Polν and Polθ have specialized functions in immunoglobulin gene rearrangements and only contribute to DNA repair when other homologous recombination–related DNA polymerases are absent.
Abstract
The chicken DT40 B lymphocyte line diversifies its immunoglobulin (Ig) V genes through translesion DNA synthesis–dependent point mutations (Ig hypermutation) and homologous recombination (HR)–dependent Ig gene conversion. The error-prone biochemical characteristic of the A family DNA polymerases Polν and Polθ led us to explore the role of these polymerases in Ig gene diversification in DT40 cells. Disruption of both polymerases causes a significant decrease in Ig gene conversion events, although POLN−/−/POLQ−/− cells exhibit no prominent defect in HR-mediated DNA repair, as indicated by no increase in sensitivity to camptothecin. Polη has also been previously implicated in Ig gene conversion. We show that a POLH−/−/POLN−/−/POLQ−/− triple mutant displays no Ig gene conversion and reduced Ig hypermutation. Together, these data define a role for Polν and Polθ in recombination and suggest that the DNA synthesis associated with Ig gene conversion is accounted for by three specialized DNA polymerases.
Introduction
The chicken DT40 B lymphocyte cell line provides a unique opportunity to analyze the role of individual DNA polymerases in homologous recombination (HR) and translesion DNA synthesis (TLS) because DT40 cells diversify Ig V genes through HR (Ig gene conversion) and nontemplated single-nucleotide substitutions (Ig hypermutation) during in vitro culture (Buerstedde et al., 1990; Sale et al., 2001). Ig gene conversion introduces tracts of templated mutations derived from an array of pseudo-Vλ (ΨVλ) regions, located upstream of rearranged VJλ, to the Vλ segment of the rearranged VJλ (Reynaud et al., 1987). Because donor and recipient segments have an ∼10% sequence divergence, sequential Ig gene conversion events are able to substantially diversify the Ig V gene. However, Ig hypermutation is performed by TLS past abasic sites in DT40 cells (Simpson and Sale, 2003; Arakawa et al., 2006; Saberi et al., 2008).
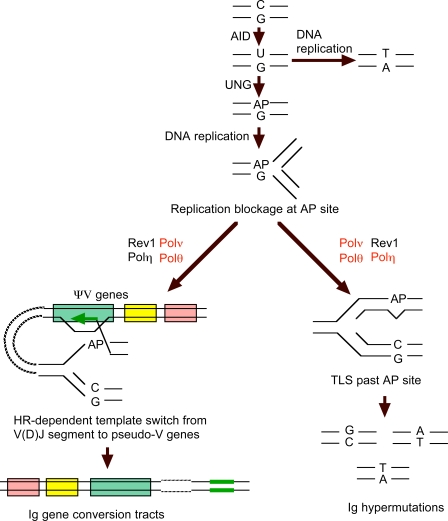
Activation-induced deaminase (AID) is responsible for triggering Ig hypermutation and Ig gene conversion (Fig. 1; Arakawa et al., 2002; Harris et al., 2002). AID catalyses deoxycytidine to generate uracil followed by elimination of uracil by uracil glycosylase to induce abasic sites (Di Noia and Neuberger, 2002; Petersen-Mahrt et al., 2002). Replication blockages at abasic sites generated at the V(D)J segment and subsequent release from the blockage by HR and TLS may cause Ig gene conversion and Ig hypermutation, respectively, in DT40 cells (Fig. 1; Simpson and Sale, 2003; Saberi et al., 2008; Nakahara et al., 2009). In Ig gene conversion, replication blockage may induce template switch from the V(D)J segment to pseudo-V segments. Subsequent DNA synthesis using pseudo-V segments as a template may lead to gene conversion from the pseudo-V segments to the V(D)J segment (Buerstedde and Arakawa, 2006). Collectively, determination of Ig V nucleotide sequences in DT40 cells provides a unique opportunity to identify both the gene conversion tracts and the spectrum of TLS-dependent mutations. This allows identification of the DNA polymerases involved in these Ig V diversification reactions.
Figure 1.
Molecular mechanisms for Ig gene diversification, yielding substitutions at C/G pairs and Ig gene conversion in DT40 cells. AID-mediated deamination of C generates a U/G mispair. Uracil DNA glycosylase (UNG) can excise the U residue to generate an abasic site (AP), which causes replication blockage. (bottom right) Release of replication blockage by TLS causes mutations at C/G pairs, depending on which nucleotide is inserted opposite the abasic site. (bottom left) Alternatively, HR-dependent template switch from V(D)J segment to pseudo-V genes induces Ig gene conversion. Colored boxes indicate pseudo-V genes. DNA polymerases with a previously identified role in Ig gene diversification are shown in black (Kawamoto et al., 2005; Okada et al., 2005; Ross and Sale, 2006; Saberi et al., 2008), and those identified in this study are shown in red. This figure is based on previously described results (Sale, 2004; Buerstedde and Arakawa, 2006).
HR is a multistep process that repairs double-strand breaks (DSBs) and releases replication blockage using intact homologous sequences as a template (Pâques and Haber, 1999; Wyman and Kanaar, 2006; Takeda et al., 2007). DSBs are processed during the early steps of HR, leading to the formation of 3′ single-strand overhangs, which associate with polymerized Rad51. The resulting complex, including the 3′ overhangs and Rad51, invades intact homologous duplex DNA to form a D loop structure. DNA synthesis from the invading 3′ overhang, followed by the recapture of the newly synthesized DNA strand by the other end of the DSB, completes DSB repair. This type of HR is called synthesis-dependent strand annealing and does not cause the generation of crossover DNA. Because the D loop is unstable, efficient DNA synthesis may significantly increase the rate of gene conversion (Pâques et al., 1998). DNA synthesis can be performed by DNA polymerases η and ζ (Polη and Polζ; Sonoda et al., 2003; Kawamoto et al., 2005; McIlwraith et al., 2005), although other DNA polymerases may also contribute to HR. Another unresolved question concerns the nature of the DNA polymerases that are involved in HR-dependent release of replication blockage.
Computational analysis of the human genome revealed genes encoding two A-type DNA polymerases, POLN (Marini et al., 2003) and POLQ (Seki et al., 2003, 2004), in addition to the POLG gene, a unique DNA polymerase found in mitochondria. POLQ, but not POLN, contains a helicase domain near its N terminus, as does MUS308, the prototype orthologue of POLN/POLQ in Drosophila melanogaster (Boyd et al., 1990). Subsequent biochemical studies have shown that Polν and Polθ, which lack intrinsic exonucleolytic proofreading activity, can indeed perform TLS past abasic sites, undergo DNA synthesis with very low fidelity, and extend from mismatches (Seki et al., 2004; Takata et al., 2006; Arana et al., 2007, 2008; Seki and Wood, 2008). Genetic studies have addressed the function of POLN and POLQ using D. melanogaster, mice, and chicken DT40 cells (Boyd et al., 1990; Shima et al., 2004; Yoshimura et al., 2006). D. melanogaster deficient in MUS308 are hypersensitive to chemical cross-linkers, indicating the critical role played by the A-type polymerases in DNA damage response (Boyd et al., 1990). However, phenotypic analysis of both POLN−/−/POLQ−/− DT40 cells and POLQ−/− mice cells shows that Polν and Polθ do not play the same critical role as Mus308 in cellular responses to chemical cross-linkers, and Polθ makes significant contributions to the DNA repair of base damage and probably to TLS (Shima et al., 2004; Yoshimura et al., 2006).
