Depletion of the Nup188–Nup93 nuclear pore complex results in enlarged nuclei due to unrestricted protein import.
Abstract
All transport across the nuclear envelope (NE) is mediated by nuclear pore complexes (NPCs). Despite their enormous size, ∼60 MD in vertebrates, they are comprised of only ∼30 distinct proteins (nucleoporins or Nups), many of which form subcomplexes that act as building blocks for NPC assembly. One of these evolutionarily conserved subcomplexes, the Nup93 complex, is a major structural component linking the NPC to the membranes of the NE. Using in vitro nuclear assembly assays, we show that two components of the Nup93 complex, Nup188 and Nup205, are dispensable for NPC formation. However, nuclei lacking Nup188 increase in size by several fold compared with wild type. We demonstrate that this phenotype is caused by an accelerated translocation of integral membrane proteins through NPCs, suggesting that Nup188 confines the passage of membrane proteins and is thus crucial for the homeostasis of the different nuclear membranes.
Introduction
The defining attribute of eukaryotes is the compartmentalization of genetic material inside the cell’s nucleus. The physical boundary of the nuclear envelope (NE) has allowed eukaryotic cells to separate transcription and translation both spatially and temporally to achieve a level of regulatory complexity unprecedented in prokaryotes. Two lipid bilayers form the NE, the outer nuclear membrane (ONM) and the inner nuclear membrane (INM; for review see Hetzer et al., 2005). The ONM is continuous with the ER and is thought to have a rather similar protein composition (Mattaj, 2004; Lusk et al., 2007). The INM is characterized by a set of integral membrane proteins, although little is currently known as to how this specific protein composition is established and maintained (Mattaj, 2004; Zuleger et al., 2008).
Nuclear pore complexes (NPCs) are incorporated into the NE at sites where the INM and the ONM are fused. They function as gatekeepers of the nucleus, performing the essential cellular role of mediating an enormous exchange of proteins, nucleic acids, and other factors between the nucleoplasm and the cytoplasm. Transport of soluble factors has attracted much interest in previous decades: ions and small metabolites can diffuse through the NPCs; however, molecules >2.5 nm in radius (Mohr et al., 2009) are actively translocated by a large family of transport receptors (for reviews see Weis, 2003; Cook et al., 2007).
In addition to soluble cargoes, membrane-anchored proteins must also reach the nuclear interior. In the interphasic cell, passage through the NPC via the facing pore membrane is the most likely mechanism by which this could occur (Ohba et al., 2004). Transmembrane proteins of the NE are thought to be synthesized in the ER. They might reach the ONM and the cytoplasmic portion of the NPC by diffusion, although the existence of a directed transport mechanism cannot currently be ruled out. As INM proteins pass along the pore membrane, both transmembrane segments and hydrophilic domains encounter elements of the NPC. A recent study in mammalian cells has demonstrated that transport of integral membrane proteins to the INM is energy dependent (Ohba et al., 2004), and in budding yeast, INM targeting has been found to require transport receptors (King et al., 2006).
NPCs are huge assemblies of ∼45 MD in budding yeast and 60 MD in vertebrates (for review see Brohawn et al., 2008). They are formed by about 30 different nuclear pore proteins referred to as nucleoporins or Nups. Although most, if not all, nucleoporins have now been identified, it is not understood how the individual nucleoporins interact to form this large complex (Antonin et al., 2008; D’Angelo and Hetzer, 2008).
The majority of nucleoporins are organized in discrete subcomplexes each present in multiple copies in the NPC. The best characterized of these is the heptameric Nup84 complex in yeast and the vertebrate counterpart, the nonameric Nup107–Nup160 complex, which is essential for NPC assembly and one of the major structural components of the pore (Fabre and Hurt, 1997; Boehmer et al., 2003; Galy et al., 2003; Harel et al., 2003; Walther et al., 2003). The second major structural unit of the NPC is the Nup93 subcomplex in vertebrates or the Nic96 complex in yeast. In vertebrates, it is comprised of five nucleoporins: Nup205, Nup188, Nup155, Nup93, and Nup53. Both Nup155 and Nup53 have been found to be essential for NE and NPC assembly (Franz et al., 2005; Hawryluk-Gara et al., 2008), and loss of the two budding yeast Nup155 homologues Nup170p and Nup157p blocks NPC assembly (Makio et al., 2009). Interestingly, Nup170p and Nup157p bind to Nup53p and Nup59p, which in turn associate, like the corresponding Nup53 in vertebrates, with the transmembrane nucleoporin Ndc1p (Mansfeld et al., 2006; Onischenko et al., 2007), potentially anchoring the NPC in the membrane of the NE. In vertebrates, the three remaining nucleoporins Nup205, Nup188, and Nup93 are thought to exist in a complex (Meier et al., 1995). Immunodepletion of Nup93 from Xenopus laevis extracts followed by in vitro nuclear assembly reactions resulted in nuclei with reduced NPC staining, which suggests that NPC formation is impaired in the absence of Nup93 (Zabel et al., 1996). Similarly, the budding yeast Nup93 homologue Nic96p is required for NPC formation and interacts with both Nup188p and Nup192 (Nehrbass et al., 1996; Zabel et al., 1996; Gomez-Ospina et al., 2000), which in metazoans correspond to Nup188 and Nup205, respectively.
In this study, we characterize vertebrate Nup188 and Nup205. We show that both interact with Nup93 but not with each other and form two distinct complexes that can be functionally distinguished. Depletion of the Nup188–Nup93 complex from Xenopus egg extracts produces in vitro assembled nuclei that increase several fold in size compared with control nuclei and that contain NPCs, which allow an elevated flow of INM proteins across the pore membrane. This suggests that Nup188–Nup93 can restrict the passage of membrane proteins from the ONM to the INM and is thus an important factor in the establishment of these two membrane subcompartments of the NE.
Results
Nup93 is part of two different complexes
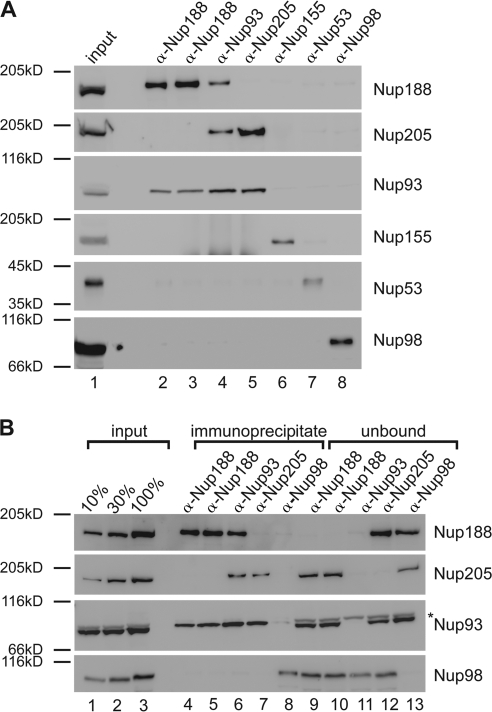
To understand the protein interaction network of the Nup93 complex, we performed immunoprecipitations using antibodies against each of the five members of the complex from the cytosol of Xenopus egg extracts (Fig. 1 A). Antibodies against Nup53 and Nup155 precipitated the respective antigens but not major proportions of other members of the Nup93 complex (Fig. 1 A, lane 6 and 7), as observed previously (Franz et al., 2005; Hawryluk-Gara et al., 2008). Unexpectedly, the three remaining members of the complex did not form a trimeric complex: antibodies against Nup188 from two different rabbits coprecipitated Nup93 but not Nup205 (Fig. 1 A, lane 2 and 3), and antibodies against Nup205 precipitated Nup93 but not Nup188 (Fig. 1 A, lane 5). Consistent with this, antibodies against Nup93 coprecipitated both Nup188 and Nup205 (Fig. 1 A, lane 4). As a negative control, no interactions were seen between Nup98 and Nup205, Nup188, Nup155, Nup93 or Nup53.
Figure 1.
Nup93 forms two distinct complexes. (A) Immunoprecipitations were performed from Xenopus egg cytosol with antibodies against Nup188 (two antibodies raised in different rabbits), Nup93, Nup205, Nup155, and Nup53 and, as a control, Nup98. The indicated proteins were identified by Western blotting with the respective antisera. 10% of starting material (input) was loaded on the left. (B) Quantitative immunoprecipitations were performed from limited amounts of Xenopus egg cytosol with antibodies against Nup188, Nup93, and Nup205 and, as a control, Nup98. Immunoprecipitates and unbound material as well as 10, 30, and 100% of the starting material (input) were analyzed by Western blotting. The Nup93 antibody also recognized a cross-reactivity (asterisk) in the starting and unbound material migrating slightly slower.
