mTOR induces MyoD-dependent miR-1 expression, leading to follistatin-mediated myocyte fusion.
Abstract
Mammalian target of rapamycin (mTOR) has emerged as a key regulator of skeletal muscle development by governing distinct stages of myogenesis, but the molecular pathways downstream of mTOR are not fully understood. In this study, we report that expression of the muscle-specific micro-RNA (miRNA) miR-1 is regulated by mTOR both in differentiating myoblasts and in mouse regenerating skeletal muscle. We have found that mTOR controls MyoD-dependent transcription of miR-1 through its upstream enhancer, most likely by regulating MyoD protein stability. Moreover, a functional pathway downstream of mTOR and miR-1 is delineated, in which miR-1 suppression of histone deacetylase 4 (HDAC4) results in production of follistatin and subsequent myocyte fusion. Collective evidence strongly suggests that follistatin is the long-sought mTOR-regulated fusion factor. In summary, our findings unravel for the first time a link between mTOR and miRNA biogenesis and identify an mTOR–miR-1–HDAC4–follistatin pathway that regulates myocyte fusion during myoblast differentiation in vitro and skeletal muscle regeneration in vivo.
Introduction
Skeletal myoblast fusion, which results in the formation of multinucleated myofibers, is critical for embryonic muscle development, adult muscle regeneration and maintenance, and muscle hypertrophy under certain conditions. The molecular mechanisms underlying myoblast fusion represent one of the central questions in skeletal muscle biology (Wakelam, 1985; Jansen and Pavlath, 2008). Two molecularly separable stages of fusion have been identified in mammalian muscle cells (Jansen and Pavlath, 2008). After an early stage of differentiation, including cell cycle withdrawal, myogenin expression, and contractile protein expression, mononucleated myoblasts fuse to form nascent myofibers/myotubes. Subsequently, growth and maturation of the muscle cells are achieved through a second-stage fusion, which occurs between the nascent myofibers/myotubes and myoblasts. Although many regulators of these fusion processes have been revealed in recent years (Jansen and Pavlath, 2008), a better understanding of the regulation is still needed.
The Ser/Thr protein kinase mammalian target of rapamycin (mTOR) mediates signaling in response to nutrient availability, cellular energy sufficiency, mitogenic signals, and various types of stress signals. mTOR signaling regulates a wide range of biological processes, including cell growth, various types of cellular differentiation, and metabolism (Erbay et al., 2005; Sarbassov et al., 2005; Wullschleger et al., 2006). mTOR assembles two biochemically and functionally distinct protein complexes, mTORC1 (mTOR complex 1) and mTORC2, which are sensitive and insensitive to rapamycin, respectively (Sarbassov et al., 2005). Rapamycin-sensitive mTORC1 signaling has emerged as a key regulator of skeletal muscle differentiation and remodeling. Rapamycin inhibits myoblast differentiation in vitro (Coolican et al., 1997; Cuenda and Cohen, 1999; Erbay and Chen, 2001), insulin-like growth factor (IGF)–induced myotube hypertrophy in vitro (Rommel et al., 2001; Park et al., 2005), compensatory myofiber hypertrophy in vivo, and regrowth of myofibers after atrophy (Bodine et al., 2001). mTORC1 is also involved in the mechanical stimulation of skeletal muscle ex vivo (Hornberger et al., 2006). The regulation of skeletal myocyte differentiation by mTORC1 occurs at two stages via distinct mechanisms. mTORC1 controls the initiation of myoblast differentiation by regulating IGF-II expression (Erbay and Chen, 2001; Erbay et al., 2003), whereas a late-stage myocyte fusion leading to myotube maturation is regulated by mTORC1 through a yet to be identified secreted factor (Park and Chen, 2005). These regulatory mechanisms are also recapitulated by mTOR functions in muscle regeneration in vivo (Ge et al., 2009).
Micro-RNAs (miRNAs) are a class of small noncoding RNAs that regulate protein expression mainly by targeting the 3′ untranslated region of messenger RNAs (Bartel, 2004, 2009). Intensive studies in recent years have revealed miRNA as a principal regulatory mechanism of gene expression, governing numerous biological processes across the species (Bushati and Cohen, 2007; Bartel, 2009). Several miRNAs have been recognized as important modulators in the development of skeletal and cardiac muscle (van Rooij et al., 2008). Among them, miR-1 is a conserved muscle-specific miRNA that is essential for myogenesis. In Caenorhabditis elegans, miR-1 regulates synaptic functions at the neuromuscular junctions (Simon et al., 2008). In Drosophila melanogaster, deletion of miR-1 leads to defects in muscle differentiation or maintenance, presumably by removing the inhibition on the Notch ligand Delta (Sokol and Ambros, 2005). In mammals, deletion of miR-1-2 in mice causes dysregulation of cardiogenesis (Zhao et al., 2005, 2007), and inhibiting miR-1 impairs the differentiation of skeletal myoblasts (Chen et al., 2006).
Despite the well-recognized importance of both miRNAs and mTOR in myogenesis, a possible connection between the two has never been implicated or examined. In this study, we present evidence revealing for the first time the regulation of a miRNA by mTORC1 signaling. Furthermore, we have identified follistatin as the fusion factor regulated by mTORC1 signaling through miR-1 and histone deacetylase 4 (HDAC4) in myoblast differentiation and muscle regeneration.
Results
mTORC1 regulates miR-1 levels in skeletal muscle
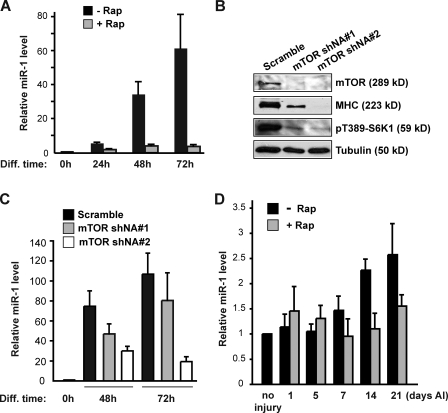
To examine a potential link between miRNA and mTORC1 signaling in skeletal myogenesis, we performed miRNA microarray analyses. We compared miRNA expression profiles of C2C12 cells at differentiation time 0 and 72 h, the latter with or without rapamycin treatment. Among the miRNAs up-regulated during differentiation (Fig. S1 and Table S1), miR-1 expression was drastically inhibited by rapamycin. Quantitative RT-PCR (qRT-PCR) results confirmed that miR-1 increased dramatically during C2C12 differentiation (Fig. 1 A), which is consistent with a previous study (Chen et al., 2006), and the increase was almost completely blocked by rapamycin treatment. As expected, rapamycin abolished myotube formation (not depicted) and drastically inhibited the expression of myogenic markers, including myogenin, myosin heavy chain (MHC), MEF2A, and MEF2C (Fig. S2). To directly confirm mTOR’s role in controlling miR-1 levels, we knocked down mTOR in C2C12 cells using lentivirus-delivered small hairpin RNA (shRNA) with two independent target sequences, which inhibited S6K1 phosphorylation and suppressed MHC expression (Fig. 1 B). At the same time, the miR-1 level during differentiation was suppressed by mTOR knockdown (Fig. 1 C).
Figure 1.
mTOR controls miR-1 levels during myogenesis both in vitro and in vivo. (A) C2C12 cells were induced to differentiate in the absence or presence of 50 nM rapamycin (Rap). RNA was isolated at the indicated time points of differentiation (Diff), and miR-1 levels were measured by qRT-PCR. (B) C2C12 cells were transduced with lentiviruses expressing two independent mTOR shRNAs or scrambled shRNA, selected with puromycin, and induced to differentiate. At 72 h of differentiation, cells were lysed and subjected to Western analysis. 59-kD S6K1 has an apparent molecular mass of 70 kD on SDS-PAGE. (C) Cells treated as in B were harvested at the indicated times of differentiation, and miR-1 levels were measured by qRT-PCR. In both A and C, relative levels are shown as fold increase compared with the level at 0 h. (D) TA muscles in mice hind limbs were injured by intramuscular injection of BaCl2 followed by daily intraperitoneal injection of rapamycin. On various days AI, injected muscles were isolated and homogenized for RNA isolation followed by quantitative PCR to determine relative miR-1 levels, shown as fold increase compared with uninjured muscles. Data are presented as mean ± SD (n = 3).
