D may have replaced genotype A in the Mediterranean area, including North Africa. If this replacement occurred after the spread of HBV strains to America, it would explain the important presence of genotype A in Argentina. The observed HBV genotype A dominance when HIV is concomitantly present deserves future research to elucidate whether direct viral interactions among HBV-A and HIV reciprocally improve viral fitness. Three out of four HBV isolates characterized from patients with an anti-HBc alone profile were ascribed to genotype A; the remaining was classified as belonging to genotype D.
Meticulous analysis of the translated S gene sequence from strains characterized from patients with an anti-HBc alone profile did not show any amino acid change into the major antigenic determinant “a.” Considering that these patients maintain the anti-HBc alone serological pattern a year later (data not shown), the occult HBV infection may reflect an unresolved chronic infection with low-grade, possibly intermittent virus production.
The efficacy of lamivudine therapy is limited by the emergence of resistance strains containing mutations that typically occur in the tyrosine–methionine–aspartate–aspartate (YMDD) motif of the catalytic domain (domain C) of the polymerase (P) coding region.20 We also sequenced an HBV viral DNA fragment targeted on the P region reverse transcriptase (rt) domain of the HBV genome following the protocol of Ono et al.19 A final 341-bp fragment, corresponding to amino acids 465 to 562 of the HBV DNA polymerase gene, was obtained. In the present study, the prevalence and characterization of point mutations related to lamivudine resistance were done by direct analysis of the deduced amino acid sequences by comparison with the published sequences for the HBV polymerase.21 Among the 24 HBV DNA-positive HIV-coinfected individuals, six out of them were naive of antiretroviral treatment. Among the 18 patients exposed to 3TC at any time, the observed drug resistance emerged with high rates in those patients infected with genotype A, as described elsewhere22, 23 (Table 2).
Four out of 24 individuals with viral detectable DNA exhibited genotypic resistance, with all of them showing a methionine-to-valine substitution at codon 204 (rtM204V). This mutation was found in association with an upstream B-domain change at rtL180M. Additionally, two of them exhibited the additional rtV173L mutation. These four patients have received treatment with lamivudine during a period >2 years [median time of 3TC therapy: 70.5 (60–94) months], which would contribute to the appearance of resistance24 (Table 2). Three out of four HBV-resistant isolates were ascribed to genotype A, with the remaining one belongs to genotype F. Among the HBV genome sequences exhibiting no lamivudine resistance-associated mutations, were sequences isolated from naive patients (n = 6) or from patients under viral therapy for a shorter period of time [n = 14, median time of 3TC therapy: 23 (1–76) months]. By comparing the values between those two groups (Mann-Whitney test), a significant difference was found (p = 0. 0015).
In our study, individuals infected with HBV genotype A and with lamivudine resistance had high levels of HBV replication, measured as plasma viral load level. It suggests that at least in this HIV-HBV coinfected population replication efficiency may be comparable to wild-type virus with a potential risk to develop significant and progressive liver disease. This fact could be explained by taking into account that the negative impact on viral fitness by the rtM204V/I mutation could be partly compensated (partially restoring the replication fitness of the virus) by subsequent mutations in the A (rtV173L) and B (rtL180M) domains upstream from the altered domain of the polymerase.25 But considering the small size of the sample, further studies of the effects of these other mutations on viral fitness are clearly needed. In this cross-sectional study a range of HBV viral loads was found for patients with the resistance mutations rtM204V, with rtL180M being higher for genotype A HBV isolates than for the F (>4. 0 × 107 versus 4. 4 × 103 copies/ml). This wide range reflects the divergence of HBV DNA levels found in chronic hepatitis B, and indicates that for HBV there is no defined setpoint as is seen in HIV.26
The triple mutant rtV173L plus rtL180M plus rtM204V seen in two individuals with genotype A HBV infection causes concomitant amino acid substitutions at positions 164 and 195 (sE164D plus sI195M) in the HBsAg protein as a result of the overlapping reading frames of the envelope and polymerase genes.27 In vitro anti-HBs binding assays of this mutant have demonstrated significantly reduced affinity of anti-HBs antibody, comparable to the hepatitis B immunoglobulin vaccine escape mutants such as sG145R.28 These data raise the possibility that the selection of antigenically distinct HBV mutants during lamivudine therapy could potentially behave as HBV vaccine escape mutants. Primary infection of an HIV-infected individual with lamivudine-resistant HBV has been reported,29 demonstrating the ability of resistant HBV to be transmitted. These HIV-infected individuals had not been immunized against HBV. Unfortunately, there now exists the possibility for lamivudine-resistant HBV such as the rtV173L plus rtL180M plus rtM204V Pol mutant to be similarly transmitted, even in the setting of protective levels of anti-HBs in vaccinated individuals. Further studies are urgently required to determine the clinical and public health relevance of these findings, as well as the importance of the other changes identified in the envelope protein of HBV found as a consequence of lamivudine therapy.