To exclude the possibility that the lack of interaction is caused by precipitating only a subpopulation of Nup188 or Nup205, we performed the experiment in a quantitative manner (Fig. 1 B). Even when all Nup188 was immunoprecipitated and therefore removed from the unbound (Fig. 1 B, lane 9 and 10), Nup205 was not detected in the precipitate (Fig. 1 B, lane 4 and 5) and vice versa (Fig. 1 B, lane 7 and 12). Both Nup188 and Nup205 immunoprecipitated ∼45% of Nup93 (Fig. S1). These data indicate that Nup93 forms two distinct complexes in Xenopus egg extracts of approximate equal abundance, one with Nup188 and one with Nup205.
Nuclei lacking Nup188–Nup93 enlarge in size
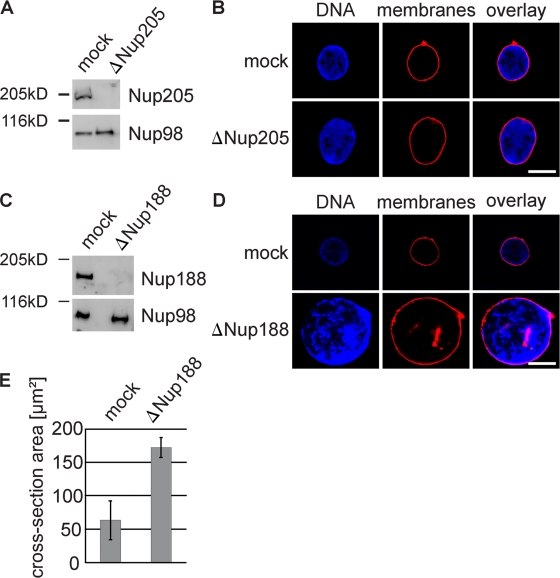
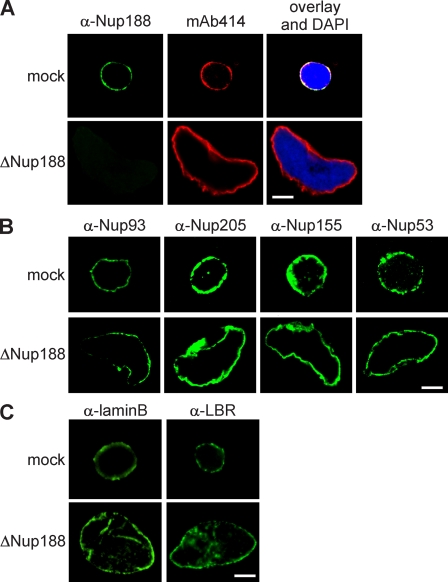
To understand the functional significance of the two Nup93 complexes for NPC assembly and function, we depleted them separately from Xenopus egg extracts using antibodies specific for Nup205 or Nup188. Extracts were efficiently depleted of the respective Nup93 complexes, as determined by Western blotting (Fig. 2, A and C) and immunofluorescence (Fig. 3 A and Fig. S2 A). Nuclei formed in extracts depleted of the Nup205–Nup93 complex for 90 min were normal in appearance, as visualized by the membrane and chromatin stain (Fig. 2 B), and contained NPCs (Fig. S2 A), as determined by mAb414 staining, which recognizes FG repeat–containing nucleoporins, and with Nup93, Nup188, Nup53, and Nup155, the other members of the Nup93 complex (Fig. S2 B). In contrast, nuclei lacking Nup188–Nup93 were much larger than control nuclei (Fig. 2, D and E). These nuclei also contained NPCs (Fig. 3 A), as shown by mAb414 immunofluorescence. We found threefold more NPCs on Nup188–Nup93-depleted (18,480 ± 4,224 NPCs per nucleus) compared with control nuclei (6,137 ± 1,080; see Fig. 7 B). However, the NPC density per membrane area remained approximately constant (8.5 ± 1.2 NPCs/µm2 in Nup188–Nup93 vs. 9.1 ± 1.2 NPCs/µm2 in mock depletion). The NPCs contained Nup93 as well as Nup205, Nup53, and Nup155 (Fig. 3 B). Except for their increased size, Nup188–Nup93-depleted nuclei did not exhibit morphological alterations of the nuclear membranes or NPCs (Fig. S2 C), as determined by transmission electron microscopy. Also, marker proteins like B-type lamins and lamin B receptor (LBR) localized to the INM (Fig. 3 C). In a time course, chromatin substrates in mock- and Nup188–Nup93-depleted extracts were undistinguishable until a closed NE was formed at 50 min, after which Nup188–Nup93-depleted nuclei grew rapidly in size (Fig. S3). It should be noted that control nuclei also grow after having formed a closed NE as long as reentry into mitosis is blocked; however, they do this at a significantly slower rate.
Figure 2.
Removal of Nup188–Nup93 enlarges nuclei. (A and C) Western blot analysis of mock- and Nup205–Nup93 (A)- or Nup188–Nup93 (C)-depleted extracts. (B and D) Nuclei were assembled in mock- and Nup205–Nup93 (B)- or Nup188–Nup93 (D)-depleted extracts for 90 min, fixed with 4% PFA and 0.5% glutaraldehyde, and analyzed for chromatin and membrane staining (blue, DAPI; red, DiIC18). (E) Quantitation of the cross-sectional area of mock- and Nup188–Nup93-depleted nuclei of experiments performed as in D. More than 100 randomly chosen chromatin substrates were counted per reaction. The mean of three independent experiments is shown, and error bars represent the total variation. Bars, 20 µm.
Figure 3.
Nup188–Nup93-depleted nuclei contain NPCs and INM proteins. (A) Nuclei were assembled in mock- or Nup188–Nup93-depleted extracts for 90 min, fixed with 4% PFA, and analyzed with Nup188-specific antibodies (green) or the monoclonal antibody mAb414, which recognizes FG-repeat nucleoporins (red). Chromatin is stained with DAPI. (B and C) Samples were prepared as in A and analyzed by immunofluorescence with the indicated antibodies. Bars, 20 µm.
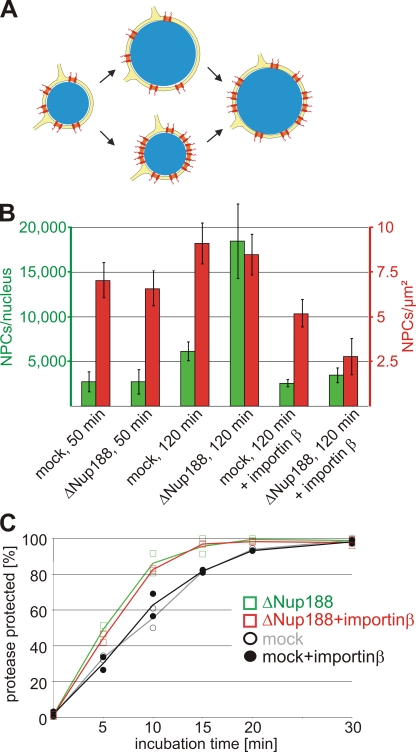
Figure 7.
The nuclear growth phenotype and the faster INM targeting are independent of the increased NPC number upon Nup188–Nup93 depletion. (A) Two possible scenarios for the relationship of the nuclear membrane expansion and the increase in NPC numbers upon Nup188–Nup93 depletion. In the top route, Nup188–Nup93 depletion causes a faster growth of the NE, which in turn allows more NPCs (red) to be integrated. In the bottom route, NPC assembly increases on Nup188–Nup93-depleted nuclei. The resulting higher total transport capacity of these allows accelerated nuclear volume and envelope expansion. (B) Nuclei were assembled in mock- and Nup188–Nup93-depleted extracts for 50 min and 120 min. Where indicated, de novo NPC assembly was blocked by the addition of 2 µM importin β after 50 min, i.e., after a closed NE had formed, and nuclei were further incubated for 70 min. NPC numbers per nucleus (green bars) and NPC density (red bars) were quantified. (C) Nuclei were assembled in mock- and Nup188–Nup93-depleted extracts for 50 min. Then, where indicated, de novo NPC assembly was blocked by the addition of 2 µM importin β. INM targeting of the reporter was analyzed as in Fig. 6. Squares are from Nup188–Nup93-depleted samples, and circles from mock-treated samples. Lines mark the mean of two independent experiments.