To validate the dependence of miR-1 levels on mTORC1 in vivo, we examined miR-1 expression in mouse regenerating skeletal muscle. Muscle regeneration was induced upon injury elicited by barium chloride (BaCl2) injection into the tibialis anterior (TA) muscle (Caldwell et al., 1990; Ge et al., 2009). On various days after injury (AI), the TA muscle was isolated, and miR-1 levels were measured by qRT-PCR. miR-1 was found to increase during regeneration, and the increase was blocked by daily rapamycin administration to the mice (Fig. 1 D), accompanied by inhibition of regeneration as we reported previously (Ge et al., 2009). It should be mentioned that others reported a decrease of miR-1 levels in injured muscles (Yuasa et al., 2008; Greco et al., 2009), which was not observed by us at any time point from day 1 to day 21 AI (Fig. 1 D). It has been proposed that the temporary decrease of miR-1 during muscle injury is associated with the loss of myofibers and induction of fibrosis upon injury, rather than muscle regeneration (Greco et al., 2009). Thus, it is possible that miR-1 levels do not decline in our experimental system, owing to less extensive tissue damage induced by BaCl2 compared with the methods used by others (Yuasa et al., 2008; Greco et al., 2009). miR-1 expression in regenerating muscles was also confirmed by in situ hybridization, which showed accumulation of miR-1 signals adjacent to both centrally and peripherally localized nuclei (Fig. S3, enlarged image) and dramatic decrease of signals upon rapamycin administration. Collectively, our observations suggest that mTORC1 controls the levels of miR-1 in skeletal myocytes both in vitro and in vivo.
mTORC1 regulates transcription of miR-1 through an upstream enhancer
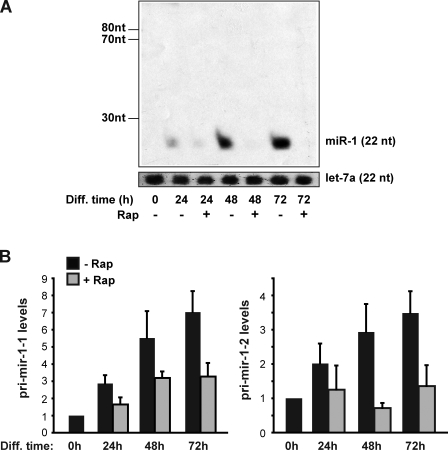
miRNAs are transcribed as pri-miRNAs, cleaved by Drosha to form pre-miRNAs, followed by another cleavage by Dicer, which results in mature miRNAs. A blockage by rapamycin at any of those steps would result in a decrease of mature miR-1 levels. As revealed by Northern blotting (Fig. 2 A), rapamycin treatment of differentiating C2C12 cells drastically decreased the level of mature miR-1 without inducing pre–miR-1 (73 and 78 nucleotides), indicating that rapamycin is unlikely to block the processing of pre–miR-1 to miR-1. The lack of detectable pre–miR-1 by Northern analysis is consistent with rapid processing by Dicer observed for most miRNAs. To test whether the decrease of mature miR-1 could be attributed to a blockage in the processing of pri–miR-1 to pre–miR-1, we measured pri–miR-1 levels by qRT-PCR. MiR-1 is encoded at two chromosomal loci, resulting in two distinct primary transcripts. As shown in Fig. 2 B, rapamycin inhibited rather than enhanced the levels of both forms of pri–miR-1, which excludes the possibility that suppression of pri–miR-1 processing is responsible for the decreased mature miR-1 levels. It is noteworthy that the steady-state level of pri-miRNA does not directly reflect the actual transcription rate because of potentially rapid processing of the transcripts. Nevertheless, the increase of both pri–miR-1 during differentiation and its sensitivity to rapamycin (Fig. 2 B) are consistent with transcriptional regulation of the miR-1 genes. Collectively, our observations suggest that rapamycin suppresses miR-1 levels by impacting the transcription rather than maturation of miR-1.
Figure 2.
miR-1 maturation is not affected by rapamycin. C2C12 cells were induced to differentiate in the absence or presence of 50 nM rapamycin (Rap). RNA was isolated at the indicated time points of differentiation (Diff). (A) Northern blotting was performed to examine miR-1 (22 nucleotides) and pre–miR-1 (73 and 78 nucleotides). Let-7a was blotted as a loading control. (B) qRT-PCR was performed to measure the relative levels of pri–miR-1-1 and pri–miR-1-2. Data shown are the representative results of four independent experiments (A) and the mean ± SD of three independent experiments (B).
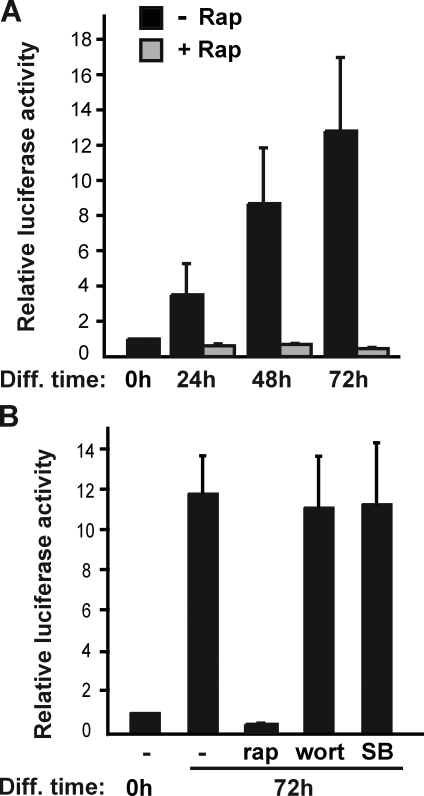
To further investigate the possibility that mTORC1 signaling might regulate the transcription of miR-1, we considered the upstream enhancers for the two miR-1 genes that had been found to regulate the expression of miR-1 in cardiac muscle (Zhao et al., 2005). We set out to examine luciferase reporters for these enhancers in C2C12 cells. As shown in Fig. 3 A, the miR-1-2 enhancer reporter activity increased during C2C12 differentiation, indicating that the enhancer regulating miR-1 in cardiac muscle is also functional in skeletal muscle. Remarkably, rapamycin treatment abolished the increase of the enhancer activity (Fig. 3 A, gray bars). A reporter for the miR-1-1 enhancer displayed similar up-regulation during differentiation and inhibition by rapamycin (unpublished data). To ask whether the effect of rapamycin on miR-1 enhancer was direct or a consequence of rapamycin inhibition of differentiation, we examined the effect of inhibiting phosphatidylinositol-3-kinase and p38, two pathways required for C2C12 differentiation (Kaliman et al., 1996; Cuenda and Cohen, 1999). Strikingly, prolonged (3 d) treatment of C2C12 cells with either wortmannin or SB203580, specific inhibitors of phosphatidylinositol-3-kinase and p38-MAPK, respectively, had no impact on the miR-1 enhancer reporter activity (Fig. 3 B), although both drugs drastically inhibited the differentiation of C2C12 cells from which reporter assays were performed. These observations, together with the fact that not all miRNAs regulated during differentiation were sensitive to rapamycin (Fig. S1 and Table S1), suggest that mTORC1 signaling may directly regulate miR-1 expression through the upstream enhancer.
Figure 3.
Rapamycin inhibits miR-1 enhancer activity. (A) C2C12 cells were transfected with the miR-1-2 enhancer reporter or the corresponding enhancerless reporter, induced to differentiate (Diff) in the absence or presence of 50 nM rapamycin (Rap), and lysed for luciferase assays at the times indicated. (B) Cells were transfected as in A and induced to differentiate for 72 h in the presence of 50 nM rapamycin, 100 nM wortmannin (Wort), or 1 µM SB203580 (SB) followed by cell lysis and luciferase assays. The enhancerless reporter activity was subtracted from the enhancer reporter activity for each condition, and the data shown have been normalized with 0 h of activity as 1. Data shown are the mean ± SD of three independent experiments.
mTORC1 regulates miR-1 through MyoD
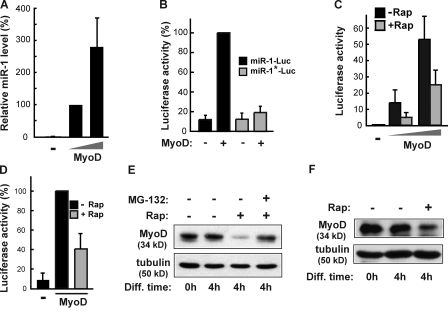
Next, we set out to probe the mechanism underlying mTORC1 regulation of miR-1 transcription. MyoD is a transcription factor that is essential and specific for skeletal myogenesis. Furthermore, putative MyoD-binding sites have been found in both miR-1-1 and miR-1-2 enhancers (Zhao et al., 2005). Indeed, expression of recombinant MyoD in C3H10T1/2 cells, which lack endogenous MyoD expression, drastically stimulated the expression of miR-1 (Fig. 4 A). The expression of recombinant MyoD also markedly activated the miR-1 enhancer reporter in the same cells (Fig. 4 B). A reporter with the putative MyoD-binding site on the enhancer mutated was found to be almost completely insensitive to the expression of MyoD (Fig. 4 B), suggesting that the effect of MyoD on the enhancer is most likely direct, rather than a consequence of MyoD induction of differentiation in C3H10T1/2 cells (Davis et al., 1987).
