3D reconstruction by cryo-EM provides the first structural description of a ribosomal biogenesis factor (Nmd3) in complex with the 60S ribosomal subunit.
Abstract
The nucleocytoplasmic shuttling protein Nmd3 is an adaptor for export of the 60S ribosomal subunit from the nucleus. Nmd3 binds to nascent 60S subunits in the nucleus and recruits the export receptor Crm1 to facilitate passage through the nuclear pore complex. In this study, we present a cryoelectron microscopy (cryo-EM) reconstruction of the 60S subunit in complex with Nmd3 from Saccharomyces cerevisiae. The density corresponding to Nmd3 is directly visible in the cryo-EM map and is attached to the regions around helices 38, 69, and 95 of the 25S ribosomal RNA (rRNA), the helix 95 region being adjacent to the protein Rpl10. We identify the intersubunit side of the large subunit as the binding site for Nmd3. rRNA protection experiments corroborate the structural data. Furthermore, Nmd3 binding to 60S subunits is blocked in 80S ribosomes, which is consistent with the assigned binding site on the subunit joining face. This cryo-EM map is a first step toward a molecular understanding of the functional role and release mechanism of Nmd3.
Introduction
Eukaryotic ribosomes are produced in the nuclear subcompartment, the nucleolus, in a complex series of precise RNA processing and protein assembly steps. After nucleolar assembly and nucleolar and nucleoplasmic maturation, preribosome subunits are exported to the cytoplasm through the nuclear pore complex (Johnson, 2009; for reviews see Fromont-Racine et al., 2003; Hage and Tollervey, 2004; Zemp and Kutay, 2007; Henras et al., 2008). In order for transport substrates to partition into the hydrophobic lumen of the nuclear pore complex, they must recruit specialized receptor proteins that have affinity for nucleoporins (Fried and Kutay, 2003; Pemberton and Paschal, 2005; Köhler and Hurt, 2007; Tran et al., 2007). In yeast, the large ribosomal subunit utilizes three receptors for export: Crm1, recruited by the adaptor protein Nmd3 (Ho et al., 2000b; Gadal et al., 2001), the heterodimeric mRNA export factor Mex67-Mtr2 (Yao et al., 2007), and the noncanonical receptor Arx1 (Bradatsch et al., 2007; Hung et al., 2008). Among these factors, only Nmd3 appears to have a conserved role in ribosome export in vertebrates (Thomas and Kutay, 2003; Trotta et al., 2003).
Nmd3 is a highly conserved protein found throughout eukaryotes and archaea. The N-terminal 35 kD of the protein contains two zinc-binding centers and comprises the ribosome-binding domain (Hedges et al., 2006). The nucleocytoplasmic shuttling of Nmd3 is driven by a nuclear localization sequence and a nuclear export sequence, both of which are located in the C terminus of the protein. The leucine-rich nuclear export sequence (aa 496–505) is predicted to form an amphipathic helix, with the hydrophobic residues critical for Crm1 interaction aligned on one surface. The presence of Nmd3 orthologues in archaea suggests a role in ribosome biogenesis that predates the evolution of the nuclear envelope. Currently, no high-resolution structural information is available on this protein.
Genetic and biochemical experiments indicate that the release of Nmd3 from 60S subunits in the cytoplasm requires the loading of the large ribosomal subunit protein Rpl10 and the conserved cytoplasmic GTPase Lsg1p (Hedges et al., 2005; West et al., 2005). Yeast Rpl10 belongs to the L10e family of ribosomal proteins and is orthologous to bacterial L16. The crystal structure of the Haloarcula marismortui large subunit reveals that L10e is located in the deep cleft created by the central protuberance (CP) and the GTPase-associated center (GAC; Klein et al., 2004), corresponding to the position of L16 in Escherichia coli (Schuwirth et al., 2005).
To address the molecular mechanism by which the adaptor protein Nmd3 interacts with the large subunit, we have generated a 3D reconstruction of a 60S subunit in complex with Nmd3, determined at 16-Å resolution by cryo-EM using the single particle approach. We identify the helix 95 region at the intersubunit surface of the large subunit as the anchoring site of Nmd3. An extended part of the protein reaches close to the ribosomal protein Rpl10. However, no direct interaction with the Rpl10 site is detected. We provide supporting biochemical data that corroborate the structural results. This study provides the first structural description of a biogenesis factor in complex with the large subunit.