Nup188–Nup93-depleted nuclei have normal nuclear functions
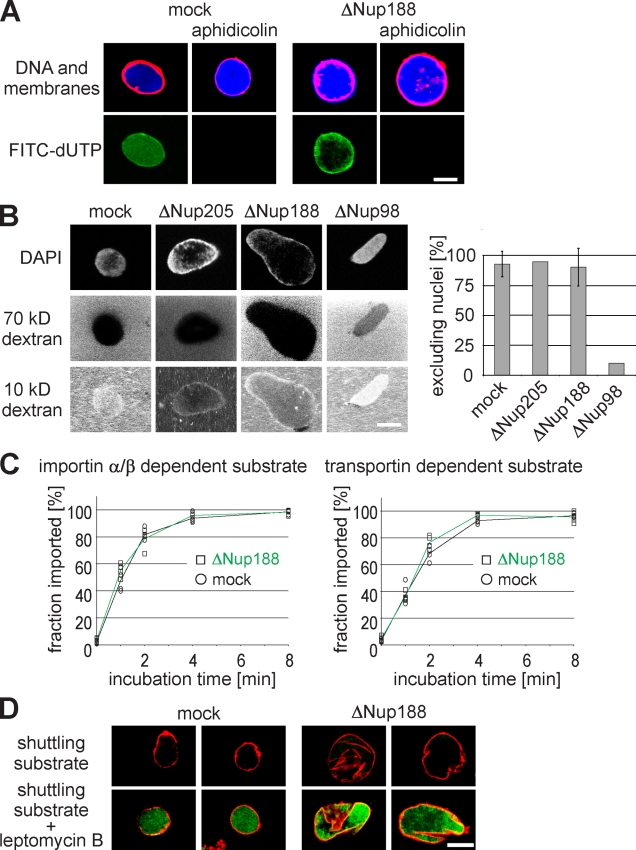
Given that Nup188–Nup93-depleted nuclei grow to an increased size after a closed NE is formed, we sought to determine which nuclear function is responsible for the observed phenotype. Upon nuclear assembly in Xenopus egg extracts, DNA can undergo one round of replication. Replication licensing ensures that replication happens exactly once during each round of the cell cycle (Blow and Sleeman, 1990). We wanted to know whether Nup188–Nup93 depletion impairs replication licensing, causing more than one round of DNA replication, which would in turn lead to increased DNA content in the nuclei. To test this idea, we assembled nuclei in mock- and Nup188–Nup93-depleted extracts in the presence of 16 µM of the DNA polymerase inhibitor aphidicolin. These conditions blocked DNA replication, as confirmed by the failure of DNA to incorporate fluorescently labeled nucleotides (Fig. 4 A). When DNA replication was blocked, the size of Nup188–Nup93-depleted nuclei still enlarged, indicating that the volume increase is not caused by a higher DNA content.
Figure 4.
Nuclear functions are unaffected by Nup188–Nup93 depletion. (A) Nuclei were assembled in mock- or Nup188–Nup93-depleted extracts. After 50 min, fluorescently labeled dUTP (green in overlay) and, where indicated, aphidicolin were added. After 90 min, replication was analyzed by confocal microscopy. Membranes were stained with DiIC18 (red), and chromatin was stained with DAPI (blue). (B) Nuclei were assembled in mock-, Nup205–Nup93-, Nup188–Nup93-, and Nup98-depleted extracts. After 90 min, fluorescently labeled 2-MD (not depicted), 70-kD (middle), and 10-kD (bottom) dextrans were added. DNA was stained with DAPI, and samples were analyzed by confocal microscopy. For quantitation, only nuclei with an intact NE (as judged by the exclusion of 2-MD dextrans; >95% of nuclei in each sample) were analyzed. For mock- and Nup188–Nup93-depleted samples, seven independent experiments were analyzed (error bars show the standard deviation), and for Nup205–Nup93- and Nup98-depleted samples, two independent experiments were analyzed. (C) Nuclei were assembled as in A. After 50 min, i.e., when a closed NE had formed, importin α/β– or transportin-dependent reporter substrates containing a TEV cleavage site were added. To assay the nuclear import kinetics of the reporter, NusA-fused TEV protease was added at the indicated time points. Protease cleavage was stopped after 1 min by the addition of SDS sample buffer and boiling. Lines mark the mean of four independent experiments. (D) Nuclei were assembled as in A, and an EGFP-fused shuttling substrate was added. Nuclear export function was inhibited by the addition of 300 nM leptomycin B. Nuclei were isolated and analyzed by confocal microscopy. Membranes are stained with DiIC18 (red). Bars, 20 µm.
NPCs constitute a diffusion barrier between the cytoplasm and the nucleoplasm for substances >30 kD (Mohr et al., 2009), which can be translocated only by signal-mediated transport. We were wondering whether depletion of Nup188–Nup93 would impair this diffusion barrier, causing an uncontrolled accumulation of material in the nucleus and in turn lead to the observed size increase. Indeed, the yeast homologue Nup188p was shown to be important for the diffusion barrier (Shulga et al., 2000). To test the integrity of the diffusion barrier of the NPC, we added fluorescently labeled dextrans of different but defined sizes to in vitro assembled nuclei that were lacking either Nup205 or Nup188 (Fig. 4 B). Both, Nup205–Nup93- and Nup188–Nup93-depleted nuclei were still able to exclude 70-kD dextrans, indicating that the diffusion barrier in these nuclei was intact. Smaller sized 10-kD dextrans could diffuse through the NPC as expected. As a control for an impaired diffusion barrier, nuclei depleted for Nup98 did not exclude the 70-kD dextran. These data indicate that a malfunction of the diffusion barrier of the NPC is unlikely to cause the enlargement of Nup188–Nup93-depleted nuclei.
NPCs mediate signal-dependent import and export across the NE. To check whether these functions are affected by Nup188–Nup93 depletion, we first tested nuclear accumulation of substrates containing an importin α/β– or transportin-based import signal, respectively. Both substrates were fused to an N-terminal EGFP linked to a tobacco etch virus (TEV) protease recognition site. They were added to a nuclear assembly reaction at 50 min, when an NE had formed and Nup188–Nup93- and mock-depleted nuclei were of similar size with approximately the same NPC content (2,737 ± 1,160 NPCs per nucleus in mock-depleted nuclei vs. 2,739 ± 1,437 in Nup188–Nup93-depleted nuclei; see Fig. 7 B). At the indicated time points, a TEV protease fused to a bacterial protein (NusA) was added. NusA increased the size of the protease fusion protein to 90 kD, which prevented its diffusion through the NPC. Nuclear import of the substrates rendered them protease protected and they could thus be quantified by Western blotting. Import kinetics were unchanged upon Nup188–Nup93 depletion for both substrates (Fig. 4 C).
To check for nuclear export, we used an EGFP-fused shuttling substrate with a nuclear import and export signal. The substrate did not accumulate in the nucleus both in mock- and Nup188–Nup93-depleted nuclei, indicating that nuclear export was not impaired (Fig. 4 D). To verify the function of the shuttling substrate, we added leptomycin B, which blocks crm1-mediated nuclear export (Fornerod et al., 1997). As expected, under these conditions, the shuttling substrate accumulated in the nucleus. Interestingly, leptomycin B–treated control nuclei did not increase in size, showing that a block in nuclear export does not phenocopy Nup188–Nup93 depletion. These data show that nuclei lacking Nup188–Nup93 have unchanged nuclear transport capabilities for the tested proteins. Together with the fact that the Nup188–Nup93-depleted nuclei replicate their DNA and grow, both processes requiring nuclear import, it is unlikely that an impairment of nuclear transport of soluble cargoes causes the observed nuclear growth phenotype upon Nup188–Nup93 depletion.
Xenopus egg extracts are largely transcriptionally inactive. Thus, an accumulation of mRNA in the nucleus is unlikely to cause nuclear enlargement. Coherent with this, addition of the transcriptional inhibitors 5,6-dichloro-1-β-d-ribobenzimidazole or actinomycin D did not block nuclear growth of Nup188–Nup93-depleted nuclei (unpublished data).