Figure 4.
mTORC1 regulates miR-1 through MyoD. (A) C3H10T1/2 cells were transfected with MyoD for 24 h followed by extraction of total RNA and qRT-PCR to measure relative miR-1 levels. (B) C3H10T1/2 cells were cotransfected with MyoD and the miR-1-2 enhancer reporter (miR-1–Luc) or the MyoD site-mutated reporter (miR-1*–Luc) and induced to differentiate for 24 h followed by luciferase assays. (C) C3H10T1/2 cells were cotransfected with the miR-1-2 enhancer reporter and two different doses of MyoD and induced to differentiate in the absence or presence of 50 nM rapamycin (Rap) for 24 h followed by luciferase assays. (D) C3H10T1/2 cells were transfected as in C but with a single dose of MyoD, induced to differentiate for 1 d, and treated with 50 nM rapamycin for 4 h followed by luciferase assays. For the data in A–D, relative values are shown, with the following samples as references: (A) lower amount of MyoD transfected (100%), (B) miR-1–Luc with MyoD (100%), (C) without MyoD and without Rap (1), and (D) with MyoD without Rap (100%). (E) At the induction of differentiation (Diff; 0 h), confluent C2C12 cells were treated with 50 nM rapamycin with or without 1 µM MG-132 for 4 h followed by cell lysis and Western analysis for endogenous MyoD. (F) C3H10T1/2 cells were transfected with MyoD, grown to confluence, and treated with 50 nM rapamycin in differentiation medium for 4 h followed by Western analysis of the cell lysates for recombinant MyoD. Tubulin is shown as a loading control in E and F. Error bars indicate mean ± SD.
Consistent with a role of mTORC1 in regulating miR-1 transcription, rapamycin significantly diminished MyoD-induced miR-1 enhancer activity during 24 h of differentiation in C3H10T1/2 cells (Fig. 4 C). Furthermore, we observed that a short period (4 h) of rapamycin treatment was as effective as 24-h treatment in suppressing MyoD-induced miR-1 enhancer activity (Fig. 4 D), once again confirming that the regulation of miR-1 expression by mTORC1 through MyoD is direct. The incomplete rapamycin inhibition of the reporter activity may suggest the existence of additional pathways in the regulation of MyoD activity, but an equally likely possibility is that overexpression of MyoD partially overrides regulation by mTORC1, rendering MyoD somewhat constitutively active and thus partially resistant to rapamycin.
To gain further insight into the mechanism by which mTOR may control MyoD activity, we examined the endogenous MyoD protein upon rapamycin treatment. As shown in Fig. 4 E, rapamycin treatment induced reduction of MyoD protein levels in C2C12 cells. The rapid response to rapamycin exposure (4 h) is consistent with a direct effect of rapamycin on MyoD levels. In addition, the proteasome inhibitor MG-132 completely reversed the effect of rapamycin on MyoD, suggesting that the stability of MyoD is regulated in a proteasome-dependent manner. A similar effect of rapamycin was found on the level of recombinant MyoD in C3H10T1/2 cells (Fig. 4 F), further confirming that the decrease of MyoD protein is posttranslational. Indeed, rapamycin treatment has no effect on the mRNA levels of MyoD in C2C12 cells (Fig. S4 A) or of recombinant MyoD in C3H10T1/2 cells (Fig. S4 B). Collectively, our data suggest that mTORC1 controls MyoD by suppressing its proteasome-dependent degradation.
miR-1 regulates skeletal myogenesis through HDAC4 and follistatin
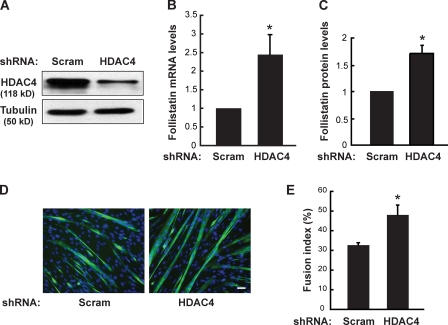
To delineate the pathway downstream of mTOR and miR-1 in myogenesis, we first considered the targets of miR-1. Several potential targets of miR-1 in skeletal and cardiac muscles have been reported (Zhao et al., 2005, 2007; Chen et al., 2006; Yang et al., 2007). Specifically, miR-1 was shown to promote the differentiation of skeletal myoblast by suppressing the expression of HDAC4, a class II HDAC (Chen et al., 2006). It was also reported that HDAC inhibitors induced myocyte fusion through the production of follistatin (Iezzi et al., 2004), but the identity of the HDAC was not known. We hypothesized that miR-1 might promote skeletal myogenesis by suppressing HDAC4 and subsequently inducing follistatin expression. To examine this hypothesis, we first asked whether HDAC4 could be involved in follistatin production and myogenesis. To this end, we knocked down HDAC4 in C2C12 cells with a lentivirus-delivered shRNA (Fig. 5 A). Depletion of HDAC4 led to an increase in follistatin mRNA levels (Fig. 5 B) as well as the amount of secreted follistatin protein (Fig. 5 C). Meanwhile, HDAC4 knockdown enhanced myocyte differentiation (Fig. 5 D) with an increase in fusion index, defined as the percentage of nuclei in multinucleated cells (Fig. 5 E). These data suggest that HDAC4 suppresses follistatin production and is a negative regulator of myogenic fusion.
Figure 5.
Knockdown of HDAC4 enhances follistatin expression and myocyte fusion. C2C12 cells were transduced with lentiviruses expressing shRNA for HDAC4 or a scrambled sequence as negative control (Scram). (A–E) After puromycin selection, the cells were induced to differentiate for 72 h followed by Western analysis of cell lysates (A), qRT-PCR assays to determine the relative levels of follistatin mRNA (B), ELISA to measure relative levels of secreted follistatin protein (C), immunostaining with anti-MHC (green) and DAPI (blue; D), and measurement of fusion index (E). Bar, 50 µm. One-sample t tests were performed in B and C, and a paired two-tailed t test was performed in E. *, P ≤ 0.02. Error bars indicate mean ± SD.
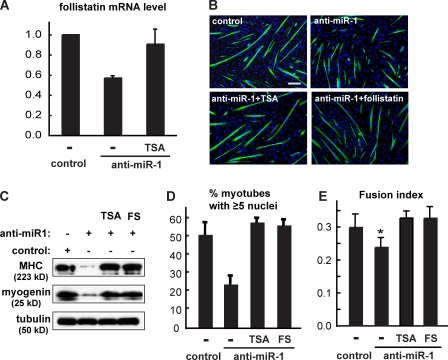
Next, we examined the requirement of miR-1 for follistatin expression. Inhibition of miR-1 function was achieved by a locked nucleic acid (LNA)–containing antisense oligonucleotide approach that had been reported to be effective in blocking the actions of the targeted miRNAs (Naguibneva et al., 2006a). As shown in Fig. 6 A, anti–miR-1 significantly suppressed the mRNA levels of follistatin, suggesting that miR-1 is required for the expression of follistatin. In addition, the inhibition of follistatin by anti–miR-1 was almost completely reversed when cells were treated with the HDAC inhibitor trichostatin A (TSA; Fig. 6 A). These data are consistent with a cascade in which miR-1 regulates follistatin expression through impairing an HDAC. At the same time, a negative effect of anti–miR-1 was found on C2C12 myogenic differentiation (Fig. 6 B), reflected by suppressed expression of myogenin and MHC (Fig. 6 C). The percentage of myotubes containing five or more nuclei was significantly reduced by anti–miR-1 (Fig. 6 D), and the overall fusion index was also lower in anti–miR-1–expressing cells (Fig. 6 E). More importantly, addition of recombinant follistatin to the cell medium fully rescued the myogenic protein expression (Fig. 6 C), myotube size (Fig. 6 D), and fusion index (Fig. 6 E) in the presence of anti–miR-1. Inhibition of HDAC by TSA had a similar rescue effect (Fig. 6, C–E).
Figure 6.
miR-1 is required for myocyte fusion through HDAC and follistatin. C2C12 cells were transfected with LNA anti–miR-1 or a scrambled LNA oligo as control and induced to differentiate. To inhibit HDAC activity, 25 nM TSA was added to the growth medium when cell density was ∼60% and removed the next day upon switching to differentiation medium. Where indicated, 0.2 µg/ml recombinant follistatin was added to the medium during the last 2 d of differentiation. (A) Relative follistatin mRNA levels after 2 d of differentiation were determined by qRT-PCR. (B) C2C12 cells after 3 d of differentiation were immunostained with anti-MHC (green) and DAPI (blue). Bar, 100 µm. (C) Myogenic marker expression was examined by Western analysis in 3-d differentiated C2C12 cells. (D and E) The percentage of myotubes containing five or more nuclei (D) and the fusion index (E) were calculated. In E, paired two-tailed t tests were performed to compare each data with the control. FS, follistatin. *, P < 0.05. Error bars indicate mean ± SD.