We examined Ig V diversification in POLN−/−, POLQ−/−, and POLN−/−/POLQ−/− DT40 clones. Compared with wild-type cells, POLN−/−/POLQ−/− DT40 cells exhibited a significant reduction in the rate of Ig gene conversion, which was associated with increased length of gene conversion but not Ig hypermutation rate. This observation is in marked contrast to the absence of prominent defects in POLN−/−/POLQ−/− cells in general HR, including gene targeting and camptothecin sensitivity. Moreover, POLN−/−/POLQ−/− clones showed significant reduction in the number of C to G mutations, indicating that Polν and Polθ play a role in TLS past the abasic site. Because Polη is involved in Ig hypermutation and Ig gene conversion (Kawamoto et al., 2005), we disrupted the POLH gene in the POLN−/−/POLQ−/− background. Ig hypermutation rate was significantly decreased in the resulting POLH−/−/POLN−/−/POLQ−/− cells. Remarkably, no Ig gene conversion events were detectable in POLH−/−/POLN−/−/POLQ−/− cells, indicating that the three DNA polymerases were responsible for Ig gene diversification (Fig. 1). Only POLH−/−/POLN−/−/POLQ−/− cells showed increased sensitivity to camptothecin, a topoisomerase I inhibitor. In conclusion, Polν and Polθ may have adapted to perform a specialized function for Ig V diversification and can contribute to HR-dependent repair only when other HR-related DNA polymerases are absent.
Results
Cells deficient in POLN and POLQ exhibit a decrease in the rate of Ig gene conversion
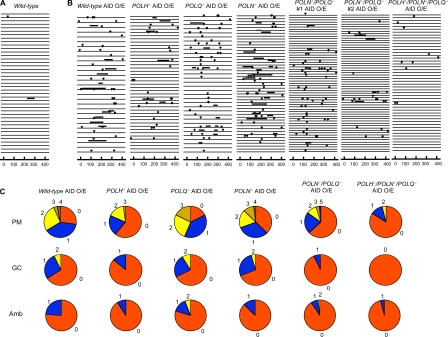
To investigate the role of Polν and Polθ in Ig gene diversification, we induced Ig gene conversion and Ig hypermutation by ectopically expressing AID in POLN−/−, POLQ−/−, POLN−/−/POLQ−/−, and wild-type DT40 cells through retrovirus infection (Shinkura et al., 2004; Saberi et al., 2008). We determined the VJλ nucleotide sequences of more than three clonally expanded populations from each genotype. The overexpression of the AID transgene increased the rate of Ig gene conversion in wild-type cells by 17-fold to 9.8 × 10−3 per nucleotide per division (Fig. 2, A and B; and Fig. 3 C). POLH−/− cells showed a decrease in Ig gene conversion rate as we have previously shown (Kawamoto et al., 2005). Wild-type, POLN−/−, and POLQ−/− clones exhibited indistinguishable rates of Ig gene conversion, whereas the rate of Ig gene conversion in POLN−/−/POLQ−/− cells is 4.9 times lower than in wild-type cells (P < 0.0002; Fig. 2 C and Fig. 3 C). This finding reveals an unexpected function of Polν and Polθ in HR (Fig. 1). Moreover, the synergistic effect of POLN and POLQ mutations on the Ig gene conversion rate indicates that Polν and Polθ may complement each other in HR-dependent Ig V diversification.
Figure 2.
Nucleotide sequence analysis of VJλ segments to detect Ig gene conversions and nontemplated point mutations. (A) VJλ segments isolated from wild-type cells, clonally expanded for 3 wk. Each horizontal line represents the analyzed 430 bp VJλ, showing point mutations (lollipop shape) and Ig gene conversion tracts (horizontal bar above line). The numbers at the bottom indicate the number of nucleotides from the 76th base of the intronic sequence between leader and variable exons. (B) VJλ segments from clonally expanded cells carrying the indicated genotype at 2 wk after AID virus infection. O/E, ectopic overexpression of AID. Data are displayed as in A. Numbers of analyzed VJλ sequences are as follows: wild type, 29; POLH−/−, 43; POLQ−/−, 34; POLN−/−, 48; POLN−/−/POLQ−/− #1, 59; POLN−/−/POLQ−/− #2, 60; and POLH−/−/POLN−/−/POLQ−/−, 49. (C) Proportion of VJλ sequences indicating the number of Ig hypermutations (PM), Ig gene conversions (GC), and mutations of ambiguous origin (Amb; either hypermutations or gene conversions; see Materials and methods) at 2 wk after AID virus infection. Segment sizes are proportional to the number of sequences, indicated by the number of mutations (numbers around the perimeter of the pie charts). Data from POLN−/−/POLQ−/− #1 and POLN−/−/POLQ−/− #2 clones were combined and shown as a POLN−/−/POLQ−/− AID-overexpressing sample.
Figure 3.
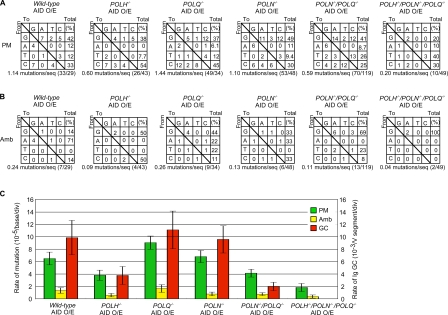
Nucleotide substitution preferences and rate of Ig hypermutation, Ig gene conversion, and mutations of ambiguous origin at 2 wk after AID virus infection in wild-type and mutant cells. (A) Nucleotide substitution preferences deduced from Ig hypermutation in the VJλ sequences (seq) of cells carrying the indicated genotype at 2 wk after AID virus infection. O/E, ectopic overexpression of AID. (B) AID-induced nucleotide substitution preferences from ambiguous (Amb) mutations, which are attributable to either Ig hypermutation or Ig gene conversion, are shown. (C) Rate of Ig hypermutation (PM), Ig gene conversion (GC), and mutations of ambiguous origin at 2 wk after AID virus infection. Error bars indicate SEM.
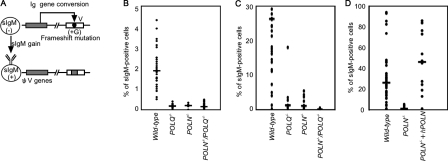
We next evaluated the contribution of Polν and Polθ to Ig gene conversion without overexpressing AID. We generated POLN−/−, POLQ−/−, and POLN−/−/POLQ−/− cells from surface IgM (sIgM)–negative wild-type cells carrying a +G frameshift mutation in the rearranged VJλ segment at the same site as in the CL18 clone (Fig. 4 A and Fig. 5 B; Buerstedde et al., 1990). We monitored the gain of sIgM expression resulting from the Ig gene conversion–mediated elimination of the frameshift mutation. We measured sIgM gain fluctuation using subclones derived from a single sIgM-negative cell after 3 wk of clonal expansion. Because sIgM gain was hardly detectable in the POLN−/− or POLQ−/− cells (Fig. 4 B), we augmented the rate of Ig gene conversion by treating cells with trichostatin A (TSA), a histone deacetylase inhibitor. TSA increases Ig gene conversion 50-fold without inducing Ig hypermutation, probably by relaxing the local chromatin structure specifically at the ψV segments and thereby promoting the interaction between the ψV donor segments and V(D)J segments (Seo et al., 2005; Nakahara et al., 2009). When compared with wild-type cells, the POLN−/− and POLQ−/− cells exhibited a significant decrease in the appearance of sIgM-positive revertant fractions (Fig. 4 C). It should be noted that such a decrease was not observed in AID-overexpressing cells (Fig. 3 C). This apparent discrepancy might be explained by the idea that the rate-limiting step of Ig gene conversion may be the interaction between the ψV segments and V(D)J segments in AID-overexpressing cells, whereas a subsequent DNA synthesis step may be rate limiting in TSA-treated cells. Thus, a compromised DNA synthesis step in POLN−/− and POLQ−/− cells may cause significant decreases in the rate of gene conversion only in TSA-treated cells but not in AID-overexpressing cells. In addition to significantly reduced Ig gene conversion in POLN−/− and POLQ−/− cells, remarkably, the reversion fraction was nearly undetectable in the POLN−/−/POLQ−/− cells (Fig. 4 C).
Figure 4.