The faster disease progression, increased rates of lamivudine resistance, higher HBV loads, as well as elevated serum ALT levels associated with different HBV genotypes emphasize the need for appropriate HBV molecular monitoring including both HBV viral genomic characterization and genotypic resistance profiles in HIV–HBV-coinfected individuals in order to guide patient management adequately.
ACKNOWLEDGMENTS
Natalia Laufer was supported by a Fogarty International Center/NIH grant through the AIDS International Training and Research Program at Mount Sinai School of Medicine, Argentina Program (Grant 5D43 TW0010137).
Footnotes
The GenBank accession numbers of the sequences reported here are DQ249922 to DQ249941.
REFERENCES
- 1.Lok A, Heathcote E, Hoofnagle J. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology. 2001;120:1828–1853. doi: 10.1053/gast.2001.24839. [DOI] [PubMed] [Google Scholar]
- 2.Puoti M, Airoldi M, Bruno R. Hepatitis B virus co-infection in HIV-infected subjects. AIDS Rev. 2002;4:27–35. [PubMed] [Google Scholar]
- 3.Thio C, Seaberg E, Skolasky R, Phair J, Visscher B, Muñoz A, et al. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS) Lancet. 2002;360:1921–1926. doi: 10.1016/s0140-6736(02)11913-1. [DOI] [PubMed] [Google Scholar]
- 4.Di Martino V, Thevenot T, Colin J, Boyer N, Martinot M, Degos F, et al. Influence of HIV infection on the response to interferon therapy and the long-term outcome of chronic hepatitis B. Gastroenterology. 2002;123:1812–1822. doi: 10.1053/gast.2002.37061. [DOI] [PubMed] [Google Scholar]
- 5.Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J Viral Hepat. 2005;12(5):456–464. doi: 10.1111/j.1365-2893.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 6.Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47(6):289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 7.Simmonds P, Midgley S. Recombination in the genesis and evolution of hepatitis B virus genotypes. J Virol. 2005;79(24):15467–15476. doi: 10.1128/JVI.79.24.15467-15476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fainboim H, Gonzalez J, Fassio E, Martinez A, Otegui L, Eposto M, Cahn P, Marino R, Landeira G, Suaya G, Gancedo E, Castro R, Brajterman L, Laplume H. Prevalence of hepatitis viruses in an anti-human immunodeficiency virus-positive population from Argentina. A multicentre study. J Viral Hepat. 1999;6(1):53–57. doi: 10.1046/j.1365-2893.1999.t01-1-6120135.x. [DOI] [PubMed] [Google Scholar]
- 9.Weissenbacher M, Rossi D, Radulich G, Sosa-Estani S, Vila M, Vivas E, Avila MM, Cuchi P, Rey J, Peralta LM. High seroprevalence of bloodborne viruses among street-recruited injection drug users from Buenos Aires, Argentina. Clin Infect Dis. 2003;37 Suppl. 5:S348–S352. doi: 10.1086/377560. [DOI] [PubMed] [Google Scholar]
- 10.Pando M, Biglione MM, Toscano MF, Rey JA, Russell KL, Negrete M, Gianni S, Martinez-Peralta L, Salomon H, Sosa-Estani S, Montano SM, Olson JG, Sanchez JL, Carr JK, Avila MM. Human immunodeficiency virus type 1 and other viral co-infections among young heterosexual men and women in Argentina. Am J Trop Med Hyg. 2004;71(2):153–159. [PubMed] [Google Scholar]
- 11.Zeng GB, Wen SJ, Wang ZH, Yan L, Sun J, Hou JL. A novel hepatitis B virus genotyping system by using restriction fragment length polymorphism patterns of S gene amplicons. World J Gastroenterol. 2004;10(21):3132–3136. doi: 10.3748/wjg.v10.i21.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Franca PH, Gonzalez JE, Munne MS, Brandao LH, Gouvea VS, Sablon E, Vanderborght BO. Strong association between genotype F and hepatitis B virus (HBV) e antigen-negative variants among HBV-infected argentinean blood donors. J Clin Microbiol. 2004;42(11):5015–5021. doi: 10.1128/JCM.42.11.5015-5021.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Telenta PF, Poggio GP, Lopez JL, Gonzalez J, Lemberg A, Campos RH. Increased prevalence of genotype F hepatitis B virus isolates in Buenos Aires, Argentina. J Clin Microbiol. 