Results and discussion
Nmd3 binds stoichiometrically with the 60S subunit
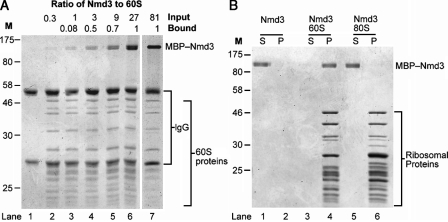
To examine the interaction of Nmd3 with 60S subunits, we used a rapid coimmunoprecipitation technique. A fixed amount of epitope-tagged subunits (60S-Rpl25-13xmyc) was bound to protein A beads and incubated with increasing amounts of maltose-binding protein (MBP)–Nmd3. After binding, the beads were washed extensively, and bound proteins were eluted and separated by SDS-PAGE. As the ratio of Nmd3 to 60S was increased, the amount of Nmd3 bound to 60S increased accordingly, reaching a maximum of 1:1, even at 81-fold excess of Nmd3 relative to 60S (Fig. 1 A). This result suggests that Nmd3 binds to the 60S subunit as a monomer and to a single site on the subunit.
Figure 1.
In vitro binding of MBP-Nmd3 to 60S but not 80S ribosomes. (A) Increasing amounts of MBP-Nmd3 were incubated with Rpl25-myc–containing 60S subunits and immunoprecipitated with anti-myc antibody and protein A beads. Bound proteins were eluted in Laemmli sample buffer, separated on 12% SDS-PAGE, and stained with Coomassie brilliant blue. See Materials and methods for details. Lane 1 shows MBP-Nmd3 without 60S; lanes 2–7 show 60S-myc with increasing amounts of MBP-Nmd3 as indicated. The molar ratio of Nmd3 to 60S subunits is given for the input and bound samples. (B) Nmd3 does not bind 80S ribosomes. MBP-Nmd3 was incubated alone (lanes 1 and 2), with 60S (lanes 3 and 4), or with 80S subunits (lanes 5 and 6). Samples were layered over 60% sucrose cushions and centrifuged. Supernatants (S) and pellets (P) were separated on a 12% SDS-PAGE, and proteins were visualized by Coomassie staining. (A and B) The positions of molecular mass markers (M) are given in kilodaltons.
The Nmd3 protein used in this work was expressed as a fusion to MBP. MBP-Nmd3 fully complemented an nmd3 deletion mutant (unpublished data), indicating that the fusion protein is functional in vivo. We found that cleavage of MBP from Nmd3 destabilized the protein and reduced 60S binding. Consequently, all work was performed with the intact fusion protein. Previously, we showed reconstitution of the Nmd3–60S complex using a GST-Nmd3 fusion (Ho et al., 2000a). However, because this protein dimerizes 60S subunits (unpublished data), we used it in this work only as a control for specificity of RNase footprinting.
Nmd3 binds to free 60S subunits but not to 40S subunits or 80S complexes in vivo, suggesting that its binding site may be blocked by the presence of the 40S subunit (Ho and Johnson, 1999). To test whether this result could be recapitulated in vitro, we compared the binding of Nmd3 to purified 60S subunits versus 80S ribosomes. MBP-Nmd3 was incubated alone, with 60S subunits, or with preformed 80S ribosomes, and reactions were separated by centrifugation through a sucrose cushion. Under these conditions, free Nmd3 remained entirely in the supernatant (Fig. 1 B, lanes 1 and 2), whereas in the presence of 60S subunits, Nmd3 quantitatively cosedimented with the subunits (Fig. 1 B, lanes 3 and 4). In contrast, Nmd3 did not cosediment with preformed 80S ribosomes, but rather remained in the supernatant fraction (Fig. 1 B, lanes 5 and 6).