Analysis of Ig gene conversion rate measured by sIgM gain. (A) Diagram of Ig gene conversion assay for DT40 cells. The frameshift mutation caused by the insertion of G in the rearranged Vλ segment in the parent cells is indicated by the circle. Gene conversion events using an upstream ΨV donor segment can eliminate the frameshift mutation, resulting in a gain in sIgM expression. The ΨV segment is indicated by the shaded box. (B) The kinetics of Ig gene conversion is evaluated by measuring the frequency of sIgM gain. The frequency of sIgM gain revertants of the indicated genotype was determined at 3 wk without TSA. We determined the percentage of sIgM gain variants from the following number of sIgM-negative subclones: wild type, 35; POLQ−/−, 22; POLN−/−, 13; and POLN−/−/POLQ−/−, 34. Wild-type data were described previously (Kawamoto et al., 2005; Nakahara et al., 2009). The horizontal bars in the panel indicate median percents. (C) The frequency of sIgM gain revertants of the indicated genotype was determined at 3 wk after addition of 1.25 ng/ml TSA. We determined the percentage of sIgM gain variants from the following number of sIgM-negative subclones: wild type, 14; POLQ−/−, 9; POLN−/−, 10; and POLN−/−/POLQ−/−, 6. (D) The frequency of sIgM gain revertants was determined at 3 wk after addition of 1.25 ng/ml TSA in POLN−/− clones reconstituted with human POLN cDNA. We determined the percentage of sIgM gain variants from the following number of sIgM-negative subclones: wild type, 39; POLN−/−, 10; and POLN−/− + hPOLN, 12.
Figure 5.
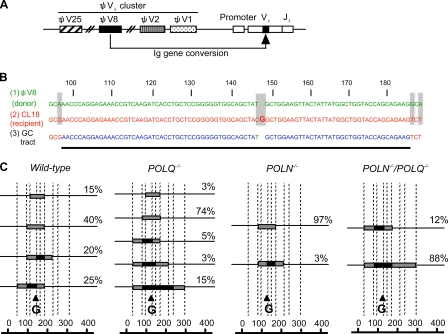
Increased length of Ig gene conversion tracts in POLN−/−/POLQ−/− cells. (A) Primary structure of the chicken Ig Vλ gene is shown. There are 25 ΨVλ segments upstream of a single functional VJλ segment. (B) Nucleotide sequences around the frameshift (bolded G nucleotide) of the recipient Vλ segment. The maximum gene conversion tract in sIgM-positive revertant cells is indicated by the bolded line. Shaded letters indicate mismatches between donor ΨV8 and recipient ΨVλ. The donor ΨV8 and recipient VJ are green and red, respectively. In the converted sequences, donor- and recipient-derived nucleotides are also colored green and red, respectively, whereas sequences derived from either donor or recipient are blue. Thus, gene conversion is initiated and terminated somewhere within the blue sequences flanking the green 146th T nucleotide. The numbers shown above the sequences indicate the nucleotide numbers in the Vλ exon. (C) Comparison of gene conversion tract spectra in the ΨVλ segment rearranged with ΨV8 in sIgM-positive revertants from three independent clones derived from wild-type, POLQ−/−, POLN−/−, and POLN−/−/POLQ−/− cells at 3 wk after TSA treatment. Conversion tract length can be estimated by the distance between the nucleotides at the mismatched sites in the donor and recipient DNA. The sequence shared by both the donor ΨV8 and the recipient gene is indicated by the shaded boxes. Crossover sites must be within these boxes. The dotted vertical lines indicate the position of mismatches between donor ΨV8 and recipient ΨVλ (these positions are 48, 97, 108, 145, 172, 182, 222, 237, and 304 bases from the 76th base of the intron between the leader and the Vλ exons). Black triangles indicate the position of the frameshift (corresponding to the bolded G in B) of the recipient VJ segment. The number of analyzed ΨVλ segments rearranged with the ΨV8 donor in wild-type, POLQ−/−, POLN−/−, and POLN−/−/POLQ−/− cells was 20, 39, 30, and 17, respectively.
To confirm the role of Polν in Ig gene conversion, we reconstituted POLN−/− cells with the human POLN transgene (Takata et al., 2006). The POLN transgene rescued the rate of Ig gene conversion to a level comparable with that of wild-type cells (Fig. 4 D), confirming an important role for Polν in Ig gene conversion. When taken together, nucleotide sequence analysis of Ig Vλ in AID-overexpressing cells (Fig. 3) and measurement of sIgM gain (Fig. 4) indicate the involvement of both Polν and Polθ in Ig gene conversion (Fig. 1).
Increased length of gene conversion tracts in POLN−/−/POLQ−/− cells
To assess the nature of Ig gene conversion events, we examined the type of Ig gene conversion that causes sIgM gain. We sorted sIgM-positive cells from three clonally expanded populations from each genotype. To measure the length of the gene conversion tracts, we analyzed those tracts where the +G frameshift mutation of the rearranged VJλ segment (Buerstedde et al., 1990) is replaced by the ψV8 donor segment (Fig. 5 A). To achieve this, we subcloned each genotype, cultured individual subclones in the presence of TSA, sorted sIgM-positive revertants at 3 wk, and determined the nucleotide sequence in the VJλ segment. The sequences around the parental frameshift mutation were replaced by ψV8 in most of the VJλ segments analyzed, presumably because the VJλ segment around the frameshift mutation has the highest sequence similarity (92.4%) to the ψV8 segment (Buerstedde et al., 1990).
The length of the gene conversion tract involving ψV8 was estimated as reported previously for the analysis of POLH−/− cells (Kawamoto et al., 2005). Conducting a sequence comparison between (a) a pseudo-V8 (donor), (b) the VJλ segment containing the frameshift mutation (recipient), and (c) their converted sequences (Fig. 5 B) enabled us to determine which sequences in the recipient were replaced by which donor in each product. For example, sequence analysis of tracts in wild-type cells indicated that the 5′-TACGGCT-3′ sequences in the recipient were replaced by ψV8-derived 5′-TATGCT-3′ sequences in all analyzed products (Fig. 5 B; note that the bolded G causes the frameshift and corresponds to the black triangle in Fig. 5 C). Thus, the CG of the recipient is replaced by the T of the donor through Ig gene conversion, which restores the reading frame of the Ig Vλ gene. These converted sequences were flanked by identical sequences that were shared by all donor, recipient, and converted sequences (Fig. 5 B, blue). Outside this region of ∼80 nucleotides, there are occasional mismatches, indicated by the gray shading in Fig. 5 B and by the dotted lines in Fig. 5 C.
We were able to define a minimum tract length (distance between the furthest mismatched nucleotides in the Ig gene conversion product and recipient) and a maximum tract length (distance between the closest mismatched nucleotides in the Ig gene conversion product and donor; Fig. 5 C). We confirmed that the starting sequences of the frameshift in wild-type and each mutant were identical (Fig. S1). We next calculated the maximum length of the gene conversion tract for each genotype. 25% of the analyzed gene conversion tracts contained the donor sequences at the two mismatches, the 48th and 182nd nucleotides in the wild-type clones. This result defines the maximum length of the conversion tract in wild-type cells (134 nucleotides). Likewise, 3% of the gene conversion tracts exhibited a maximum length of 125 nucleotides in the POLN−/− cells. Remarkably, the POLN−/−/POLQ−/− clones exhibited a dramatic increase in the length of the gene conversion tract compared with cells carrying the other genotypes, with 88% of the gene conversion tracts exhibiting a maximum length of 256 nucleotides. Tract length in POLN−/−/POLQ−/− clones is statistically longer than that of POLQ−/− clones, and POLN−/−/POLQ−/− clones have the longest tract (242 vs. 116; P < 0.01). Moreover, all of the gene conversion tracts exhibited a longer minimum length in three subclones derived from the POLN−/−/POLQ−/− cells than in the wild-type cells. These results suggest that other unidentified polymerases may carry out DNA synthesis with higher processivity in the absence of Polν and Polθ and thereby significantly increase the length of the gene conversion tracts in POLN−/−/POLQ−/− cells (Fig. 5 C).