Nup188–Nup93 restricts transport of membrane proteins through the NPC
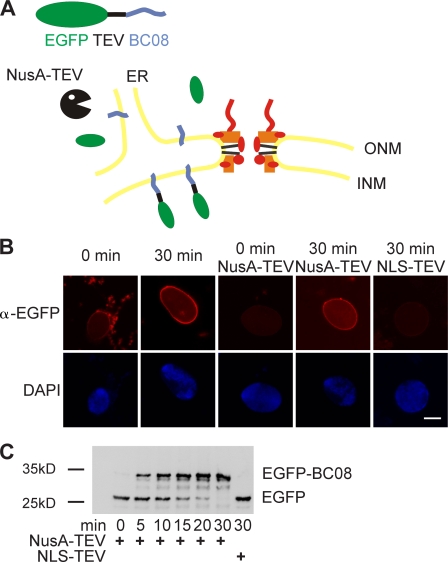
Having established that both diffusion and active transport of soluble substrates is unlikely to account for the increase in nuclear size upon Nup188–Nup93 depletion, we focused on the transport of membrane components. As described in the section Nuclei lacking Nup188–Nup93 enlarge in size, nuclei assembled in Xenopus egg extracts did enlarge upon further incubation provided that reentry into mitosis was prevented. However, this nuclear growth was much slower in untreated or mock-depleted extracts compared with the Nup188–Nup93-depleted nuclei (Fig. S3 and not depicted). We reasoned that as nuclear growth requires expansion of the NE, it must also involve an acquisition of nuclear membrane components. As the ER is continuous with the ONM, it is easy to envision how membrane components (both lipids and proteins) can be delivered to the ONM. The INM and ONM are connected by the membrane facing the NPC, the pore membrane. Thus, if the INM expands, membrane components need to pass through the NPC via the pore membrane. We speculated that pore membrane passage could limit and restrict nuclear growth and that this restriction might be attenuated upon Nup188–Nup93 depletion. To investigate this, we developed an assay to follow the transport of integral membrane proteins to the INM. BC08, an INM protein with a single C-terminal transmembrane region (Ulbert et al., 2006), was fused to an N-terminal EGFP followed by a recognition site for the TEV protease. The fusion protein was expressed and purified in Escherichia coli and reconstituted into liposomes. Liposomes were then added to a nuclear assembly reaction at time points when the NE had already formed around chromatin and under conditions in which they readily fused with the ER (Fig. 5, A and B). The reporter was distributed immediately throughout the membranes of the ER, including the NE, as observed by immunofluorescence (Fig. 5 B). After 30 min, the reporter was enriched at the NE. To distinguish between INM and ONM (including bulk ER) localization, we added the TEV protease fused to NusA, which prevents its diffusion through the NPC. Initially, the reporter was protease sensitive, indicating that it was localized to the ER and the ONM. However, after 30 min, the reporter was protease protected, which is indicative of INM localization. At the same time point, a TEV protease coupled to an NLS-bearing peptide (which is therefore localized to the interior of the nucleus) was able to cleave the reporter. This confirms that the reporter remains cleavable when it is accessible to the protease, and therefore, it is localization to INM that renders it protease protected in this assay. Cleavage of the reporter and its protease protection can be also followed by Western blotting over time, which indicates that about half of the reporter was protease protected after 10 min (Fig. 5 C).
Figure 5.
An assay for the transport of membrane proteins to the INM. (A) Schematic representation of the assay. The transmembrane-containing INM protein BC08 is fused to an EGFP domain followed by a recognition site for the TEV protease (top). The fusion protein is reconstituted into liposomes that rapidly fuse with the ER and ONM in the nuclear assembly reaction. INM localization of the reporter can be distinguished from ONM and ER localization by the reporter’s protection from TEV protease fused with a large NusA domain to prevent its diffusion through the NPC. (B) Nuclei were assembled in Xenopus egg extracts. After 50 min, i.e., when a closed NE had formed, proteoliposomes containing the reporter were added. At the indicated time points, buffer, NusA-fused TEV protease, or TEV protease linked to NLS peptides was added. Protease cleavage was stopped after 5 min by fixation, and samples were processed for immunofluorescence and analyzed by confocal microscopy. α-EGFP immunofluorescence is shown in red, and DAPI is shown in blue. (C) Reactions were performed as in B. Protease cleavage was stopped by the addition of SDS sample buffer and boiling. Protease protection of the reporter was analyzed by Western blotting with the position of the uncleaved (EGFP-BC08) and the cleaved reporter with only the EGFP visible indicated. Bar, 20 µm.
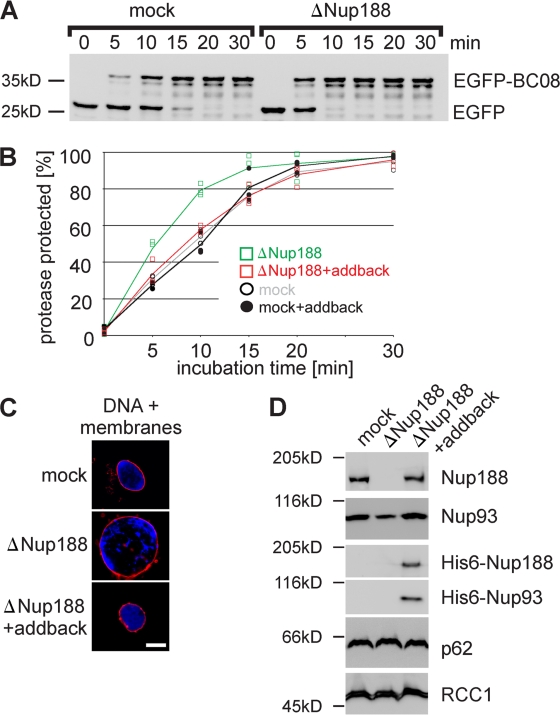
Having established a functional assay for INM translocation, we asked whether the increased size of Nup188–Nup93-depleted nuclei is the result of accelerated membrane delivery to the INM. Nuclei were assembled in Nup188–Nup93- or mock-depleted extracts for 50 min, at which point the NE was closed but the nuclei had not increased in size (Fig. S3, 50-min time points). Liposomes containing the reporter were then added, and protease protection followed over time. In Nup188–Nup93-depleted nuclei, ∼50% of the reporter was protease protected after 5 min, significantly earlier than in control-treated nuclei (Fig. 6, A and B). Similar results were obtained when a reporter containing the first transmembrane region of the LBR, sufficient for INM targeting (Soullam and Worman, 1995), was used (Fig. S4 A). To confirm that the effect of Nup188–Nup93 depletion was specific, an addback experiment was performed. Xenopus Nup93 and mouse Nup188 were cotranslated in vitro (Fig. S4 B) and added to mock- or Nup188–Nup93-depleted extracts. Nuclei assembled in these extracts grew to a similar size as mock-treated extracts (Fig. 6 C and Fig. S4 C) and, importantly, did not show accelerated delivery of the BC08 and LBR reporter to the INM (Fig. 6 B and Fig. S4 A). As the antibody against Xenopus Nup188 does not detect the mammalian orthologue by immunofluorescence, nuclei assembled in Nup188–Nup93-depleted extracts supplemented with the recombinant proteins were isolated, and incorporation of recombinant Nup188–Nup93 into the nuclei was confirmed by Western blotting (Fig. 6 D). Notably, addition of neither Nup188 nor Nup93 alone rescued the observed phenotypes, indicating that the complex of Nup188 and Nup93 is indeed required (unpublished data). Collectively, these data indicate that both the increase in nuclear size and the accelerated targeting of integral membrane proteins to the INM are specific for Nup188–Nup93 depletion. Interestingly, nuclei depleted for Nup205–Nup93 showed the same rate of INM targeting of the two reporters as mock controls (Fig. S4 D), which is consistent with the fact that they do not show the nuclear growth phenotype.
Figure 6.