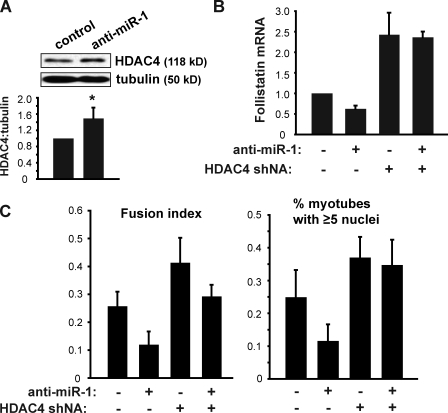
We further probed the specific role of HDAC4 in mediating miR-1 regulation of follistatin. We found that anti–miR-1 indeed enhanced the expression of HDAC4 modestly but consistently (Fig. 7 A). Furthermore, knockdown of HDAC4 completely overrode the negative effect of anti–miR-1 on follistatin expression (Fig. 7 B) and, at the same time, restored myocyte fusion as indicated by both the fusion index and the percentage of large myotubes (Fig. 7 C). Collectively, these observations strongly support the existence of a miR-1–HDAC4–follistatin cascade in the regulation of myogenesis.
Figure 7.
HDAC4 functions downstream of miR-1. (A) C2C12 cells were transfected with LNA anti–miR-1 or a scrambled LNA oligo as control and induced to differentiate for 72 h. HDAC4 protein levels in cell lysates were examined by Western blotting and quantified by densitometry using ImageJ (National Institutes of Health). The ratio of HDAC4 to tubulin was normalized with scrambled LNA as 1. A one-sample t test was performed. *, P < 0.05. (B and C) C2C12 cells were transduced with lentiviruses expressing shRNA for HDAC4 or a scrambled hairpin sequence, puromycin selected, and transfected with anti–miR-1 or scrambled LNA followed by induction of differentiation for 72 h. The cells were subjected to RNA isolation and qRT-PCR to measure follistatin mRNA (B) or characterization of myotube fusion (C) as described in Fig. 6. Error bars indicate mean ± SD (n = 3).
mTORC1 regulates myocyte fusion through follistatin
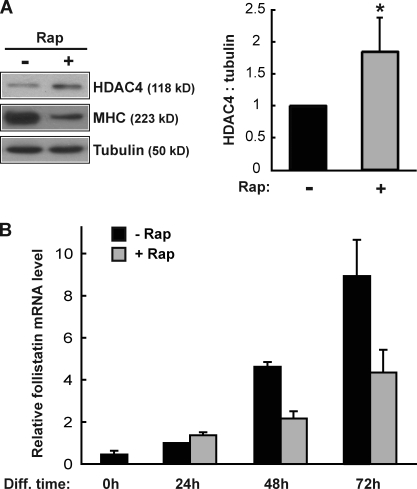
Now that we have established a regulatory pathway from miR-1 to follistatin via HDAC4, the immediate question is whether mTORC1 controls this pathway, in other words, whether inhibition of mTORC1 enhances HDAC4 expression and in turn suppresses follistatin expression. Indeed, we found that rapamycin treatment increased HDAC4 protein levels (Fig. 8 A) and at the same time markedly suppressed the mRNA level of follistatin (Fig. 8 B). Notably, rapamycin’s inhibitory effect on follistatin was only obvious at a later stage during differentiation (days 2 and 3) when myocyte fusion takes place, supporting the idea that follistatin is a myogenic factor that promotes fusion rather than the initiation of differentiation. The comparable kinetics of miR-1 and follistatin expression (dramatic increase of both at 48 and 72 h of differentiation; Fig. 1 A and Fig. 8 B) are also consistent with miR-1 regulation of follistatin.
Figure 8.
Rapamycin leads to increased HDAC4 levels and decreased follistatin levels. (A) C2C12 cells were differentiated for 2 d in the presence or absence of 50 nM rapamycin (Rap), and the lysates were analyzed by Western blotting. Western blots were quantified using ImageJ, and the relative ratio of HDAC4 to tubulin is shown. A one-sample t test was performed. *, P < 0.01. (B) C2C12 cells were induced to differentiate in the absence or presence of 50 nM rapamycin, and total RNA was isolated at the indicated time points of differentiation (Diff). Relative follistatin mRNA levels were measured by qRT-PCR with the 24-h sample without rapamycin as 1. Data shown are mean ± SD of three independent experiments.
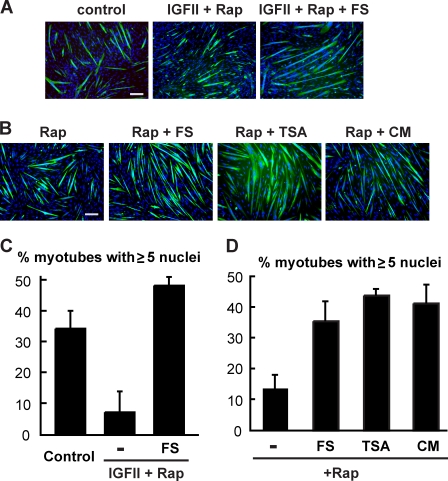
Previously, we demonstrated that mTORC1 regulates at least two distinct processes of myogenesis and that its regulation of the second-stage myocyte fusion leading to myotube maturation or growth is through a yet to be identified secreted factor (Park and Chen, 2005). In light of the discovery of a connection between mTORC1 and follistatin through miR-1 and HDAC4, we wondered whether follistatin might be the long-sought second-stage fusion factor under the control of mTOR. To this end, we tested whether follistatin could rescue myogenic differentiation from rapamycin inhibition. When C2C12 cells were induced to differentiate in the presence of rapamycin, follistatin alone failed to rescue differentiation (unpublished data). This outcome was not unexpected, as rapamycin would block the production of both the fusion factor and IGF-II, the latter required for the initiation of differentiation (Erbay et al., 2003). To bypass the initiation of differentiation, thus allowing examination of myocyte fusion specifically, we added recombinant IGF-II to the differentiation medium, which would fully support the initial differentiation and formation of nascent myotubes in the presence of rapamycin (Erbay et al., 2003; Park and Chen, 2005). In the presence of both IGF-II and rapamycin, C2C12 cells differentiated into myotubes but arrested at a small myotube size with fewer myonuclei than mature myotubes (Fig. 9, A and C). Strikingly, the addition of recombinant follistatin in this system led to a complete rescue of mature myotube size (Fig. 9, A and C), suggesting that follistatin is a strong candidate for an mTORC1-regulated second-stage fusion factor.
Figure 9.
mTORC1 regulates myocyte fusion through follistatin. (A) C2C12 cells were differentiated in the absence or presence of 300 ng/ml IGF-II and 50 nM rapamycin (Rap) for 3 d. Where indicated, 0.2 µg/ml follistatin (FS) was present during the last 2 d of differentiation. Differentiated cells were immunostained with anti-MHC (green) and DAPI (blue). (B) C2C12 cells stably expressing RR/KI mTOR were induced to differentiate in the presence of 50 nM rapamycin for 3 d. 0.2 µg/ml follistatin was added to the medium for the last 2 d of differentiation. 25 nM TSA was added when the cells were ∼60% confluent and removed the next day upon induction of differentiation. Conditioned medium (CM) was collected daily from parental C2C12 cells that had been induced to differentiate 1 d earlier than the RR/KI mTOR cells and fed to the latter with 50 nM rapamycin added. The cells were immunostained as in A. Bars, 100 µm. (C and D) Percentage of myotubes containing at least five nuclei was calculated from the experiments in A and B, respectively. All data shown are mean ± SD of at least three independent experiments.
Previously, we had reported that C2C12 cells stably expressing a rapamycin-resistant (RR) and kinase-inactive (KI) mTOR differentiated in the presence of rapamycin but arrested at the nascent myotube stage (Park and Chen, 2005). To confirm that follistatin was the missing factor in those cells, we added follistatin to the medium of the RR/KI mTOR cells containing rapamycin. Remarkably, this resulted in a dramatic increase of myotube size, as indicated by the higher percentage of myotubes containing five or more nuclei (Fig. 9, B and D). The effectiveness of follistatin was comparable with that of conditioned medium from fully differentiated C2C12 cells. In addition, inhibiting HDAC by TSA had a similar effect (Fig. 9, B and D). Collectively, these data strongly suggest that follistatin is a fusion factor regulated by mTORC1.