1997;35:1873–1875. doi: 10.1128/jcm.35.7.1873-1875.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mbayed VA, López JL, Telenta PFS, Palacios G, Badía I, Ferro A, Galoppo C, Campos R. Distribution of hepatitis B virus genotypes in two different pediatric populations from Argentina. J Clin Microbiol. 1998;36:3362–3365. doi: 10.1128/jcm.36.11.3362-3365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pérez-Olmeda M, Nunez M, Garcia-Samaniego J, Rios P, Gonzalez-Lahoz J, Soriano V. Distribution of hepatitis B virus genotypes in HIV-infected patients with chronic hepatitis B: Therapeutic implications. AIDS Res Hum Retroviruses. 2003;19(8):657–659. doi: 10.1089/088922203322280874. [DOI] [PubMed] [Google Scholar]
- 18.Magnius LO, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38(1–2):24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 19.Ono SK, Kato N, Shiratori Y, Kato J, Goto T, Schinazi RF, Carrilho FJ, Omata M. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J Clin Invest. 2001;107:449–455. doi: 10.1172/JCI11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuyver LJ, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, Schinazi RF. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33:751–757. doi: 10.1053/jhep.2001.22166. [DOI] [PubMed] [Google Scholar]
- 21.Bartholomeusz A, Schinazi R, Locarnini S. Significance of mutations in the hepatitis B virus polymerase selected by nucleoside analogues and implications for controlling chronic disease. Viral Hepatitis Rev. 1998;4:167–187. [Google Scholar]
- 22.Buti M, Cotrina M, Valdes A, Jardi R, Rodriguez-Frias F. Is hepatitis B virus subtype testing useful in predicting virological response and resistance to lamivudine? J Hepatol. 2002;36:445–446. doi: 10.1016/s0168-8278(01)00283-5. [DOI] [PubMed] [Google Scholar]
- 23.Zollner B, Petersen J, Schroter M, Laufs R, Schoder V, Feucht HH. 20-Fold increase in risk of lamivudine resistance in hepatitis B virus subtype adw. Lancet. 2001;357:934–935. doi: 10.1016/S0140-6736(00)04219-7. [DOI] [PubMed] [Google Scholar]
- 24.Leung NW, Lai CL, Chang TT, Guan R, Lee CM, Ng KY, Lim SG, Wu PC, Dent JC, Edmundson S, Condreay LD, Chien RN. on behalf of the Asia Hepatitis Lamivudine Study Group: Extended lamivudine treatment in patients with chronic hepatitis B enhances hepatitis B e antigen seroconversion rates: Results after 3 years of therapy. Hepatology. 2001;33:1527–1532. doi: 10.1053/jhep.2001.25084. [DOI] [PubMed] [Google Scholar]
- 25.Delaney WE, 4th, Yang H, Westland CE, Das K, Arnold E, Gibbs CS, Miller MD, Xiong S. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J Virol. 2003;77(21):11833–11841. doi: 10.1128/JVI.77.21.11833-11841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loeb KR, Jerome KR, Goddard J, Huang M, Cent A, Corey L. High-throughput quantitative analysis of hepatitis B virus DNA in serum using the TaqMan fluorogenic detection system. Hepatology. 2000;32:626–629. doi: 10.1053/jhep.2000.9878. [DOI] [PubMed] [Google Scholar]
- 27.Locarnini S. Hepatitis B virus surface and polymerase gene variants: Potential virological and clinical significance. Hepatology. 1998;27:294–297. doi: 10.1002/hep.510270144. [DOI] [PubMed] [Google Scholar]
- 28.Torresi J, Earnest-Silveira L, Civitico G, Walters TE, Lewin SR, Fyfe J, Locarnini SA, Manns M, Trautwein C, Bock TC. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the ‘Fingers’ subdomain of the viral polymerase selected as a consequence of mutations in the overlapping s gene. Virology. 2002;299:88–99. doi: 10.1006/viro.2002.1448. [DOI] [PubMed] [Google Scholar]
- 29.Thibault V, Aubron-Olivier C, Agut H, Katlama C. Primary infection with a lamivudine-resistant hepatitis B virus. AIDS. 2002;16:131–133. doi: 10.1097/00002030-200201040-00020. [DOI] [PubMed] [Google Scholar]