No detectable Ig gene conversion events in POLH−/−/POLN−/−/POLQ−/− cells
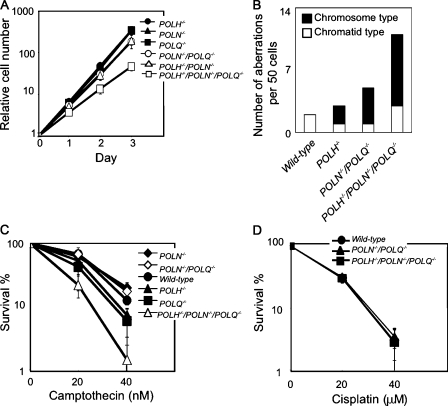
Because both POLN−/−/POLQ−/− and POLH−/− DT40 clones exhibited a significant decrease in the rate of Ig gene conversion, we wanted to explore the functional redundancy between the two A-type DNA polymerases and Polη. To this end, we disrupted the POLH gene in two independently isolated POLN−/−/POLQ−/− clones. The resulting three POLH−/−/POLN−/−/POLQ−/− clones consistently showed a marked retardation of cellular proliferation (Fig. 6 A) accompanied by increased levels of spontaneous chromosomal aberrations (Fig. 6 B). Thus, the three DNA polymerases functionally overlap in terms of the maintenance of chromosomal DNA. We next measured Ig gene conversion in POLH−/−/POLN−/−/POLQ−/− clones. Remarkably, we were not able to detect any Ig V gene conversion tracts in POLH−/−/POLN−/−/POLQ−/− clones (Fig. 2 C and Fig. 3 C). Moreover, ambiguous A/T mutations, which are products of HR (Saberi et al., 2008), were abolished in the POLH−/−/POLN−/−/POLQ−/− cells (Fig. 3 B). These observations demonstrate that these three error-prone DNA polymerases are sufficient for all Ig gene conversion events in DT40 cells.
Figure 6.
POLH−/−/POLN−/−/POLQ−/− cells displayed growth retardation, chromosomal aberrations, and hypersensitivity to chemotherapeutic agent. (A) Growth curve for cells of the indicated genotype. (B) Spontaneous chromosomal aberrations per 50 cells in the indicated genotype. (C) Cells carrying the indicated genotype were exposed to camptothecin. (D) Cells carrying the indicated genotype were exposed to cisplatin. Doses are displayed on the x axes on a linear scale, and the fractions of surviving colonies are displayed on the y axes on a logarithmic scale. Error bars show the standard error of the mean for at least three independent experiments.
HR-dependent repair of POLN−/−/POLQ−/− and POLH−/−/POLN−/−/POLQ−/− cells
Although HR plays a key role in cellular tolerance to cisplatin (Nojima et al., 2005), POLN−/−/POLQ−/− cells exhibit a normal tolerance to chemical cross-linking agents (Yoshimura et al., 2006). The apparent discrepancy between this observation and the important role played by Polν and Polθ in Ig gene conversion led us to reevaluate the efficiency of HR reactions in cells deficient in POLN and POLQ. To this end, we measured the frequency of gene targeting (Table I) and cellular tolerance to cisplatin and camptothecin, a topoisomerase I poison (Fig. 6, C and D), and the HR-dependent repair of ISceI restriction enzyme–induced DSBs (Fig. 7).
Table I.
Targeted integration frequencies
| Genotype | OVALBMIN locus | POLK locus |
| Wild type | 18/19 (95%) | 2/23 (8.7%) |
| POLQ−/− | 20/21 (95%) | ND |
| POLN−/− | 22/23 (96%) | 8/51 (16%) |
| POLN−/−/POLQ−/− | 29/31 (94%) | 1/10 (10%) |
Wild-type and knockout cells were transfected with targeting constructs of the indicated genotype. The number of targeted clones/number of drug-resistant clones analyzed is shown.
Figure 7.
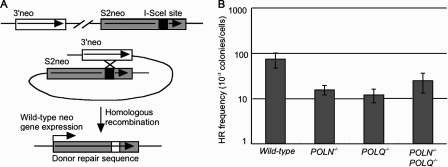
The role of Polν and Polθ in HR-dependent repair of ISceI-mediated DSBs. (A) The diagram shows the method of measuring the frequency of HR in an artificial substrate DNA, SCneo. The expression vector encoding the ISceI restriction enzyme was transiently transfected into cells carrying SCneo in the OVALBUMIN locus. Shaded and white boxes represent the S2neo and 3′neo genes, respectively (not to scale). Successful gene conversion from 3′neo to S2neo reconstitutes the functional neomycin resistance gene. (B) Results of an ISceI-induced gene conversion assay for the indicated genotypes are shown. The histogram shows the frequency of HR-dependent DSB repair, which is estimated by measuring the frequency of neomycin-resistant colonies per cell. Error bars show the standard error of the mean for at least three independent experiments.
Camptothecin stabilizes a complex of topoisomerase I covalently linked to nicked DNA. This complex interrupts replication and causes DSBs in one of the sister chromatids. These DSBs are repaired exclusively by HR using the intact sister chromatid (Hochegger et al., 2006; Pommier, 2006; Saberi et al., 2007). We found that neither POLN−/−, POLQ−/−, nor POLN−/−/POLQ−/− clones exhibited an increased sensitivity to camptothecin (Fig. 6 C). Likewise, the frequency of targeted integration into the OVALBUMIN and the POLK locus was not diminished in any of the mutant clones (Table I). Collectively, although Polν and Polθ play a critical role in Ig gene conversion, these DNA polymerases have little, if any, contribution to HR-mediated repair. Alternatively, the functional overlap between the two A-type DNA polymerases and other DNA polymerases may mask the contribution of Polν and Polθ to HR-dependent repair.
We next explored the functional relationship of Polν and Polθ with Polη in HR-mediated repair. To this end, we measured cellular sensitivity to camptothecin and cisplatin. Intriguingly, the POLH−/−/POLN−/−/POLQ−/− cells exhibited an increase in sensitivity to camptothecin, a phenotype that was not shared by the POLH−/− or POLN−/−/POLQ−/− clones (Fig. 6 C). These observations reveal that the two A-type DNA polymerases can play a role in general HR only when Polη is deleted. In contrast, POLH−/−/POLN−/−/POLQ−/− clones showed a normal sensitivity to cisplatin (Fig. 6 D). Presumably, other polymerases may have substituted for Mus308, the prototype polymerase of D. melanogaster, which plays a critical role in providing a cellular tolerance to cross-linking reagents.
To analyze HR-dependent DSB repair in an artificial substrate, we inserted an HR substrate, SCneo, which carries the rare cutting endonuclease site, ISceI, into the OVALBUMIN locus of wild-type and all mutant clones (Johnson and Jasin, 2000; Fukushima et al., 2001). This construct carries two mutant neomycin-resistance genes (S2neo and 3′-neo), which are localized in tandem and are complementary to each other (Fig. 7 A). After transient expression of ISceI, the induced DSB in S2neo is repaired by HR, with the 3′-neo serving as the donor for gene conversion, leading to the restoration of a functional neomycin resistance gene. Thus, we can measure the frequency of HR-dependent DSB repair by counting the number of neomycin-resistant (neoR) colonies. The frequency of HR-dependent repair was decreased three to six times in POLQ−/−, POLN−/−, and POLN−/−/POLQ−/− cells in comparison with wild-type cells (Fig. 7 B). Collectively, Polν and Polθ contribute to some of the HR reactions.
Polν, Polθ, and Polη are involved in TLS-dependent hypermutation at IgV
Biochemical studies and our previous study on POLN−/−/POLQ−/− DT40 cells suggest a role for Polν and Polθ in TLS (Seki et al., 2005; Takata et al., 2006; Yoshimura et al., 2006; Arana et al., 2007). To verify this conclusion, we examined nontemplated single-base substitutions (Ig hypermutation) in cells deficient in POLH, POLN, and POLQ. AID overexpression for 2 wk increased the rate of Ig hypermutation from 1.3 × 10−6 to 6.3 × 10−5 per nucleotide per division (Fig. 2, A and B; and Fig. 3 C). The rate of Ig hypermutation exhibited only a modest change in the single- and double-mutant clones when compared with wild-type cells; 1.7 times lower for POLH−/− cells, 1.4 times higher for POLQ−/− cells, 1.1 times higher for POLN−/− cells, and 1.6 times lower for POLN−/−/POLQ−/− cells (Fig. 2 C and Fig. 3 C). On the contrary, the rate of Ig hypermutation in wild-type cells is 3.5 times higher than in POLH−/−/POLN−/−/POLQ−/− cells (Fig. 2 C and Fig. 3 C).