Follistatin is regulated by mTORC1 during muscle regeneration in vivo
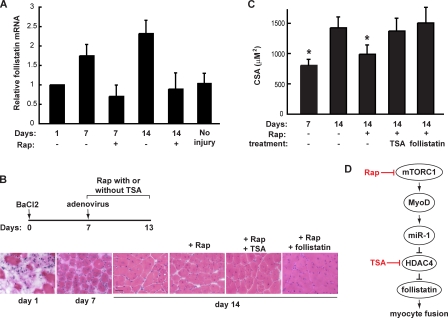
Regeneration of damaged adult skeletal muscles involves satellite cell activation and proliferation followed by myoblast differentiation and fusion. Systemic administration of rapamycin impairs the regeneration of skeletal muscle induced by BaCl2 injection in mice, suggesting that mTORC1 plays a key role in muscle regeneration (Ge et al., 2009). Significantly, follistatin mRNA levels increased by about twofold during regeneration, and this increase was abolished by rapamycin administration (Fig. 10 A), consistent with mTORC1 regulation of follistatin in vivo. We further probed the involvement of follistatin in mTORC1-regulated myocyte fusion and myofiber growth in vivo. To study myofiber maturation, which relies on myocyte fusion, BaCl2-injected muscles were allowed to regenerate for 7 d, during which time new fibers were formed but not fully matured, before rapamycin was systemically administered. As shown in Fig. 10 (B and C), rapamycin administration significantly diminished the growth of regenerating myofibers, as indicated by the smaller average cross-section area of the fibers. Most important, intramuscular injection of adenovirus-expressing recombinant follistatin alleviated the negative effect of rapamycin, resulting in normal growth of the regenerating myofibers in the presence of rapamycin (Fig. 10, B and C), whereas adenovirus expressing GFP did not have any effect (not depicted). TSA has been shown to induce the expression of follistatin in skeletal muscles in vivo (Minetti et al., 2006). Indeed, similar to intramuscular expression of recombinant follistatin, systemic administration of TSA fully rescued the growth of regenerating myofibers from rapamycin inhibition (Fig. 10, B and C). Collectively, these observations provide in vivo validation for follistatin as an mTORC1-regulated myogenic fusion factor.
Figure 10.
Follistatin is regulated by mTORC1 during muscle regeneration in vivo. (A) Mouse hind limb TA muscles were injected with BaCl2 followed by daily systemic administration of rapamycin starting 1 d AI. On various days indicated, the injected muscles were isolated and total RNA extracted followed by qRT-PCR to measure levels of follistatin mRNA. Data shown are mean ± SD (n = 3). (B) TA muscles were injected with BaCl2 followed by daily systemic administration of rapamycin (Rap) with or without TSA from day 7 to day 13 AI. For some animals, a single dose of adenovirus-expressing follistatin was injected into the injured TA muscle on day 7 AI. A schematic diagram is shown to summarize the various injections of animals. On days 1, 7, or 14 AI, the animals were sacrificed, and the injured muscles were dissected and cryosectioned followed by hematoxylin and eosin staining. Representative images are shown (n = 7 mice for each time point). Bar, 50 µm. (C) Cross-section areas of regenerating myofibers shown in B were measured. Data shown are mean ± SD (n = 7 mice for each data point). One-way analysis of variance was performed to analyze the data. Significant difference comparing each data point to that of day 14 without treatment; *, P < 0.001. (D) A schematic representation of the myogenic pathway discovered in this study. mTORC1 controls MyoD-dependent expression of miR-1 that targets HDAC4 and subsequently up-regulates the production of follistatin, which in turn governs myocyte fusion in skeletal myogenesis.
Discussion
Our study has revealed for the first time regulation of an miRNA by mTOR, a master regulator of cell growth and differentiation. We have also identified follistatin as the long-sought myogenic fusion factor under the regulation of mTORC1 signaling (Park and Chen, 2005). We propose a myogenic pathway (Fig. 10 D) in which rapamycin-sensitive mTORC1 controls MyoD-dependent transcription of miR-1, which in turn suppresses HDAC4 and subsequently up-regulates the production of follistatin, stimulating skeletal myocyte fusion in vitro and in vivo. The linearity of this pathway presented in this study would be an oversimplification of the actual regulatory network, as miR-1 and HDAC4 would most likely have multiple targets in myogenesis, and mTOR certainly regulates other myogenic pathways independent of miR-1. Nevertheless, our study identifies a functional pathway important for myocyte fusion and muscle growth, providing a new target for therapeutic intervention in muscle repair and regeneration.
mTOR regulation of miRNA
Best known as a regulator of protein synthesis through its effectors S6K1 and 4E-BP1 (Gingras et al., 2001; Hay and Sonenberg, 2004), the rapamycin-sensitive mTORC1 also regulates RNA polymerase I–dependent ribosomal DNA transcription through the initiation factors TIF-IA, SL-1, and upstream-binding factor (Mayer and Grummt, 2006). Regulation of mRNA expression has been implicated for mTORC1 as well, although the mechanisms remain to be deciphered. Our discovery that mTORC1 regulates biogenesis of miRNA further expands the repertoire of this master regulator for mammalian cellular and developmental processes.
There are two distinct chromosomal loci for the miR-1 gene, miR-1-1 and miR-1-2. An upstream enhancer has been found at each locus to mediate the regulation of miR-1 expression in cardiac muscle, and regulatory sites for serum response factor, MyoD and MEF2, have been identified in the enhancers (Zhao et al., 2005). Our results have indicated that the miR-1 enhancer activity is indeed up-regulated during skeletal myocyte differentiation and inhibited by rapamycin, suggesting mTORC1 regulation of miR-1 expression through this enhancer. We have further shown that MyoD mediates mTOR regulation of the miR-1 enhancer activity and that mTORC1 controls the protein stability of MyoD. mTOR regulation of MyoD and miR-1 is most likely direct, based on the following observations: (a) the effects of rapamycin on MyoD degradation and miR-1 enhancer activity were acute and not dependent on myogenic differentiation (Fig. 4, D–F), and (b) although rapamycin abolished miR-1 enhancer activity, other drugs that block differentiation did not have any effect (Fig. 3). In addition to the upstream enhancers of miR-1 genes, an intragenic enhancer has been reported to activate muscle-specific transcription of the bicistronic primary transcript encoding miR-1-2 and miR-133a-1 (Liu et al., 2007). Interestingly, we found that a reporter of this intragenic enhancer was activated during C2C12 cells differentiation and inhibited by rapamycin (Fig. S5). Because MyoD also regulates this enhancer (Liu et al., 2007), it raises the possibility that the mTORC1–MyoD axis controls miR-1 transcription at multiple levels.
In contrast to mTOR regulation of ribosomal DNA transcription, which requires S6K1 (Mayer et al., 2004), mTOR regulation of miR-1 expression does not seem to involve S6K1, as knockdown of S6K1 has no effect on miR-1 expression during myogenesis (unpublished data). It is conceivable that mTORC1 phosphorylates MyoD and, in turn, stabilizes MyoD. We have indeed observed that mTOR phosphorylates MyoD in vitro (unpublished data). The reported MyoD phosphorylation sites, Ser5 and Ser200, do not conform to this model, as their phosphorylation correlates with degradation of MyoD (Song et al., 1998; Tintignac et al., 2004). The exact mechanism by which mTORC1 regulates MyoD stability is currently under investigation.
miRNA profiling of differentiating C2C12 cells has been reported by other groups (Chen et al., 2006; Wong and Tellam, 2008). Although there is some degree of agreement within the results obtained by different groups including ours, our data have revealed more myogenically regulated miRNAs than the other studies, possibly because of the difference in the miRNA chips used, robustness of cell differentiation, and/or methods of data analysis (e.g., we used statistical significance rather than fold change to set the threshold for data selection). In any event, our array results have confirmed the up-regulation during differentiation of almost all reported regulators of myogenesis in addition to miR-1, including miR-24, -26a, -27b, -133, -206, -181, -214, and -499 (Table S1; Naguibneva et al., 2006b; Sun et al., 2008; Wong and Tellam, 2008; Crist et al., 2009; Juan et al., 2009; van Rooij et al., 2009; Williams et al., 2009). Many miRNAs not previously reported to be involved in myogenesis are found to be up- or down-regulated during differentiation (Table S1), potentially representing novel regulators of myogenic pathways that are worthy of future investigation. Intriguingly, among ∼50 miRNAs that were differentially expressed during myogenesis (out of ∼500 total), 24 miRNAs other than miR-1 displayed various degrees of rapamycin sensitivity (Fig. S1 and Table S1). Although some of these miRNAs may lie far downstream of mTORC1 in myogenesis, it is conceivable that other miRNAs in addition to miR-1 may be directly regulated by mTORC1. Further characterization of those candidate miRNAs will likely enhance our understanding of the myogenic regulatory network.
Follistatin as an mTOR-regulated fusion factor
Previously, we reported the existence of a fusion factor under the control of mTORC1 signaling in an mTOR kinase-dependent manner, which promoted maturation of myotubes arrested at the nascent myotube stage by the treatment of rapamycin (Park and Chen, 2005). Our current study has revealed follistatin as this long-sought fusion factor regulated by mTORC1 through an miR-1–mediated pathway. Several other secreted factors have been found in the conditioned medium of normally maturing myotubes but not of rapamycin-arrested nascent myotubes (unpublished data), including the established second-stage fusion factor IL-4 (Horsley et al., 2003) and the NF-kB–induced myogenic stimulator IL-6 (Baeza-Raja and Muñoz-Cánoves, 2004). However, none of them are able to rescue rapamycin-inhibited myotube maturation (unpublished data). The capacity of exogenous follistatin to fully rescue from rapamycin inhibition myotube maturation in vitro (Fig. 9) and muscle regeneration in vivo (Fig. 10, B and C), together with the inhibitory effect of rapamycin on follistatin expression both in vitro and in vivo (Fig. 8 B and Fig. 10 A), makes follistatin the major, if not the only, fusion factor regulated by mTORC1.