Nup188–Nup93 depletion causes a faster delivery of integral membrane proteins to the INM. (A) Nuclei were assembled in mock- or Nup188–Nup93-depleted Xenopus egg extracts and analyzed by Western blotting. Positions of the uncleaved (EGFP-BC08) and the cleaved (EGFP) reporter are indicated. (B) Quantification from three independent experiments performed as in A. Squares are from Nup188–Nup93-depleted samples (green without and red with the addition of in vitro translated Nup188–Nup93 [addback]), and circles are from mock-treated samples. Lines mark the mean of three independent experiments. (C) The addition of recombinant Nup188–Nup93 rescues the Nup188–Nup93 depletion phenotype. Nuclei were assembled for 120 min in mock- or Nup188–Nup93-depleted extracts without or with the addition of in vitro translated Nup188–Nup93 (addback), fixed with 4% PFA and 0.5% glutaraldehyde, and analyzed for chromatin and membrane staining (blue, DAPI; red, DiIC18). (D) In vitro translated Nup188–Nup93 is incorporated into in vitro assembled nuclei. Nuclei were assembled for 1 h to ensure approximately equal NPC numbers per nucleus, isolated, and analyzed by Western blotting. In vitro translated Nup188 and Nup93 could be also detected using an anti-His6 antibody (third and fourth row). The nucleoporin p62 was detected with mAb414, and the chromatin-binding protein RCC1 serves as a control for equal loading of nuclei. Bar, 20 µm.
As INM proteins reached the nuclear interior significantly faster in Nup188–Nup93-depleted nuclei compared with controls, we wondered whether the effect was specific to proteins destined for the INM or whether the segregation of the INM from the ONM and connected ER was lost in the absence of Nup188–Nup93. To test this idea, we replaced the INM reporter with bona fide ER membrane proteins: SPC18, a membrane-spanning subunit of the signal peptidase complex, or a C-terminal fragment (including the transmembrane region) of the ER-localized chaperone protein calnexin. Both reporters were sensitive to TEV protease cleavage even after 60 min, indicating that they were not localized to the INM (Fig. S5 A). Notably, the cytoplasmic domains for the SPC18 (31 kD)- and calnexin (41 kD)-based reporters fall within the size range of the nucleoplasmic regions of the BC08 and LBR reporters described in the preceding experiments. Therefore, Nup188–Nup93-depleted NPCs continue to act as a diffusion barrier preventing uncontrolled intermixing of INM and ONM (and ER) components.
The nucleoplasmic domains of most INM proteins are limited in size to ∼40 kD. It has been shown that increasing this size to 47 kD prevents membrane protein targeting to the INM (Ohba et al., 2004). We tested whether depletion of Nup188–Nup93 would impair the stringency of this size limit. For this, the size of the BC08 reporter was increased to 94 kD and 64 kD by fusing an additional NusA or GST moiety, respectively (Fig. S5 B). In all cases, the reporter did not target to the INM, both in mock- and Nup188–Nup93-depleted nuclei.
Up to now, we have demonstrated that upon Nup188–Nup93 depletion, the passage of INM proteins through the pore is enhanced and that the transfer of membrane components could be rate limiting for nuclear growth. Two possible scenarios can be envisioned. Nup188–Nup93 could restrict the passage of transmembrane or membrane proteins through the pore. Depletion of Nup188–Nup93 might abrogate this restriction, causing an increased flow of membrane components through the pore, leading to an enlargement of the NE in which more NPCs can integrate (Fig. 7 A, top route). Alternatively, Nup188–Nup93 depletion could facilitate NPC formation, giving rise to more NPCs with an overall increased total transport capacity (Fig. 7 A, bottom route). In this scenario, the increase in NE area would be a secondary effect. To distinguish the two possibilities, we blocked de novo NPC assembly into intact NEs by an excess of importin β (D’Angelo et al., 2006), which inhibits the Nup107–Nup160 complex, an essential early component for NPC formation (Harel et al., 2003; Walther et al., 2003; Rasala et al., 2006; Franz et al., 2007). When 2 µM importin β was added to mock- and Nup188–Nup93-depleted extracts after 50 min, i.e., when a closed NE with intact NPCs was formed, the NPC number did not increase in the next 70 min (Fig. 7 B, green bars). Despite approximately the same number of NPCs at the time of importin β addition and a subsequent block in de novo NPC assembly, Nup188–Nup93-depleted nuclei grew within the next 70 min significantly faster than mock-depleted nuclei so that the NPC density at the end of the incubation time was more decreased in Nup188–Nup93-depleted compared with mock-depleted nuclei (2.8 NPCs/µm2 NE membrane vs. 5.2 NPCs/µm2; Fig. 7 B, red bars). These data indicate that the accelerated growth of the nuclei and the nuclear membrane expansion upon Nup188–Nup93 depletion cannot only be attributed to an increased NPC number. Importantly, in Nup188–Nup93-depleted nuclei treated with importin β, the EGFP-BC08 reporter was still protease protected significantly earlier than in mock-treated nuclei (Fig. 7 C). Therefore, we could rule out that an increased NPC number was the primary cause of the enhanced translocation of the reporter to the INM. Collectively, this demonstrates that Nup188–Nup93 depletion results in increased passage of proteins destined for the INM across the NPC. This suggests that Nup188–Nup93 is a crucial component within the NPC that restricts the flux of integral membrane proteins and likely other membrane components along the pore membrane.
Discussion
In this study, we have shown that the nucleoporin Nup93 is part of two distinct complexes. It interacts either with Nup205 or -188. Unexpectedly, nuclei depleted of Nup188–Nup93 grew to enormous sizes, a phenotype previously not observed upon depletion of a nucleoporin. Integral membrane proteins and probably other membrane components are delivered faster to the INM upon Nup188–Nup93 depletion. We suggest that this causes rapid expansion of the NE as a primary effect and in turn facilitates increased integration of NPCs. The higher NPC number would have a higher overall transport rate both for soluble cargoes and membrane components, which would then further augment the nuclear growth effect.
Transport of soluble cargo via the NPC has been intensely studied in previous decades (for reviews see Weis, 2003; Cook et al., 2007). However, transmembrane proteins must also access the INM, where several of them bind lamin and/or chromatin. Little is currently known about how transmembrane proteins are delivered to the INM of the interphasic nucleus (Zuleger et al., 2008). Although a scenario can be envisioned in which the ONM and INM periodically fuse to provide transient connections allowing diffusional exchange between these membrane domains, there is at present no evidence supporting this. Alternatively, membrane vesicle budding from the ONM into the perinuclear lumen and subsequent fusion with the INM could also explain how membrane proteins are delivered to the INM. However, aside from the observation that the reverse process occurs during herpesvirus egress from the nucleus (Mettenleiter et al., 2009), there is little proof for this model. A more likely scenario involves transmembrane protein transit from the ONM to the INM via the NPC along the plane of the pore membrane. Indeed, antibodies to the transmembrane nucleoporin gp210 as well as WGA, a lectin which cross-links several nucleoporins, are both able to block the movement of integral membrane proteins from the ONM to the INM, most likely by sterically occluding the passageways (Ohba et al., 2004). In yeast, deletion of Nup170, one of the Nup155 homologues, causes delocalization of two integral membrane proteins, Heh1p and Heh2p, from the INM (King et al., 2006). Together with the fact that delivery of Heh1p and Heh2p to the INM requires classical transport receptors (importin α and β), these data argue strongly in favor of NPC involvement in the movement of integral membrane proteins from the ONM to the INM. Indeed, when depleting the Nup107–Nup160 complex, which leads to the formation of nuclei without NPCs (Harel et al., 2003; Walther et al., 2003), no INM targeting of the reporter constructs was observed (Fig. S5 C).
In this study, we show that Nup188–Nup93 restricts the passage of integral membrane proteins from the ONM to the INM. Although proteins destined to the INM reach these sites faster in Nup188–Nup93-depleted nuclei, bona fide ER proteins are not mislocalized to the INM. In yeast, depletion of Nup188p or the integral membrane nucleoporin Pom152p was found to reduce INM localization of the ubiquitin ligase Doa10p (Deng and Hochstrasser, 2006). Normally, the majority of cellular Doa10p resides in the ER, although it is also found at the INM, where it drives the degradation of intranuclear targets. During excessive nuclear membrane proliferation, Doa10p is enriched at the INM. Under these conditions, deletion of Nup188 causes the redistribution of Doa10p to the ER, probably via enhanced passage through the NPC.
Nucleoporins containing FG-repeat domains contribute to the transport properties of the NPC by establishing a permeability barrier and promoting transport receptor–mediated NPC passage. They are mainly localized to the interior center of the NPC (Rout et al., 2000; Alber et al., 2007). In contrast, Nup188p and Nup170p do not contain FG repeats and are found comparatively more proximal to the pore membrane. Interestingly, depletion of Nup188–Nup93 did not reduce mAb414 staining, which recognizes FG-containing nucleoporins (Fig. 3 A), and did not destroy the diffusion barrier of the NPC or affect nuclear import and export of soluble reporter cargoes (Fig. 4). This suggests that the transport of soluble and membrane proteins requires distinct features within the NPC and uses different pathways within the pore.