The POLN−/−/POLQ−/− cells showed a marked reduction in the number of C to G mutations when compared with wild-type cells, indicating that Polν and Polθ play a role in TLS past the abasic site (P < 0.0095; Fig. 3 A). The C to G, C to T, and G to C mutations tended to also be decreased in POLH−/−/POLN−/−/POLQ−/− clones when compared with wild-type cells. These data support the notion that A family DNA polymerases Polν and Polθ, together with Y family DNA polymerase Polη, contribute to TLS-dependent Ig hypermutation in DT40 cells (Fig. 1).
Discussion
We previously demonstrated that Polθ can play a role in base excision repair and probably in TLS (Yoshimura et al., 2006). In this study, we demonstrate a dominant role played by Polθ and Polν in HR-dependent Ig V gene conversion and in TLS-dependent Ig hypermutation. These DNA polymerases make only a small contribution to HR, as indicated by the normal range of cellular tolerance to the anticancer agents cisplatin and camptothecin and by normal efficiency of gene targeting (Fig. 6, C and D; and Table I). Nonetheless, the hypersensitivity of POLH−/−/POLN−/−/POLQ−/− cells but not of POLH−/− or POLN−/−/POLQ−/− cells to camptothecin reveals that the two A-type DNA polymerases can contribute to HR-dependent DNA repair if other HR-related DNA polymerases are not available (Fig. 6 C). In addition, reduced HR-dependent DSB repair in POLN−/− and POLQ−/− cells as well as in Polν-depleted human cells (Zietlow et al., 2009; Moldovan et al., 2010) indicates a contribution of Polν and Polθ preferentially to HR in DSB repair (Fig. 7 B). Conceivably, the choice of DNA polymerases in the DNA synthesis step of HR may be dependent on the type of DNA damage, which may explain why POLN−/− and POLQ−/− cells showed defective HR-dependent DSB repair without displaying sensitivity to cisplatin or camptothecin.
Accumulating evidence indicates that individual DNA polymerases can perform multiple roles, e.g., Polκ for nucleotide excision repair and TLS (Ogi and Lehmann, 2006; Ogi et al., 2010), Polη for HR and TLS (Kawamoto et al., 2005), and Polζ for HR and TLS (Sonoda et al., 2003). Thus, we can also add the role played by Polν and Polθ in HR and TLS to the list of multiple functions performed by individual DNA polymerases.
Polν and Polθ may promote DNA synthesis in Ig gene conversion
Our results support the premise that Polν and Polθ play a role in the DNA synthesis step of HR. First, Ig gene conversion was significantly decreased in POLN−/−/POLQ−/− cells (Fig. 2 C, Fig. 3 C, and Fig. 4 C). Second, this decrease may be associated with a marked increase in the length of the gene conversion tract (Fig. 5 C). Third, no Ig gene conversion events were detectable in POLH−/−/POLN−/−/POLQ−/− clones (Fig. 2 C and Fig. 3 C). Therefore, Polη may be responsible for gene conversion events having the longer gene conversion tract. Purified Polη can undergo DNA synthesis using a D loop structure as a template (McIlwraith et al., 2005). Thus, like Polη, the two A-type DNA polymerases may contribute to the DNA synthesis step of Ig gene conversion.
During metazoan evolution, Polν and Polθ appear to have obtained a specialized HR function (HR-dependent Ig V diversification in B lymphocyte precursors), whereas other DNA polymerases may have substituted for Mus308, the prototype DNA polymerase, which plays a critical role in cellular tolerance to DNA-damaging agents. It should be noted that Ig gene conversion is distinctly different from other HR reactions because it allows for recombination between two diverged sequences. In fact, although a 0.1% sequence divergence strongly suppresses HR reactions (te Riele et al., 1992), Ig gene conversion is efficiently performed between homeologous sequences with 10% divergence. This divergence may strongly suppress DNA synthesis in the D loop structure such that a mismatch between primer and template strands interferes with DNA synthesis from the primer and also destabilizes the D loop (Johnson and Jasin, 2000; Biet et al., 2003). Accordingly, during evolution, Polν and Polθ may have acquired a specialized character suitable for this extremely difficult task, HR-dependent Ig V diversification. Ig gene conversion plays an important role in diversifying Ig V gene not only in chickens but also in the rabbit, whereas primates and rodents rely on V(D)J recombination to diversify Ig V (Lundqvist et al., 2006; Mage et al., 2006). Evidence that Ig gene conversion contributes to Ig V diversification in other mammals such as pig, cattle, sheep, and horses is less convincing (Butler et al., 2006; Zhao et al., 2006). Because performing effective DNA synthesis in Ig gene conversion is a difficult challenge for DNA polymerases, this study suggests the critical role played by Polη, Polν, and Polθ in Ig V diversification in some domesticated animals as well as in DT40 cells. Conceivably, vertebrates are faced with more complex tasks, such as acquired immune reactions, in addition to the maintenance of genomic DNA (Fig. 6 B). In addition to new DNA polymerases evolving to perform DNA damage responses, the function of existing polymerases appears to have shifted during metazoan evolution from involvement in the DNA damage response to the diversification of Ig V genes.
Contribution of Polη, Polν, and Polθ to TLS past abasic sites
In addition to the DNA synthesis step of Ig gene conversion, the following data suggest that Polν and Polθ may carry out TLS past abasic sites in Ig V hypermutation in DT40 cells. POLN−/−/POLQ−/− cells showed a significant reduction in the number of C to G mutations in comparison with wild-type cells. In addition, POLH−/−/POLN−/−/POLQ−/− cells showed 3.5 times lower total number of mutations in comparison with wild-type cells. Therefore, we conclude the involvement of these three DNA polymerases in nontemplated Ig V hypermutation in DT40 cells (Fig. 1). It should be noted that the role of TLS in Ig V hypermutation in human and mouse B lymphocytes is unclear. In other words, although several studies demonstrate the role of TLS polymerases in murine Ig V hypermutation (Masuda et al., 2006; Martomo et al., 2008; Faili et al., 2009; Schenten et al., 2009), it is unclear whether TLS polymerases carry out DNA synthesis on intact template strands or bypass unknown DNA lesions to accumulate mutations.
DT40 cells deficient in Rev1, another TLS polymerase, exhibit a 75% drop in the number of Ig V hypermutation events in comparison with wild-type cells, indicating a higher contribution of Rev1 to Ig V hypermutation compared with Polη, Polν, and Polθ (Ross and Sale, 2006). Intriguingly, Polθ contains the Rev1-interacting motif x-x-x-F-F-y-y-y-y (x, no specific residue; y, no specific residue but not proline), which is conserved among species (three motifs in human and chicken and four motifs in mouse; Ohashi et al., 2009). The biological significance of possible interactions between Rev1 and Polθ should be elucidated in the future.
The data of nontemplated mutations do not completely agree with biochemical studies of Polν and Polθ. Although only Polθ, but not Polν, can efficiently bypass abasic sites in vitro (Seki and Wood, 2008), our study indicates the contribution of both DNA polymerases to this bypass reaction in vivo (Fig. 3 A). Furthermore, although in vitro studies predict that a defect in Polθ causes reduction in C to T and G to A transition mutations (Seki et al., 2004), this prediction is not verified by the spectrum of nontemplated mutations in our study (Fig. 3 A). This is also the case with POLQ-deficient mice, which show ambiguous results on Ig hypermutation, making it difficult to conclude whether Polθ is involved in Ig hypermutation (Zan et al., 2005; Masuda et al., 2006; Martomo et al., 2008). Purified Polν is an extremely low fidelity enzyme incorporating T opposite template G with a frequency of 0.45 (Takata et al., 2006; Arana et al., 2007), whereas C to T or G to A transition mutations are not significantly decreased in the absence of Polν in this study (Fig. 3 A). However, it should be noted that some C to T and G to A transitions might be derived from nonprocessed uracil (simply generated by AID), which instructs template T and may be bypassed by many polymerases and not just Polν and Polθ. In contrast with these discrepancies, Polη-mediated preferential incorporation of A opposite abasic sites in vitro (Kokoska et al., 2003; Zhao et al., 2004) is in agreement with the decreased number of C to T or G to A transition mutations caused by the loss of Polη in POLN−/−POLQ−/− cells (Fig. 3 A). Further studies are required to discuss the relevance of in vitro studies to TLS in vivo because of complex functional interactions among multiple polymerases.