Follistatin, an activin-binding protein essential for multiple aspects of mouse development (Matzuk et al., 1995), is thought to control skeletal muscle development through antagonizing the myogenic inhibitor myostatin (Lee and McPherron, 2001; Amthor et al., 2004). The stimulatory effect of follistatin on adult muscle growth has been demonstrated in the mdx mice (Minetti et al., 2006; Haidet et al., 2008; Nakatani et al., 2008). In vitro, follistatin was shown to mediate TSA- or nitric oxide/cyclic GMP–induced myoblast fusion (Iezzi et al., 2004; Pisconti et al., 2006). It is likely that follistatin plays a role in both the initial myoblast fusion and the second-stage fusion, as we have observed that differentiating C2C12 cells exposed to recombinant follistatin display increased fusion index in addition to increased size of myotubes (unpublished data). This well corroborates our observation that inhibiting miR-1 results in decreased myotube size and fusion index (Fig. 6), supporting the notion that miR-1 and follistatin act on the same pathway to regulate two stages of myocyte fusion. The new regulatory pathway for follistatin (Fig. 10 D) discovered in our study should expand the therapeutic potential of this important myogenic factor.
Materials and methods
Antibodies and other reagents
The antibodies were obtained from the following sources: anti-tubulin was obtained from Abcam, anti-MyoD (5.8A) was obtained from Imgenex, antibodies against HDAC4, mTOR, phospho–T389-S6K1, MEF2A, and MEF2C were obtained from Cell Signaling Technology, and all secondary antibodies and DAPI were obtained from Jackson ImmunoResearch Laboratories, Inc. The MF20 anti-sarcomeric MHC and F5D anti-myogenin antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development, the National Institutes of Health, and maintained by The University of Iowa Department of Biological Sciences. Rapamycin was obtained from LC Laboratories. Follistatin (mouse FS288) was obtained from R&D Systems. TSA, gelatin, polybrene, puromycin, MG-132, and custom-designed LNA oligonucleotides were obtained from Sigma-Aldrich.
Tissue culture
C2C12 and C3H10T1/2 cells were maintained in DME (1 g/liter glucose) with 10% fetal bovine serum. For differentiation, cells were seeded in plates coated with 0.2% gelatin. Differentiation was induced by switching to DME containing 2% horse serum. Transfection of LNA anti–miR-1 and luciferase reporters into C2C12 cells was performed using nucleofector (solution V; program B-032; Lonza) following the manufacturer’s recommendations. All other transfections were performed with Trans-IT (Mirus) at 50–60% cell density.
Microarray analysis
miRNA profiling was performed using LNA miRNA arrays (Exiqon), which contained capture probes for all miRNAs annotated in miRBase (version 9.2; http://www.mirbase.org/). Samples were prepared using miRVana miRNA isolation kit (Applied Biosystems) without enrichment for small RNA. 1 µg total RNA was labeled with Cy3 or Cy5 according to the manufacturer’s protocols cohybridized to arrays overnight, washed, and scanned (GenePix 4000B; MDS Analytical Technologies). Image analysis and editing were performed using GenePix Pro (version 6.0). Data were analyzed using the bioconductor packages with R software. For each comparison, F statistics were calculated with results from four independent experiments. False discovery rate–adjusted p-value at 0.05 was used as cutoff for statistical significance.
Muscle regeneration in mice
All animal experiments in this study followed protocols approved by the Animal Care and Use Committee at the University of Illinois at Urbana-Champaign. 10–12-wk-old male FVB mice were used in all experiments. Muscle injury was induced by injection of 50 µl BaCl2 (1.2% wt/vol) into the TA muscle of the hind limb. As control, saline was injected into the TA muscle of the lateral hind limb. Rapamycin (1 µg/g body weight) and/or TSA (0.6 µg/g body weight) in a carrier containing 5% Tween 80, 5% PEG-400, and 4% ethanol was administrated once daily through intraperitoneal injection. Adenovirus (AdexCA-FS288 or AdexCA-GFP) was delivered by one-time injection of 1011 viral particles into the TA muscle.
Plasmids
pCDNA3-Flag-MyoD was created in a modified pCDNA3 vector containing the Flag epitope followed by NotI and XbaI restriction sites in which MyoD cDNA was inserted. pGL3-MH100-TK luciferase reporters containing miR-1 upstream enhancers were provided by D. Srivastava (University of California, San Francisco, San Francisco, CA; Zhao et al., 2005). The enhancerless reporter was generated from the miR-1-2 enhancer reporter construct by deleting the enhancer at KpnI sites. The 2.5-kb miR-1/133a intragenic enhancer, provided by E.N. Olson (University of Texas Southwestern Medical Center, Dallas, TX; Liu et al., 2007), was subcloned into pGL2-promoter (Promega).
Lentivirus-mediated RNAi
shRNAs in the pLKO.1-puro vector for knocking down mTOR and HDAC4 were purchased from Sigma-Aldrich (MISSION TRC). For viral packaging, pLKO-shRNA, pCMV-dR8.91, and pCMV–VSV-G were cotransfected into 293T cells using FuGENE 6 at 0.5:0.45:0.05 µg (in 1 ml for a 6-well plate). Media containing viruses were collected 48 h after transfection. The clone IDs (Sigma-Aldrich) for the shRNA constructs used in this study are the following: mTOR#1, NM_020009.1-7569s1c1; mTOR#2, NM_020009.1-5493s1c1; and HDAC4, NM_207225.1-1619s1c1. C2C12 cells were transduced with lentiviruses in growth medium containing 8 µg/ml polybrene and selected in 2 µg/ml puromycin for 1 d followed by plating into 12-well plates for differentiation.
Real-time quantitative PCR for mRNA and miRNA
Total RNA was isolated from cultured cells or muscle tissues using the mirVana RNA isolation kit (Applied Biosystems). cDNA was synthesized from 2 µg total RNA with reverse transcription (SuperScript II; Invitrogen) using oligo (dT) primer (Invitrogen). Quantitative PCR was performed on an iCycler system (Bio-Rad Laboratories) using a SYBR green PCR kit (Applied Biosystems) in a 96-well reaction plate (MicroAmp; Bio-Rad Laboratories) following the manufacturer’s protocols. β-Actin was used as a reference to obtain the relative fold change for target samples using the comparative CT method. The following primers were used: mouse follistatin (forward), 5′-AAAACCTACCGCAACGAATG-3′ and (reverse) 5′-GGTCTGATCCACCACACAAG-3′; and mouse β-actin (forward), 5′-TTGCTGACAGGATGCAGAAG-3′ and (reverse) 5′-ATCCACATCTGCTGGAAGGT-3′. Primers for pri–miR-1-1 and pri–miR-1-2 were used as described previously (Liu et al., 2007). miRNAs were quantified using quantitative PCR–based miRNA assay kits (Taqman; Applied Biosystems) following the manufacturer’s recommendations, with snoRNA 202 as the internal control for normalization.
Northern blotting
Antisense DNA oligonucleotides for mature miR-1 and let-7a were end labeled with γ-[32P]ATP. The RNA Decade Maker system (Applied Biosystems) was similarly radiolabeled as size markers. 20 µg/sample RNA was separated on 12% denaturing polyacrylamide gels, transferred to nylon membranes (GE Healthcare), and UV cross-linked. Prehybridization was performed in ultrasensitive hybridization buffer (ULTRAhyb; Applied Biosystems) for 2 h at 42°C followed by hybridization with probe in the same buffer (106 CPM/ml) overnight at 42°C. The membranes were washed with 2× SSC and 0.1% SDS at 42°C followed by exposure to x-ray films overnight at −80°C.
Luciferase assays
Cells transfected with various enhancer reporters were treated as described in Results and Figs. 3 and 4 and lysed in Passive Lysis buffer (Promega). Luciferase assays were performed using the Luciferase Assay Systems kit (Promega) following the manufacturer’s protocol.
Immunofluorescence microscopy and quantitative analysis of myocytes
C2C12 cells differentiated in 12-well plates were fixed in 3.7% formaldehyde (in PBS), permeabilized in 0.1% Triton X-100, and incubated with MF-20 (anti-MHC) antibody in 3% BSA (in PBS) followed by fluorescein isothiocyanate-conjugated anti–mouse IgG in 3% BSA (in PBS) with 4 µg/ml DAPI. The stained cells were examined with a fluorescence microscope (DMI 4000B; Leica), and the fluorescent images were captured using a camera (RETIGA Exi; QImaging). The images were processed as 24-bit images using Photoshop (CS2; Adobe). MHC and DAPI signals were pseudocolored green and blue, respectively. The fusion index was calculated as the ratio of nuclei number in myocytes with two or more nuclei versus the total number of nuclei. Each data point was generated from at least 200 randomly chosen MHC-positive cells or myotubes.