It has been proposed that NPCs accommodate not only a large central channel for active and transport receptor–mediated import and export but also eight peripheral channels that mediate passive exchange of metabolites and ions (Hinshaw and Milligan, 2003; Kramer et al., 2007; Naim et al., 2007). Although recent cryoelectron tomography of NPCs from Dictyostelium discoideum confirmed the existence of peripheral channels ∼9 nm in diameter (Beck et al., 2007), the idea of separate passageways for soluble factors is in contrast to the prevailing model in which FG-repeat domains form a common barrier for both facilitated and passive exchange (Ribbeck and Görlich, 2001; Rout and Aitchison, 2001). Passive diffusion is sensitive to many of the same ligands and inhibitors that target active transport across the NE. This argues strongly for a single permeability barrier for soluble molecules passing through the NPC either by active transport or passive diffusion (Mohr et al., 2009). Alternatively, the peripheral channels could be important for membrane proteins passing the NPC (Powell and Burke, 1990; Zuleger et al., 2008). Although the nucleoplasmic domains of most INM proteins are limited in size to ∼40 kD and therefore might be small enough to fit in the cavities of the channels, we propose that structural rearrangements within the NPC are necessary to allow for integral membrane passage without disrupting the diffusion barrier. Depletion of Nup188–Nup93 may enhance conformational flexibility within the NPC, resulting in the increased passage of membrane proteins through the NPC that we have observed in this study without affecting the size limit for these proteins.
The metazoan Nup93 complex and the corresponding Nic96 complex in yeast is one of the major building blocks of the NPC. However, compared with the well-characterized Nup107–Nup160 complex, which can be purified from Xenopus egg extracts (Vasu et al., 2001; Walther et al., 2003), the association of Nup93 complex components is remarkably less stable. Nup155 and Nup53 can be precipitated mainly as individual proteins, as observed previously (Franz et al., 2005; Hawryluk-Gara et al., 2008). Additionally, Nup205, Nup188, and Nup93 do not exist in a single complex in Xenopus egg extracts as previously suggested (Meier et al., 1995). Formerly, immunoprecipitations were performed using antibodies against Nup93, which, as we have shown, actually precipitates two different Nup93-containing complexes. However, the Nup188–Nup93 and the Nup205–Nup93 interactions seem to be rather stable. Interestingly, in the addback experiment, we could only restore the phenotype if Nup188 and Nup93 were coexpressed and added despite the fact that after Nup188–Nup93 depletion, ∼50% of Nup93 remains in the extracts (most in a complex with Nup205 and a smaller proportion not bound to Nup205; Fig. 1 and Fig. S1), suggesting that Nup93 cannot exchange between the different complexes under the given conditions.
Whereas depletion of Nup155 and Nup53 from Xenopus egg extracts prevents NPC and NE formation (Franz et al., 2005; Hawryluk-Gara et al., 2008), extracts depleted of Nup205–Nup93 or Nup188–Nup93 assemble nuclei with seemingly intact NPCs and a closed NE. Currently, we cannot exclude the possibility that the Nup188–Nup93 and Nup205–Nup93 complexes have partially redundant functions during NPC formation. Double depletion of both complexes (with antibodies against Nup205 and Nup188) followed by nuclear assembly reactions was for technical reasons unfeasible, requiring, in total, four rounds of incubation of extracts with antibody beads. Interestingly, depletion of Nup93-containing complexes from Xenopus egg extracts with α-Nup93 antibodies blocked NPC assembly (Grandi et al., 1997; unpublished data). However, we cannot exclude the possibility that Nup93 is of functional importance outside of its role in the Nup188–Nup93 and Nup205–Nup93 complexes. Indeed, a subfraction of 5–10% of Nup93 has been shown to interact with the FG repeat–containing nucleoporin p62 (Zabel et al., 1996), as does the yeast homologue Nic96p with Nsp1p (Zabel et al., 1996). This could be a subpopulation distinct from the Nup188–Nup93 and Nup205–Nup93 complexes. Notably, we found that ∼5% of Nup93 cannot be depleted with a combination of antibodies against Nup205 and Nup188 (Fig. S1)
Depletion of neither Nup188–Nup93 nor Nup205–Nup93 impaired the exclusion limit of the NPCs for inert substances (Fig. 4 B). For Nup205–Nup93, this was particularly unexpected as depletion of Caenorhabditis elegans Nup205 by RNAi causes failure of nuclear exclusion of 70-kD dextrans (Galy et al., 2003). Interestingly, up to now, no C. elegans Nup188 orthologue has been identified. It is possible that the Nup205–Nup93 and Nup188–Nup93 complexes in Xenopus have partially redundant functions in establishing the exclusion limit of NPCs. Because of the absence of Nup188 in C. elegans, Nup205–Nup93 could still accommodate this function, which is lost upon Nup205 depletion.
In summary, our work establishes that members of the Nup93 complex, which is regarded as one of the structural building blocks of the NPC, can be depleted without blocking NPC formation. Intriguingly, Nup188–Nup93 is rather important for controlling the passage of transmembrane proteins and probably other membrane components through the NPC. Thus, in addition to regulating the composition of the nucleoplasm by restricting access of soluble factors to the nuclear interior, NPCs are also important in the establishment of the two membrane subcompartments of the NE.
Materials and methods
Materials
An EST containing the C-terminal region of Xenopus Nup188 (GenBank/EMBL/DDBJ accession number BQ725804) was identified by BLAST search using the mouse sequence as bait. ESTs for Xenopus Nup98, Nup205, BC08, LBR, and mouse Nup188 were under GenBank/EMBL/DDBJ accession numbers CF286969, CA986989, BC082226, BC124994, BG861579, respectively. Xenopus Nup53, NPM2, calnexin, and SPC18 were amplified from a Xenopus cDNA.
Leptomycin B and aphidicolin were obtained from Enzo Life Sciences, Inc., and DiIC18 (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate), fluorescently labeled dextrans, and secondary antibodies (Alexa Fluor 488 goat α-rabbit IgG and Cy3 goat α-mouse IgG) were obtained from Invitrogen. Detergents were purchased from EMD, and lipids were purchased from Avanti Polar Lipids, Inc.
Antibodies
For the generation of antibodies, a fragment of Xenopus Nup188 corresponding to aa 1573–1731 of the mouse sequence and a fragment of Nup98 (aa 1–185) were expressed as GST fusion proteins and purified using glutathione Sepharose (GE Healthcare). Fragments of Xenopus Nup53 (aa 78–310) and LBR (aa 47–216) were cloned into pET28a (EMD), expressed as a His6-tagged fusion, and purified using Ni–nitrilotriacetic acid agarose (QIAGEN). A fragment of Xenopus Nup205 (aa 1–283) was fused to NusA as a solubility tag, cloned into pET28a, and expressed as a His6-tagged fusion. Recombinant proteins were dialyzed to PBS and injected into rabbits for antibody production.
Antibodies against Nup155 and RCC1 (Franz et al., 2005) as well as Nup160 and Nup93 (Franz et al., 2007) have been described previously. mAb414 was obtained from Babco, anti–lamin B (X223) was obtained from ImmuQuest, and anti-EGFP and anti-His6 were obtained from Roche.
DNA replication, dextran exclusion, and nuclear transport function
For testing DNA replication, nuclear assembly reactions were incubated with 43.5 µM fluorescein-12-dUTP and, where indicated, 16 µM aphidicolin. Samples were fixed and processed for microscopy as described previously (Franz et al., 2005). All fluorescence microscopy images were recorded on the confocal microscope (FV1000; Olympus; equipped with a photomultiplier [model R7862; Hamamatsu]) with 405-, 488-, and 559-nm laser lines and a 60× NA 1.35 oil immersion objective lens using the Fluoview software (Olympus) at room temperature using Vectashield (Vector Laboratories) as a mounting medium.
For the size exclusion assay, nuclei were assembled for 90 min, at which point 10 µg/ml of 10-kD dextran labeled with Texas red, 30 µg/ml of 70-kD dextran labeled with fluorescein, and 40 µg/ml of 2-MD dextran labeled with tetramethylrhodamine were added. The samples were incubated for 5 min and analyzed by confocal microscopy without fixation and mounting, but otherwise as in the previous paragraph.