Materials and methods
Cell culture and DNA transfection
DT40 cells were cultured in RPMI 1640 medium supplemented with 10 µM β-mercaptoethanol, 10% fetal calf serum, and 1% chicken serum (Sigma-Aldrich) at 39.5°C (Sonoda et al., 1998). 107 cells were suspended in 0.5 ml PBS containing 10–30 µg linearized plasmid for each transfection and electroporated with a gene pulser apparatus (Bio-Rad Laboratories) at 550 V and 25 mF. After electroporation, cells were transferred into 20 ml fresh medium and incubated for 24 h. Cells were resuspended in 80 ml medium containing the appropriate drugs and divided into four 96-well plates. After 7–10 d, drug-resistant colonies were transferred to 24-well plates (Buerstedde and Takeda, 1991).
Measurement of targeted integration frequencies
To analyze integration events at the OVALBMIN locus and POLK locus, targeted construct DNAs were transfected into cells, and Southern blot analysis was performed after selection of clones resistant to the appropriate antibiotics (Buerstedde and Takeda, 1991).
Generation of gene-disrupted cells
We transiently expressed Cre-ER and exposed POLN−/− and POLN−/−/POLQ−/− cells to 4-hydroxytamoxifen (OH-TAM) to delete the POLN-bsr, POLN-hisD, POLQ-neo, and POLQ-puro marker genes, which were flanked with loxP sites. The resulting POLN−/− and POLN−/−/POLQ−/− cells were sequentially transfected with POLH-puro and POLH-bsr targeting constructs to obtain POLH−/−/POLN−/− and POLH−/−/POLN−/−/POLQ−/− clones, respectively (Kawamoto et al., 2005; Yoshimura et al., 2006). To express human POLN in POLN−/− cells, human cDNA was inserted into expression vector containing the puro selection marker gene (Takata et al., 2006).
Proliferation analysis
Cells were counted using a hematocytometer, and cells were diluted to 105/ml in 5 ml medium every 24 h. Next, we calculated the relative cell number.
Colony formation assay
Serially diluted cells were plated onto 6-well plates with 5 ml/well 1.5% (wt/vol) methylcellulose (Sigma-Aldrich) containing Dulbecco’s modified Eagle’s medium/F-12 (Invitrogen), 15% fetal calf serum (Equitech-Bio), 1.5% chicken serum (Sigma-Aldrich), and 10 µM β-mercaptoethanol and were incubated at 39.5°C for 6–7 d. For exposure of cells to cisplatin (cis-platinum [II] diaminodichloride; Nihon-Kayaku), 105 cells were incubated at 39.5°C in 1 ml complete medium containing cis-platinum (II) diaminodichloride for 1 h. Cells were plated onto 6-well plates with 5 ml/well 1.5% (wt/vol) methylcellulose-containing medium and were incubated at 39.5°C for 7–10 d. To measure the sensitivity to camptothecin, cells were plated in methylcellulose medium containing camptothecin and were incubated at 39.5°C for 7–10 d (Okada et al., 2002; Kohzaki et al., 2007).
Chromosomal aberration analysis
To measure spontaneous chromosomal aberrations, cells were incubated with 0.1 mg/ml colcemid for 3 h before fixation to enrich mitotic cells. Harvested cells were treated in 75 mM KCl for 15 min at room temperature and fixed in 5 ml freshly prepared 3:1 mixture of methanol/acetic acid. Cell suspension was dropped onto a wet glass slide with 50% ethanol and immediately flame dried. The slides were treated with 3% giemsa solution in 50 mM phophate buffer, pH 6.4 (3.36 g/liter KH2PO4 and 2.78 g/liter Na2HPO4). They were rinsed carefully with water from the opposite site of the sample, air dried, covered with cover glass, and observed on microscope oil (1,000× magnification; Sonoda et al., 1998).
Measurement of recombination frequencies using ISceI-induced DSB repair
Modified SCneo was inserted into the OVALBUMIN gene construct and targeted into the OVALBUMIN locus in wild-type, POLN−/−, POLQ−/−, and POLN−/−/POLQ−/− DT40 cells. In transient transfections, 5 × 106 cells suspended in 0.5 ml PBS were mixed with the following plasmid DNAs without linearization: 10 µg pBluescript SK plus 10 µg ISceI expression vector (pCBASce). 20 µg pBluescript II KS was used as a negative control.
Cells were grown for 7–10 d, and HR frequencies were calculated using the following equation: HR frequency (colonies/cell) = number of G418-resistant colonies/plating efficiency of transfected cells in the absence of G418 (Kikuchi et al., 2005). We obtained and counted colonies in each sample to calculate a mean value and standard error.
Generation of AID expression retrovirus and infection into DT40 cells
For retrovirus infection, the pMSCV-IRES-GFP recombinant plasmid was constructed by ligating the 5.2-kb BamHI–NotI fragment from pMSCVhyg (Takara Bio Inc.) with the 1.2-kb BamHI–NotI fragment of pIRES2-EGFP (Takara Bio Inc.). Mouse AID cDNA was inserted between the BglII and EcoRI sites of pMSCV-IRES-GFP. The plasmids were transfected into the packaging cell line by FuGENE reagent (Roche). Preactivated cells were suspended at a density of 4 × 106 cells/ml in the medium containing retrovirus supplemented with 16 mg/ml polybrene (Sigma-Aldrich), centrifuged at 3,000 rpm for 45 min at 32°C, and incubated for 48 h (Shinkura et al., 2004). The efficiency of infection was >80%, as judged by GFP expression using FACS. Cells were subcloned into 96-well plates 2 d after infection, and after 2 wk, GFP-positive clones were selected by FACS analysis.
Analysis of AID-induced Ig hypermutation and Ig gene conversion at VJλ segments
DNA was extracted from more than three clones each of wild-type, POLQ−/−, POLN−/−, and POLN−/−/POLQ−/− cells at 2 wk after subcloning (Fig. S2). PCR-amplified fragments of Vλ segments were cloned into plasmid and subjected to base sequence analysis. The rearranged VJλ was amplified using the CVLF6 (5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′) and CVLR3 (5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′) primers as previously described (Sale et al., 2001). After purification with a gel extraction kit (QIAquick; QIAGEN), PCR products were cloned into the TOPO pCR2.1 cloning vector (Invitrogen) and sequenced with the M13 forward or reverse primer and a sequencer (ABI PRISM 3100; Applied Biosystems). Sequence alignment with GENETYX-MAC (Software Development) allowed the identification of changes from the consensus sequences in each clone. Frequencies of Ig hypermutation, ambiguous mutations, and Ig gene conversion were determined as previously described (Sale et al., 2001). In brief, all sequence changes were assigned to one of three categories: Ig gene conversion, nontemplated point mutation, or an ambiguous category. This discrimination rests on the published sequences of the Vλ pseudogenes that could act as donors for gene conversion. For each mutation, the database of the Vλ pseudogene was searched for a potential donor. If no pseudogene donor containing a string of >9 bp could be found, the mutation was categorized as a nontemplated point mutation. If such a string was identified and there were further mutations that could be explained by the same donor, all of these mutations were assigned to a single long-tract gene conversion event. If there were no further mutations, it was possible that the isolated mutation could have arisen through a conversion mechanism or could have been nontemplated and was therefore categorized as ambiguous.
The rate of hypermutation and Amb mutation was calculated using the following equation: rate of mutation = mutation frequency/length of sequenced Ig light chain locus (430 bp)/sequenced sample numbers/number of cell divisions (42 cycles for wild-type, 37 cycles for POLH−/−, POLN−/−, POLQ−/− single-gene–disrupted clones, 34 cycles for POLN−/−/POLQ−/− double-mutant cells, and 26 cycles for POLH−/−/POLN−/−/POLQ−/− triple-mutant cells for 14 d). The rate of Ig gene conversion was calculated using the following equation: rate of gene conversion = gene conversion frequency/sequenced sample numbers/number of cell divisions.