ELISA for follistatin measurement
Relative follistatin protein levels in media collected from differentiating C2C12 cells were measured using the human follistatin ELISA kit (26% cross reactivity with mouse follistatin; R&D Systems) following the manufacturer’s protocols.
Adenovirus production
Adenoviruses expressing human follistatin (AdexCA-FS288) or EGFP (AdexCA-GFP) under the CAG promoter were provided by W. Vale (Salk Institute, La Jolla, CA; Leal et al., 2002). The viruses were amplified through infection of 293 cells and purified by ultracentrifugation with a cesium chloride gradient (Luo et al., 2007) followed by dialysis against PBS.
Muscle tissue cryosection and histological analysis
TA muscles were isolated by dissection, frozen in liquid nitrogen–cooled 2-methylbutane, and embedded in TBS tissue freezing medium (Thermo Fisher Scientific). Sections of 10-µm thickness were made with a cryostat (Microm HM550; Thermo Fisher Scientific) at −20°C, placed on uncoated slides, and stained with hematoxylin and eosin. The stained slides were examined with a microscope (DMI 4000B), and the images were captured with a Fluotar 20× 0.4 NA dry objective (Leica) using a camera (RETIGA Exi). The images were processed as 24-bit colored images using Photoshop. The cross-section area of myofibers was measured using Q-capture Pro software (version 6.0; QImaging). For each muscle section, all central-nucleated (regenerating) myofibers >100 µm2 within a 307,200-µm2 view in the center of injury were analyzed.
In situ hybridization
In situ hybridization of miR-1 was performed on cryosections of TA muscles prepared as described in the previous paragraph. miR-1 was detected by antisense LNA probes labeled with the Dig-3 end-labeling kit (Roche) as previously described (Naguibneva et al., 2006b) with some modifications. In brief, muscle cryosections were fixed in paraformaldehyde, deproteinized with proteinase K, and acetylated with acetic anhydride and triethanolamine followed by incubation with 20 nM labeled probes in hybridization buffer (65% formamide, 5× SSC, 0.1% Tween 20, 50 mg/ml heparin, and 500 mg/ml yeast tRNA, pH 6.0) at 50°C overnight. The slides were subsequently washed with 50% formamide/2× SSC at 50°C and with PBST at room temperature followed by blocking and incubation with anti-digoxigenin (1:1,000; Roche) at 4°C overnight. The slides were washed and developed in NBT-BCIP (Sigma-Aldrich) for 4 h, dehydrated, and mounted. A probe with a scrambled sequence was used as negative control. The following probes were used (bold letters indicate LNA): anti–miR-1 LNA probe, 5′-ATACATACTTCTTTACATTCCA-3′; and scrambled LNA probe, 5′-CATGTCATGTGTCACATCTCTT-3′. An anti–miR-1 probe purchased from Exiqon was also used, and similar in situ hybridization signal patterns were observed.
Statistical analysis
All data are presented as mean ± SD. Whenever necessary, statistical significance of the data was analyzed by performing one-sample t tests, paired two-tailed t tests, or one-way analysis of variance with Bonferroni’s posttest using Prism (version 4.0; GraphPad Software, Inc.). The specific types of tests and the p-values, when applicable, are indicated in Figs. 5–8 and 10.
Online supplemental material
Fig. S1 and Table S1 show the results of miRNA profiling in differentiating C2C12 cells. Fig. S2 shows rapamycin inhibition of myogenic markers. Fig. S3 shows in situ hybridization of miR-1 in regenerating muscles. Fig. S4 shows MyoD mRNA levels in C2C12 and C3H10T1/2 cells upon rapamycin treatment. Fig. S5 shows rapamycin inhibition of the miR-1/133a intragenic enhancer reporter. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200912093/DC1.
Acknowledgments
We thank Drs. K.V. Prasanth, A. Lal, L. Chen, J. Liu, A. Polesskaya, J. Kemper, V. Tripathi, Ms. D. Kanamaluru, Ms. Y. Wang, Ms. R. Zheng, and Ms. I. Joewono for helpful discussions, guidance, and technical assistance. We are also grateful to Drs. D. Srivastava, E. Olson, and W. Vale for their generous gifts of miR-1 enhancer reporters and follistatin-expressing adenovirus.
This work was supported by grants to J. Chen from the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (grant AR48914) and the American Diabetes Association. Y. Zhao was supported by a Scientist Development Grant from the American Heart Association.
Footnotes
Abbreviations used in this paper:
- AI
- after injury
- HDAC
- histone deacetylase
- IGF
- insulin-like growth factor
- KI
- kinase inactive
- LNA
- locked nucleic acid
- MHC
- myosin heavy chain
- miRNA
- micro-RNA
- mTOR
- mammalian target of rapamycin
- qRT-PCR
- quantitative RT-PCR
- RR
- rapamycin resistant
- shRNA
- small hairpin RNA
- TA
- tibialis anterior
- TSA
- trichostatin A
References
- Amthor H., Nicholas G., McKinnell I., Kemp C.F., Sharma M., Kambadur R., Patel K. 2004. Follistatin complexes myostatin and antagonises myostatin-mediated inhibition of myogenesis. Dev. Biol. 270:19–30 10.1016/j.ydbio.2004.01.046 [DOI] [PubMed] [Google Scholar]
- Baeza-Raja B., Muñoz-Cánoves P. 2004. p38 MAPK-induced nuclear factor-kappaB activity is required for skeletal muscle differentiation: role of interleukin-6. Mol. Biol. Cell. 15:2013–2026 10.1091/mbc.E03-08-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 116:281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine S.C., Stitt T.N., Gonzalez M., Kline W.O., Stover G.L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J.C., Glass D.J., Yancopoulos G.D. 2001. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3:1014–1019 10.1038/ncb1101-1014 [DOI] [PubMed] [Google Scholar]
- Bushati N., Cohen S.M. 2007. microRNA functions. Annu. Rev. Cell Dev. Biol. 23:175–205 10.1146/annurev.cellbio.23.090506.123406 [DOI] [PubMed] [Google Scholar]
- Caldwell C.J., Mattey D.L., Weller R.O. 1990. Role of the basement membrane in the regeneration of skeletal muscle. Neuropathol. Appl. Neurobiol. 16:225–238 10.1111/j.1365-2990.1990.tb01159.x [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z. 2006. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38:228–233 10.1038/ng1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolican S.A., Samuel D.S., Ewton D.Z., McWade F.J., Florini J.R. 1997. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J. Biol. Chem. 272:6653–6662 10.1074/jbc.272.10.6653 [DOI] [PubMed] [Google Scholar]
- Crist C.G., Montarras D., Pallafacchina G., Rocancourt D., Cumano A., Conway S.J., Buckingham M. 2009. Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. Proc. Natl. Acad. Sci. USA. 106:13383–13387 10.1073/pnas.0900210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenda A., Cohen P. 1999. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J. Biol. Chem. 274:4341–4346 10.1074/jbc.274.7.4341 [DOI] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., Lassar A.B. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 51:987–1000 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- Erbay E., Chen J. 2001. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J. Biol. Chem. 276:36079–36082 10.1074/jbc.C100406200 [DOI] [PubMed] [Google Scholar]
- Erbay E., Park I.H., Nuzzi P.D., Schoenherr C.J., Chen J. 2003. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J. Cell Biol. 163:931–936 10.1083/jcb.200307158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbay E., Kim J.E., Chen J. 2005. Amino acid-sensing mTOR signaling. In Nutrient and Cell Signaling Zempleni J., Dakshinamurti K., editors Taylor & Francis Group, Boca Raton, FL. 353–380 [Google Scholar]
- Ge Y., Wu A.L., Warnes C., Liu J., Zhang C., Kawasome H., Terada N., Boppart M.D., Schoenherr C.J., Chen J. 2009. mTOR regulates skeletal muscle regeneration in vivo through kinase-dependent and kinase-independent mechanisms. Am. J. Physiol. Cell Physiol. 297:C1434–C1444 10.1152/ajpcell.00248.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Raught B., Sonenberg N. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807–826 10.1101/gad.887201 [DOI] [PubMed] [Google Scholar]
- Greco S., De Simone M., Colussi C., Zaccagnini G., Fasanaro P., Pescatori M., Cardani R., Perbellini R., Isaia E., Sale P., et al. 2009. Common micro-RNA signature in skeletal muscle damage and regeneration induced by Duchenne muscular dystrophy and acute ischemia. FASEB J. 23:3335–3346 10.1096/fj.08-128579 [DOI] [PubMed] [Google Scholar]