TEV protease was fused to a NusA domain, cloned into pET28a, and expressed and purified as a His6-tagged fusion. TEV protease without a NusA domain was expressed and purified as a His6-tagged fusion and cross-linked to NLS peptides as described previously (Vasu et al., 2001).
To test importin α/β– or transportin-dependent nuclear import function, full-length NPM2 (nucleoplasmin) or Nplc-M9-M10 (from Englmeier et al. [1999]), respectively, was fused to an N-terminal EGFP, followed by a TEV recognition site, and cloned into a modified pET28a vector, allowing purification via a C-terminal His6 tag. After Ni–nitrilotriacetic acid chromatography, all substrates were further purified by size exclusion chromatography on a Sepharose 200 column (GE Healthcare). 50-µl nuclear assembly reactions were assembled in the volume ratio as described previously (Franz et al., 2005). 50 min after the addition of membranes, 1 µg of the respective reporter was added. At the indicated time points, 10 µl of the samples was added to 1 µl of 5 µg/µl NusA-TEV protease and incubated for 1 min at 20°C. TEV cleavage was stopped by the addition of SDS sample buffer and immediate incubation at 95°C for 5 min, followed by SDS-PAGE and Western blotting.
For testing nuclear export function, a reporter as well as a control substrate, Nplc-M9-NES and Nplc-M9-M10, respectively (from Englmeier et al. [1999]) were fused to an N-terminal EGFP, cloned into pET28a, and purified as in the previous paragraph. 0.1 mg/ml of the export reporters was added to a nuclear assembly reaction, incubated for 90 min, fixed, and processed for microscopy.
For generation of reporter constructs for INM targeting, full-length BC08, a part of the N terminus of LBR including the first transmembrane region (aa 146–258), and full-length SPC18 (all from Xenopus) were fused to an N-terminal MISTIC (membrane-integrating sequence for translation of integral membrane protein constructs) fragment (Roosild et al., 2005), followed by a thrombin cleavage site, an EGFP domain, and a TEV protease cleavage site, and cloned into a modified pET28a vector, allowing purification via a C-terminal His6 tag. For the calnexin reporter construct, the C-terminal region including the transmembrane region of calnexin (aa 485–608) was placed 3′ of the MISTIC fragment and 5′ of the TEV cleavage site and the EGFP domain and integrated into the modified pET28a vector with a C-terminal His6 tag. For the INM targeting constructs with increased nucleoplasmatic sizes, a NusA or GST domain was inserted 5′ of the EGFP moiety in the reporter constructs.
Proteins were expressed in E. coli BL21de3, purified in the presence of 1% (wt/vol) cetyltrimethylammonium bromide on magnetic Ni-loaded agarose beads (EMD), and dialyzed for 16 h against PBS containing 1 mM EDTA. The MISTIC fragments were cleaved off using thrombin, and the reporters were reconstituted into liposomes. For this, ∼10 µg of protein was mixed with 20 µl of a lipid mix containing 3 mg/ml cholesterol, 3 mg/ml Na-phosphatidylserine, 3 mg/ml Na-phosphatidylinositol, 6 mg/ml phosphatidylethanolamine, 15 mg/ml phosphatidylcholine (all solubilized in 10% octylglucopyranoside), and 2 µl of 1 mg/ml DiIC18 in DMSO. Detergent was removed by passing the sample over a G-50 column equilibrated in sucrose buffer (10 mM Hepes, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2, pH 7.5). The proteoliposome-containing fraction (identified by the color of the dye) was collected, and the liposomes were pelleted by centrifugation for 30 min at 200,000 g. The pellets were resuspended in 40 µl of sucrose buffer. Proteins were correctly oriented within the liposomes with the EGFP unit facing the exterior as judged by their sensitivity to TEV protease cleavage.
45 µl of cytosol supplemented with 1 µl of 20 mg/ml glycogen and 1 µl of energy mix (50 mM ATP, 500 mM creatine phosphate, and 10 mg/ml creatine kinase) was incubated with 7,500 sperm heads as chromatin template. 50 min after the addition of 2.5 µl of membranes, 5 µl of resuspended proteoliposomes was added. At the indicated time points, 10 µl of the samples was added to 1 µl of 5 µg/µl NusA-TEV protease in sucrose buffer and incubated for 5 min at 20°C. The cleavage reaction was stopped by fixation with 4% PFA and processed for microscopy as described previously (Franz et al., 2005). For Western blot analysis, TEV cleavage was stopped by the addition of SDS sample buffer and immediate incubation at 95°C for 5 min, followed by SDS-PAGE and Western blotting.
In vitro translation
For the in vitro translation, we generated a modified pET28a vector containing a stem loop–forming sequence derived from the 5′ untranslated region of gene 10 of the T7 bacteriophage (5′-AGGGAGACCACAACGGUUUCCCU-3′) 5′ of the in vitro transcribed/translated gene. This is known to enhance in vitro transcription (O’Connor and Dahlberg, 2001). As DNA templates, full-length mouse Nup188 and Xenopus Nup93 were cloned into this vector and used in the PURExpress in vitro synthesis kit (New England Biolabs, Inc.) according to the manufacturer’s instructions, extending the incubation time to 4 h.
Miscellaneous
Generation of affinity resins for protein depletion, preparation of sperm heads and floated membranes, nuclear assembly reactions, and transmission electron microscopy were performed as described previously (Franz et al., 2005) except that tannic acid was omitted to increase the visibility of NPCs. For depletions, high speed extracts were incubated twice with a 1:1 bead to cytosol ratio for 40 min (Franz et al., 2007). Prelabeled membranes were prepared as in Antonin et al. [2005] using DiIC18, and immunoprecipitations were performed as in Franz et al. [2007]. NPCs from at least 50 nuclei from three independent experiments were counted (D’Angelo et al., 2006) using the Imiaris 6.1.5 software (Bitplane AG) for 3D reconstruction. In vitro assembled nuclei were isolated as in Baur et al. (2007).
Online supplemental material
Fig. S1 shows the quantification of the relative amounts of the different Nup93 complexes. Fig. S2 shows immunofluorescences of nuclei lacking Nup205–Nup93 as well as transmission electron images of mock- and Nup188–Nup93-depleted nuclei. Fig. S3 shows the time course of nuclear assembly in mock- and Nup188–Nup93-depleted extracts. Fig. S4 shows INM targeting assays on nuclei lacking Nup188–Nup93 and Nup205–Nup93 and additional data of the addback experiments. Fig. S5 provides supporting experiments characterizing the INM targeting assay. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912045/DC1.
Acknowledgments
We thank Josef Redolfi and Cornelia Sieverding for excellent technical support, Virgilio Failla for help with NPC quantification, and Elisa Izaurralde, Adriana Magalska, Ruchika Sachdev, Allana Schooley, and Benjamin Vollmer for critical reading of the manuscript.