Analysis of the rate of sIgM gain
The generation frequency of sIgM gain revertants was monitored by flow cytometric analysis of cells that had been expanded for 3 wk after subcloning and stained with FITC-conjugated goat anti–chicken IgM (Bethyl Laboratories, Inc.). To enhance Ig gene conversion, 1.25 ng/ml TSA was added to the sIgM-negative cell populations, and the fraction of sIgM-positive revertants was monitored over time as described previously (Seo et al., 2005).
Nucleotide sequence analysis of rearranged VJλ segments derived from sIgM gain fractions
Fractions of three independent sIgM gain revertants were obtained from each genotype using MACS separation columns (Miltenyi Biotec; Fig. S2). Genomic DNA was amplified by PCR with Pyrobest DNA polymerase (30 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min). The rearranged VJλ was amplified using the CVLF6 (5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′) and CVLR3 (5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′) primers as previously described (Sale et al., 2001). After purification with a gel extraction kit (QIAquick), PCR products were cloned into the TOPO pCR2.1 cloning vector and sequenced with the M13 forward or reverse primer and a sequencer (ABI PRISM 3100). Sequence alignment with GENETYX-MAC allowed the identification of changes from the consensus sequences in each clone. Frequencies of Ig hypermutation and Ig gene conversion were determined as previously described (see Analysis of AID-induced Ig…).
Online supplemental material
Fig. S1 shows the Vλ sequence alignment of wild-type, POLQ−/−, POLN−/−, and POLN−/−/POLQ−/− clones before TSA treatment. Fig. S2 shows the number of subclones and sequences analyzed in individual experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912012/DC1.
Acknowledgments
We thank the members of the Takeda and Halazonetis laboratories for their help and support. We thank R.D. Wood (MD Anderson Cancer Center, The University of Texas, Houston, TX) for the expression vector of human POLN. We thank J.E. Sale, C. Rada (Medical Research Council Laboratory of Molecular Biology, Cambridge, England, UK), S.C. West (Clare Hall Laboratories, London, England, UK), J. Jiricny (University of Zurich, Zurich, Switzerland), M.L. Santiago-Raber, S. Izui (University of Geneva), H. Hochegger (University of Sussex, Brighton, England, UK), K.L. Knight (Loyola University, Chicago, IL), M. Takata, T.J. Evans (Kyoto University), K. Kikuchi (Osaka University), A. Saberi, and M. Nakahara for their helpful discussion and support. Special thanks to Y. Sato, R. Ohta, A. Noguchi, and R. Kabata for their technical assistance.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas and a Grant-in-Aid from the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, and Science of Japan and by grants from The Uehara Memorial Foundation and The Naito Foundation.
Footnotes
Abbreviations used in this paper:
- AID
- activation-induced deaminase
- DSB
- double-strand break
- HR
- homologous recombination
- sIgM
- surface IgM
- TLS
- translesion DNA synthesis
- TSA
- trichostatin A
References
- Arakawa H., Hauschild J., Buerstedde J.M. 2002. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science. 295:1301–1306 10.1126/science.1067308 [DOI] [PubMed] [Google Scholar]
- Arakawa H., Moldovan G.L., Saribasak H., Saribasak N.N., Jentsch S., Buerstedde J.M. 2006. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 4:e366 10.1371/journal.pbio.0040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M.E., Takata K., Garcia-Diaz M., Wood R.D., Kunkel T.A. 2007. A unique error signature for human DNA polymerase nu. DNA Repair (Amst.). 6:213–223 10.1016/j.dnarep.2006.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M.E., Seki M., Wood R.D., Rogozin I.B., Kunkel T.A. 2008. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 36:3847–3856 10.1093/nar/gkn310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biet E., Sun J.S., Dutreix M. 2003. Stimulation of D-loop formation by polypurine/polypyrimidine sequences. Nucleic Acids Res. 31:1006–1012 10.1093/nar/gkg195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J.B., Sakaguchi K., Harris P.V. 1990. mus308 mutants of Drosophila exhibit hypersensitivity to DNA cross-linking agents and are defective in a deoxyribonuclease. Genetics. 125:813–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerstedde J.M., Arakawa H. 2006. Immunoglobulin gene conversion or hypermutation: that’s the question. Subcell. Biochem. 40:11–24 [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M., Takeda S. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell. 67:179–188 10.1016/0092-8674(91)90581-I [DOI] [PubMed] [Google Scholar]
- Buerstedde J.M., Reynaud C.A., Humphries E.H., Olson W., Ewert D.L., Weill J.C. 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9:921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.E., Sun J., Wertz N., Sinkora M. 2006. Antibody repertoire development in swine. Dev. Comp. Immunol. 30:199–221 10.1016/j.dci.2005.06.025 [DOI] [PubMed] [Google Scholar]
- Di Noia J., Neuberger M.S. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48 10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Faili A., Stary A., Delbos F., Weller S., Aoufouchi S., Sarasin A., Weill J.C., Reynaud C.A. 2009. A backup role of DNA polymerase kappa in Ig gene hypermutation only takes place in the complete absence of DNA polymerase eta. J. Immunol. 182:6353–6359 10.4049/jimmunol.0900177 [DOI] [PubMed] [Google Scholar]
- Fukushima T., Takata M., Morrison C., Araki R., Fujimori A., Abe M., Tatsumi K., Jasin M., Dhar P.K., Sonoda E., et al. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 276:44413–44418 10.1074/jbc.M106295200 [DOI] [PubMed] [Google Scholar]
- Harris R.S., Sale J.E., Petersen-Mahrt S.K., Neuberger M.S. 2002. AID is essential for immunoglobulin V gene conversion in a cultured B cell line. Curr. Biol. 12:435–438 10.1016/S0960-9822(02)00717-0 [DOI] [PubMed] [Google Scholar]
- Hochegger H., Dejsuphong D., Fukushima T., Morrison C., Sonoda E., Schreiber V., Zhao G.Y., Saberi A., Masutani M., Adachi N., et al. 2006. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 25:1305–1314 10.1038/sj.emboj.7601015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Jasin M. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398–3407 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Araki K., Sonoda E., Yamashita Y.M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Takeda S. 2005. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 20:793–799 10.1016/j.molcel.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Taniguchi Y., Hatanaka A., Sonoda E., Hochegger H., Adachi N., Matsuzaki Y., Koyama H., van Gent D.C., Jasin M., Takeda S. 2005. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol. Cell. Biol. 25:6948–6955 10.1128/MCB.25.16.6948-6955.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohzaki M., Hatanaka A., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szüts D., Sale J.E., Shinagawa H., Watanabe M., Takeda S. 2007. Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse. Mol. Cell. Biol. 27:2812–2820 10.1128/MCB.02043-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokoska R.J., McCulloch S.D., Kunkel T.A. 2003. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 278:50537–50545 10.1074/jbc.M308515200 [DOI] [PubMed] [Google Scholar]
- Lundqvist M.L., Middleton D.L., Radford C., Warr G.W., Magor K.E. 2006. Immunoglobulins of the non-galliform birds: antibody expression and repertoire in the duck. Dev. Comp. Immunol. 30:93–100 10.1016/j.dci.2005.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mage R.G., Lanning D., Knight K.L. 2006. B cell and antibody repertoire development in rabbits: the requirement of gut-associated lymphoid tissues. Dev. Comp. Immunol. 30:137–153 10.1016/j.dci.2005.06.017 [DOI] [PubMed] [Google Scholar]
- Marini F., Kim N., Schuffert A., Wood R.D. 2003. POLN, a nuclear PolA family DNA polymerase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 278:32014–32019 10.1074/jbc.M305646200 [DOI] [PubMed] [Google Scholar]
- Martomo S.A., Saribasak H., Yokoi M., Hanaoka F., Gearhart P.J. 2008. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst.). 7:1603–1608 10.1016/j.dnarep.2008.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Ouchida R., Hikida M., Nakayama M., Ohara O., Kurosaki T., O-Wang J. 2006. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst.). 5:1384–1391 10.1016/j.dnarep.2006.06.006 [DOI] [PubMed] [Google Scholar]