- Haidet A.M., Rizo L., Handy C., Umapathi P., Eagle A., Shilling C., Boue D., Martin P.T., Sahenk Z., Mendell J.R., Kaspar B.K. 2008. Long-term enhancement of skeletal muscle mass and strength by single gene administration of myostatin inhibitors. Proc. Natl. Acad. Sci. USA. 105:4318–4322 10.1073/pnas.0709144105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N., Sonenberg N. 2004. Upstream and downstream of mTOR. Genes Dev. 18:1926–1945 10.1101/gad.1212704 [DOI] [PubMed] [Google Scholar]
- Hornberger T.A., Sukhija K.B., Chien S. 2006. Regulation of mTOR by mechanically induced signaling events in skeletal muscle. Cell Cycle. 5:1391–1396 [DOI] [PubMed] [Google Scholar]
- Horsley V., Jansen K.M., Mills S.T., Pavlath G.K. 2003. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 113:483–494 10.1016/S0092-8674(03)00319-2 [DOI] [PubMed] [Google Scholar]
- Iezzi S., Di Padova M., Serra C., Caretti G., Simone C., Maklan E., Minetti G., Zhao P., Hoffman E.P., Puri P.L., Sartorelli V. 2004. Deacetylase inhibitors increase muscle cell size by promoting myoblast recruitment and fusion through induction of follistatin. Dev. Cell. 6:673–684 10.1016/S1534-5807(04)00107-8 [DOI] [PubMed] [Google Scholar]
- Jansen K.M., Pavlath G.K. 2008. Molecular control of mammalian myoblast fusion. Methods Mol. Biol. 475:115–133 10.1007/978-1-59745-250-2_7 [DOI] [PubMed] [Google Scholar]
- Juan A.H., Kumar R.M., Marx J.G., Young R.A., Sartorelli V. 2009. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell. 36:61–74 10.1016/j.molcel.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliman P., Viñals F., Testar X., Palacín M., Zorzano A. 1996. Phosphatidylinositol 3-kinase inhibitors block differentiation of skeletal muscle cells. J. Biol. Chem. 271:19146–19151 10.1074/jbc.271.32.19146 [DOI] [PubMed] [Google Scholar]
- Leal A.M., Takabe K., Wang L., Donaldson C.J., MacConell L.A., Bilezikjian L.M., Verma I.M., Vale W. 2002. Effect of adenovirus-mediated overexpression of follistatin and extracellular domain of activin receptor type II on gonadotropin secretion in vitro and in vivo. Endocrinology. 143:964–969 10.1210/en.143.3.964 [DOI] [PubMed] [Google Scholar]
- Lee S.J., McPherron A.C. 2001. Regulation of myostatin activity and muscle growth. Proc. Natl. Acad. Sci. USA. 98:9306–9311 10.1073/pnas.151270098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Williams A.H., Kim Y., McAnally J., Bezprozvannaya S., Sutherland L.B., Richardson J.A., Bassel-Duby R., Olson E.N. 2007. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc. Natl. Acad. Sci. USA. 104:20844–20849 10.1073/pnas.0710558105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Deng Z.L., Luo X., Tang N., Song W.X., Chen J., Sharff K.A., Luu H.H., Haydon R.C., Kinzler K.W., et al. 2007. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat. Protoc. 2:1236–1247 10.1038/nprot.2007.135 [DOI] [PubMed] [Google Scholar]
- Matzuk M.M., Lu N., Vogel H., Sellheyer K., Roop D.R., Bradley A. 1995. Multiple defects and perinatal death in mice deficient in follistatin. Nature. 374:360–363 10.1038/374360a0 [DOI] [PubMed] [Google Scholar]
- Mayer C., Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 25:6384–6391 10.1038/sj.onc.1209883 [DOI] [PubMed] [Google Scholar]
- Mayer C., Zhao J., Yuan X., Grummt I. 2004. mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev. 18:423–434 10.1101/gad.285504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minetti G.C., Colussi C., Adami R., Serra C., Mozzetta C., Parente V., Fortuni S., Straino S., Sampaolesi M., Di Padova M., et al. 2006. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibitors. Nat. Med. 12:1147–1150 10.1038/nm1479 [DOI] [PubMed] [Google Scholar]
- Naguibneva I., Ameyar-Zazoua M., Nonne N., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Pritchard L.L., Harel-Bellan A. 2006a. An LNA-based loss-of-function assay for micro-RNAs. Biomed. Pharmacother. 60:633–638 10.1016/j.biopha.2006.07.078 [DOI] [PubMed] [Google Scholar]
- Naguibneva I., Ameyar-Zazoua M., Polesskaya A., Ait-Si-Ali S., Groisman R., Souidi M., Cuvellier S., Harel-Bellan A. 2006b. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat. Cell Biol. 8:278–284 10.1038/ncb1373 [DOI] [PubMed] [Google Scholar]
- Nakatani M., Takehara Y., Sugino H., Matsumoto M., Hashimoto O., Hasegawa Y., Murakami T., Uezumi A., Takeda S., Noji S., et al. 2008. Transgenic expression of a myostatin inhibitor derived from follistatin increases skeletal muscle mass and ameliorates dystrophic pathology in mdx mice. FASEB J. 22:477–487 10.1096/fj.07-8673com [DOI] [PubMed] [Google Scholar]
- Park I.H., Chen J. 2005. Mammalian target of rapamycin (mTOR) signaling is required for a late-stage fusion process during skeletal myotube maturation. J. Biol. Chem. 280:32009–32017 10.1074/jbc.M506120200 [DOI] [PubMed] [Google Scholar]
- Park I.H., Erbay E., Nuzzi P., Chen J. 2005. Skeletal myocyte hypertrophy requires mTOR kinase activity and S6K1. Exp. Cell Res. 309:211–219 10.1016/j.yexcr.2005.05.017 [DOI] [PubMed] [Google Scholar]
- Pisconti A., Brunelli S., Di Padova M., De Palma C., Deponti D., Baesso S., Sartorelli V., Cossu G., Clementi E. 2006. Follistatin induction by nitric oxide through cyclic GMP: a tightly regulated signaling pathway that controls myoblast fusion. J. Cell Biol. 172:233–244 10.1083/jcb.200507083 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rommel C., Bodine S.C., Clarke B.A., Rossman R., Nunez L., Stitt T.N., Yancopoulos G.D., Glass D.J. 2001. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3:1009–1013 10.1038/ncb1101-1009 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sabatini D.M. 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596–603 10.1016/j.ceb.2005.09.009 [DOI] [PubMed] [Google Scholar]
- Simon D.J., Madison J.M., Conery A.L., Thompson-Peer K.L., Soskis M., Ruvkun G.B., Kaplan J.M., Kim J.K. 2008. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 133:903–915 10.1016/j.cell.2008.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N.S., Ambros V. 2005. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev. 19:2343–2354 10.1101/gad.1356105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A., Wang Q., Goebl M.G., Harrington M.A. 1998. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell. Biol. 18:4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M., et al. 2008. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 36:2690–2699 10.1093/nar/gkn032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintignac L.A., Sirri V., Leibovitch M.P., Lécluse Y., Castedo M., Metivier D., Kroemer G., Leibovitch S.A. 2004. Mutant MyoD lacking Cdc2 phosphorylation sites delays M-phase entry. Mol. Cell. Biol. 24:1809–1821 10.1128/MCB.24.4.1809-1821.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E., Liu N., Olson E.N. 2008. MicroRNAs flex their muscles. Trends Genet. 24:159–166 10.1016/j.tig.2008.01.007 [DOI] [PubMed] [Google Scholar]
- van Rooij E., Quiat D., Johnson B.A., Sutherland L.B., Qi X., Richardson J.A., Kelm R.J., Jr., Olson E.N. 2009. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell. 17:662–673 10.1016/j.devcel.2009.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelam M.J. 1985. The fusion of myoblasts. Biochem. J. 228:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A.H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J.L., Bassel-Duby R., Sanes J.R., Olson E.N. 2009. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 326:1549–1554 10.1126/science.1181046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.F., Tellam R.L. 2008. MicroRNA-26a targets the histone methyltransferase enhancer of Zeste homolog 2 during myogenesis. J. Biol. Chem. 283:9836–9843 10.1074/jbc.M709614200 [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B., Zhang Y., Xu C., Bai Y., Wang H., et al. 2007. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13:486–491 10.1038/nm1569 [DOI] [PubMed] [Google Scholar]
- Yuasa K., Hagiwara Y., Ando M., Nakamura A., Takeda S., Hijikata T. 2008. MicroRNA-206 is highly expressed in newly formed muscle fibers: implications regarding potential for muscle regeneration and maturation in muscular dystrophy. Cell Struct. Funct. 33:163–169 10.1247/csf.08022 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 436:214–220 10.1038/nature03817 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. 2007. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 129:303–317 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]