Footnotes
Abbreviations used in this paper:
- INM
- inner nuclear membrane
- LBR
- lamin B receptor
- MISTIC
- membrane-integrating sequence for translation of integral membrane protein constructs
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- ONM
- outer nuclear membrane
- TEV
- tobacco etch virus
References
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007. Determining the architectures of macromolecular assemblies. Nature. 450:683–694 10.1038/nature06404 [DOI] [PubMed] [Google Scholar]
- Antonin W., Franz C., Haselmann U., Antony C., Mattaj I.W. 2005. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol. Cell. 17:83–92 10.1016/j.molcel.2004.12.010 [DOI] [PubMed] [Google Scholar]
- Antonin W., Ellenberg J., Dultz E. 2008. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 582:2004–2016 10.1016/j.febslet.2008.02.067 [DOI] [PubMed] [Google Scholar]
- Baur T., Ramadan K., Schlundt A., Kartenbeck J., Meyer H.H. 2007. NSF- and SNARE-mediated membrane fusion is required for nuclear envelope formation and completion of nuclear pore complex assembly in Xenopus laevis egg extracts. J. Cell Sci. 120:2895–2903 10.1242/jcs.010181 [DOI] [PubMed] [Google Scholar]
- Beck M., Lucić V., Förster F., Baumeister W., Medalia O. 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 449:611–615 10.1038/nature06170 [DOI] [PubMed] [Google Scholar]
- Blow J.J., Sleeman A.M. 1990. Replication of purified DNA in Xenopus egg extract is dependent on nuclear assembly. J. Cell Sci. 95:383–391 [DOI] [PubMed] [Google Scholar]
- Boehmer T., Enninga J., Dales S., Blobel G., Zhong H. 2003. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc. Natl. Acad. Sci. USA. 100:981–985 10.1073/pnas.252749899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S.G., Leksa N.C., Spear E.D., Rajashankar K.R., Schwartz T.U. 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 322:1369–1373 10.1126/science.1165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., Conti E. 2007. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76:647–671 10.1146/annurev.biochem.76.052705.161529 [DOI] [PubMed] [Google Scholar]
- D’Angelo M.A., Hetzer M.W. 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18:456–466 10.1016/j.tcb.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo M.A., Anderson D.J., Richard E., Hetzer M.W. 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 312:440–443 10.1126/science.1124196 [DOI] [PubMed] [Google Scholar]
- Deng M., Hochstrasser M. 2006. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 443:827–831 10.1038/nature05170 [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo J.C., Mattaj I.W. 1999. Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol. 9:30–41 10.1016/S0960-9822(99)80044-X [DOI] [PubMed] [Google Scholar]
- Fabre E., Hurt E. 1997. Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet. 31:277–313 10.1146/annurev.genet.31.1.277 [DOI] [PubMed] [Google Scholar]
- Fornerod M., Ohno M., Yoshida M., Mattaj I.W. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 90:1051–1060 10.1016/S0092-8674(00)80371-2 [DOI] [PubMed] [Google Scholar]
- Franz C., Askjaer P., Antonin W., Iglesias C.L., Haselmann U., Schelder M., de Marco A., Wilm M., Antony C., Mattaj I.W. 2005. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J. 24:3519–3531 10.1038/sj.emboj.7600825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I.W., Antonin W. 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 8:165–172 10.1038/sj.embor.7400889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V., Mattaj I.W., Askjaer P. 2003. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol. Biol. Cell. 14:5104–5115 10.1091/mbc.E03-04-0237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ospina N., Morgan G., Giddings T.H., Jr., Kosova B., Hurt E., Winey M. 2000. Yeast nuclear pore complex assembly defects determined by nuclear envelope reconstruction. J. Struct. Biol. 132:1–5 10.1006/jsbi.2000.4305 [DOI] [PubMed] [Google Scholar]
- Grandi P., Dang T., Pané N., Shevchenko A., Mann M., Forbes D., Hurt E. 1997. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell. 8:2017–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Orjalo A.V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D.J. 2003. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 11:853–864 10.1016/S1097-2765(03)00116-3 [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara L.A., Platani M., Santarella R., Wozniak R.W., Mattaj I.W. 2008. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol. Biol. Cell. 19:1753–1762 10.1091/mbc.E07-08-0820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M.W., Walther T.C., Mattaj I.W. 2005. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21:347–380 10.1146/annurev.cellbio.21.090704.151152 [DOI] [PubMed] [Google Scholar]
- Hinshaw J.E., Milligan R.A. 2003. Nuclear pore complexes exceeding eightfold rotational symmetry. J. Struct. Biol. 141:259–268 10.1016/S1047-8477(02)00626-3 [DOI] [PubMed] [Google Scholar]
- King M.C., Lusk C.P., Blobel G. 2006. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 442:1003–1007 10.1038/nature05075 [DOI] [PubMed] [Google Scholar]
- Kramer A., Ludwig Y., Shahin V., Oberleithner H. 2007. A pathway separate from the central channel through the nuclear pore complex for inorganic ions and small macromolecules. J. Biol. Chem. 282:31437–31443 10.1074/jbc.M703720200 [DOI] [PubMed] [Google Scholar]
- Lusk C.P., Blobel G., King M.C. 2007. Highway to the inner nuclear membrane: rules for the road. Nat. Rev. Mol. Cell Biol. 8:414–420 10.1038/nrm2165 [DOI] [PubMed] [Google Scholar]
- Makio T., Stanton L.H., Lin C.C., Goldfarb D.S., Weis K., Wozniak R.W. 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185:459–473 10.1083/jcb.200810029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfeld J., Güttinger S., Hawryluk-Gara L.A., Panté N., Mall M., Galy V., Haselmann U., Mühlhäusser P., Wozniak R.W., Mattaj I.W., et al. 2006. The conserved transmembrane nucleoporin NDC1 is required for nuclear pore complex assembly in vertebrate cells. Mol. Cell. 22:93–103 10.1016/j.molcel.2006.02.015 [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. 2004. Sorting out the nuclear envelope from the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 5:65–69 10.1038/nrm1263 [DOI] [PubMed] [Google Scholar]
- Meier E., Miller B.R., Forbes D.J. 1995. Nuclear pore complex assembly studied with a biochemical assay for annulate lamellae formation. J. Cell Biol. 129:1459–1472 10.1083/jcb.129.6.1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter T.C., Klupp B.G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 10.1016/j.virusres.2009.03.018 [DOI] [PubMed] [Google Scholar]
- Mohr D., Frey S., Fischer T., Güttler T., Görlich D. 2009. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 28:2541–2553 10.1038/emboj.2009.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim B., Brumfeld V., Kapon R., Kiss V., Nevo R., Reich Z. 2007. Passive and facilitated transport in nuclear pore complexes is largely uncoupled. J. Biol. Chem. 282:3881–3888 10.1074/jbc.M608329200 [DOI] [PubMed] [Google Scholar]
- Nehrbass U., Rout M.P., Maguire S., Blobel G., Wozniak R.W. 1996. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J. Cell Biol. 133:1153–1162 10.1083/jcb.133.6.1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor M., Dahlberg A.E. 2001. Enhancement of translation by the epsilon element is independent of the sequence of the 460 region of 16S rRNA. Nucleic Acids Res. 29:1420–1425 10.1093/nar/29.7.1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T., Schirmer E.C., Nishimoto T., Gerace L. 2004. Energy- and temperature-dependent transport of integral proteins to the inner nuclear membrane via the nuclear pore. J. Cell Biol. 167:1051–1062 10.1083/jcb.200409149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E.A., Crafoord E., Hallberg E. 2007. Phosphomimetic mutation of the mitotically phosphorylated serine 1880 compromises the interaction of the transmembrane nucleoporin gp210 with the nuclear pore complex. Exp. Cell Res. 313:2744–2751 10.1016/j.yexcr.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Powell L., Burke B. 1990. Internuclear exchange of an inner nuclear membrane protein (p55) in heterokaryons: in vivo evidence for the interaction of p55 with the nuclear lamina. J. Cell Biol. 111:2225–2234 10.1083/jcb.111.6.2225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B.A., Orjalo A.V., Shen Z., Briggs S., Forbes D.J. 2006. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 103:17801–17806 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330 10.1093/emboj/20.6.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosild T.P., Greenwald J., Vega M., Castronovo S., Riek R., Choe S. 2005. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 307:1317–1321 10.1126/science.1106392 [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593–16596 10.1074/jbc.R100015200 [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison J.D., Suprapto A., Hjertaas K., Zhao Y., Chait B.T. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–651 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Mosammaparast N., Wozniak R., Goldfarb D.S. 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J. Cell Biol. 149:1027–1038 10.1083/jcb.149.5.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soullam B., Worman H.J. 1995. Signals and structural features involved in integral membrane protein targeting to the inner nuclear membrane. J. Cell Biol. 130:15–27 10.1083/jcb.130.1.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbert S., Platani M., Boue S., Mattaj I.W. 2006. Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173:469–476 10.1083/jcb.200512078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W.H., Forbes D.J. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339–354 10.1083/jcb.200108007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B.B., Köcher T., Wilm M., Allen T., et al. 2003. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 113:195–206 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Weis K. 2003. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 112:441–451 10.1016/S0092-8674(03)00082-5 [DOI] [PubMed] [Google Scholar]
- Zabel U., Doye V., Tekotte H., Wepf R., Grandi P., Hurt E.C. 1996. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J. Cell Biol. 133:1141–1152 10.1083/jcb.133.6.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleger N., Korfali N., Schirmer E.C. 2008. Inner nuclear membrane protein transport is mediated by multiple mechanisms. Biochem. Soc. Trans. 36:1373–1377 10.1042/BST0361373 [DOI] [PubMed] [Google Scholar]