- McIlwraith M.J., Mcllwraith M.J., Vaisman A., Liu Y., Fanning E., Woodgate R., West S.C. 2005. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 20:783–792 10.1016/j.molcel.2005.10.001 [DOI] [PubMed] [Google Scholar]
- Moldovan G.L., Madhavan M.V., Mirchandani K.D., McCaffrey R.M., Vinciguerra P., D’Andrea A.D. 2010. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol. Cell. Biol. 30:1088–1096 10.1128/MCB.01124-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara M., Sonoda E., Nojima K., Sale J.E., Takenaka K., Kikuchi K., Taniguchi Y., Nakamura K., Sumitomo Y., Bree R.T., et al. 2009. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Ig v in chicken B lymphocytes. PLoS Genet. 5:e1000356 10.1371/journal.pgen.1000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima K., Hochegger H., Saberi A., Fukushima T., Kikuchi K., Yoshimura M., Orelli B.J., Bishop D.K., Hirano S., Ohzeki M., et al. 2005. Multiple repair pathways mediate tolerance to chemotherapeutic cross-linking agents in vertebrate cells. Cancer Res. 65:11704–11711 10.1158/0008-5472.CAN-05-1214 [DOI] [PubMed] [Google Scholar]
- Ogi T., Lehmann A.R. 2006. The Y-family DNA polymerase kappa (pol kappa) functions in mammalian nucleotide-excision repair. Nat. Cell Biol. 8:640–642 10.1038/ncb1417 [DOI] [PubMed] [Google Scholar]
- Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G., et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 37:714–727 10.1016/j.molcel.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Ohashi E., Hanafusa T., Kamei K., Song I., Tomida J., Hashimoto H., Vaziri C., Ohmori H. 2009. Identification of a novel REV1-interacting motif necessary for DNA polymerase kappa function. Genes Cells. 14:101–111 10.1111/j.1365-2443.2008.01255.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T., Sonoda E., Yamashita Y.M., Koyoshi S., Tateishi S., Yamaizumi M., Takata M., Ogawa O., Takeda S. 2002. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 277:48690–48695 10.1074/jbc.M207957200 [DOI] [PubMed] [Google Scholar]
- Okada T., Sonoda E., Yoshimura M., Kawano Y., Saya H., Kohzaki M., Takeda S. 2005. Multiple roles of vertebrate REV genes in DNA repair and recombination. Mol. Cell. Biol. 25:6103–6111 10.1128/MCB.25.14.6103-6111.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Haber J.E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F., Leung W.Y., Haber J.E. 1998. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol. Cell. Biol. 18:2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103 10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Pommier Y. 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 6:789–802 10.1038/nrc1977 [DOI] [PubMed] [Google Scholar]
- Reynaud C.A., Anquez V., Grimal H., Weill J.C. 1987. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell. 48:379–388 10.1016/0092-8674(87)90189-9 [DOI] [PubMed] [Google Scholar]
- Ross A.L., Sale J.E. 2006. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. 43:1587–1594 10.1016/j.molimm.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Saberi A., Hochegger H., Szuts D., Lan L., Yasui A., Sale J.E., Taniguchi Y., Murakawa Y., Zeng W., Yokomori K., et al. 2007. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol. Cell. Biol. 27:2562–2571 10.1128/MCB.01243-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi A., Nakahara M., Sale J.E., Kikuchi K., Arakawa H., Buerstedde J.M., Yamamoto K., Takeda S., Sonoda E. 2008. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol. Cell. Biol. 28:6113–6122 10.1128/MCB.00156-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale J.E. 2004. Immunoglobulin diversification in DT40: a model for vertebrate DNA damage tolerance. DNA Repair (Amst.). 3:693–702 10.1016/j.dnarep.2004.03.042 [DOI] [PubMed] [Google Scholar]
- Sale J.E., Calandrini D.M., Takata M., Takeda S., Neuberger M.S. 2001. Ablation of XRCC2/3 transforms immunoglobulin V gene conversion into somatic hypermutation. Nature. 412:921–926 10.1038/35091100 [DOI] [PubMed] [Google Scholar]
- Schenten D., Kracker S., Esposito G., Franco S., Klein U., Murphy M., Alt F.W., Rajewsky K. 2009. Pol zeta ablation in B cells impairs the germinal center reaction, class switch recombination, DNA break repair, and genome stability. J. Exp. Med. 206:477–490 10.1084/jem.20080669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Wood R.D. 2008. DNA polymerase theta (POLQ) can extend from mismatches and from bases opposite a (6-4) photoproduct. DNA Repair (Amst.). 7:119–127 10.1016/j.dnarep.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Marini F., Wood R.D. 2003. POLQ (Pol theta), a DNA polymerase and DNA-dependent ATPase in human cells. Nucleic Acids Res. 31:6117–6126 10.1093/nar/gkg814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Masutani C., Yang L.W., Schuffert A., Iwai S., Bahar I., Wood R.D. 2004. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 23:4484–4494 10.1038/sj.emboj.7600424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Gearhart P.J., Wood R.D. 2005. DNA polymerases and somatic hypermutation of immunoglobulin genes. EMBO Rep. 6:1143–1148 10.1038/sj.embor.7400582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H., Masuoka M., Murofushi H., Takeda S., Shibata T., Ohta K. 2005. Rapid generation of specific antibodies by enhanced homologous recombination. Nat. Biotechnol. 23:731–735 10.1038/nbt1092 [DOI] [PubMed] [Google Scholar]
- Shima N., Munroe R.J., Schimenti J.C. 2004. The mouse genomic instability mutation chaos1 is an allele of Polq that exhibits genetic interaction with Atm. Mol. Cell. Biol. 24:10381–10389 10.1128/MCB.24.23.10381-10389.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkura R., Ito S., Begum N.A., Nagaoka H., Muramatsu M., Kinoshita K., Sakakibara Y., Hijikata H., Honjo T. 2004. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 5:707–712 10.1038/ni1086 [DOI] [PubMed] [Google Scholar]
- Simpson L.J., Sale J.E. 2003. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. EMBO J. 22:1654–1664 10.1093/emboj/cdg161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Sasaki M.S., Buerstedde J.M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598–608 10.1093/emboj/17.2.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E., Okada T., Zhao G.Y., Tateishi S., Araki K., Yamaizumi M., Yagi T., Verkaik N.S., van Gent D.C., Takata M., Takeda S. 2003. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. EMBO J. 22:3188–3197 10.1093/emboj/cdg308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K., Shimizu T., Iwai S., Wood R.D. 2006. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J. Biol. Chem. 281:23445–23455 10.1074/jbc.M604317200 [DOI] [PubMed] [Google Scholar]
- Takeda S., Nakamura K., Taniguchi Y., Paull T.T. 2007. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol. Cell. 28:351–352 10.1016/j.molcel.2007.10.016 [DOI] [PubMed] [Google Scholar]
- te Riele H., Maandag E.R., Berns A. 1992. Highly efficient gene targeting in embryonic stem cells through homologous recombination with isogenic DNA constructs. Proc. Natl. Acad. Sci. USA. 89:5128–5132 10.1073/pnas.89.11.5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C., Kanaar R. 2006. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 40:363–383 10.1146/annurev.genet.40.110405.090451 [DOI] [PubMed] [Google Scholar]
- Yoshimura M., Kohzaki M., Nakamura J., Asagoshi K., Sonoda E., Hou E., Prasad R., Wilson S.H., Tano K., Yasui A., et al. 2006. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol. Cell. 24:115–125 10.1016/j.molcel.2006.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan H., Shima N., Xu Z., Al-Qahtani A., Evinger Iii A.J., Zhong Y., Schimenti J.C., Casali P. 2005. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 24:3757–3769 10.1038/sj.emboj.7600833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Xie Z., Shen H., Wang Z. 2004. Role of DNA polymerase eta in the bypass of abasic sites in yeast cells. Nucleic Acids Res. 32:3984–3994 10.1093/nar/gkh710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Jackson S.M., Aitken R. 2006. The bovine antibody repertoire. Dev. Comp. Immunol. 30:175–186 10.1016/j.dci.2005.06.012 [DOI] [PubMed] [Google Scholar]
- Zietlow L., Smith L.A., Bessho M., Bessho T. 2009. Evidence for the involvement of human DNA polymerase N in the repair of DNA interstrand cross-links. Biochemistry. 48:11817–11824 10.1021/bi9015346 [DOI] [PMC free article] [PubMed] [Google Scholar]