Localization of Nmd3 protein on the 60S subunit
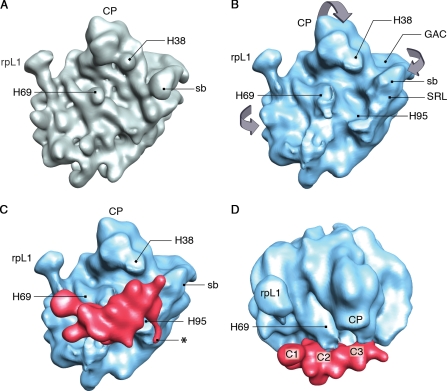
To obtain a more detailed picture of how Nmd3 interacts with the large subunit, we used cryo-EM and single particle image reconstruction. Cryo-EM maps depicting the 60S subunit alone (Fig. 2 A) and in complex with the adaptor protein (Fig. 2 C) were obtained at resolutions of 18 Å and 16 Å, respectively. A comparison of the two maps clearly shows an extra density attached to the intersubunit side of the large subunit covering the region extending from the Rpl1 stalk base to the P-protein stalk base of the 25S ribosomal RNA (rRNA; Fig. 2 C).
Figure 2.
Visualization of MBP-Nmd3 binding to the 60S subunit. (A) Intersubunit side view of the control 60S subunit. (B) Intersubunit view of the segmented 60S part of the MBP-Nmd3–60S reconstruction. Significant conformational changes are seen in the GAC, the SRL, the CP, and the region around the peptidyl-transfer center. The stalk base (sb), the L1 stalk (rpL1), and 25S rRNA helices 38, 69, and 95 (H38, H69, and H95) are also labeled. The direction of the motion of the intersubunit surface of the 60S subunit after MBP-Nmd3 binding is marked with arrows. (C) Intersubunit side view of the MBP-Nmd3–60S subunit complex. The segmented density attributed to the MBP-Nmd3 combined mass is colored red, whereas the 60S subunit is colored blue. The asterisk denotes the thread of density (see Identification of Nmd3–60S subunit interactions for details). (D) Top view of the complex showing three connections (C1, C2, and C3) of the MBP-Nmd3 mass (red) with the 60S subunit (blue).
This extra density visualized in the cryo-EM map was computationally separated by a segmentation procedure using SPIDER (system for processing image data from EM and related fields; Frank et al., 1996). Because the Nmd3 protein was purified with the MBP tag, it is expected that the extra density contains both MBP and Nmd3 as the intact fusion protein. Indeed, the molecular mass of this extra density calculated from the volume it occupies (∼110 kD at threshold value 42; mean density and variability [σ] of the MBP-Nmd3–bound 60S subunit map are 6 and 31, respectively, using 0.82 D/Å3 as protein density) is substantially larger than the known molecular mass of Nmd3 (59 kD) and close to the expected size of MBP-Nmd3 (103 kD). In addition, biochemical results (Fig. 1 A) rule out the possibility of the presence of two copies of Nmd3. Therefore, the additional mass can be attributed to the presence of the MBP tag (molecular mass ∼44 kD). These results suggest that the entire MBP-Nmd3 fusion protein is visualized in the cryo-EM map. The current resolution does not allow us to model MBP into the mass attributed to MBP-Nmd3. The position of Nmd3 at the interface of the 60S subunit is incompatible with subunit joining, which is consistent with the observation that Nmd3 does not bind to the 80S ribosome in vivo (Ho and Johnson, 1999) or in vitro (Fig. 1 B).
The conformational changes in the MBP-Nmd3–bound 60S subunit relative to the control 60S subunit are shown in Fig. 2 (A and B). Significant displacement was observed in the following regions: (a) the base of the L1 stalk, (b) the GAC and the sarcin–ricin loop (SRL; domain VI of the 25S rRNA), and (c) the CP and the region around the peptidyl-transfer center. In all of these regions, ribosome density was shifted toward the MBP-Nmd3 density. Of particular note, the cleft between the CP and the GAC was narrower in the complex than in the control map. Overall, the changes on the intersubunit side of the 60S subunit can be likened, in their tendency, to the grip of a hand around an object (MBP-Nmd3) on its palm (primary binding sites are H69 and H95 of 25S rRNA; for brevity, rRNA helices of 25S rRNA will be denoted by “H”).
The L1 stalk, containing protein Rpl1, is seen in the open position (Valle et al., 2003) in both complex and control maps in which the protein part (Rpl1) of the mushroom-shaped head is partially visible. In contrast, the extended part of the acidic P-protein stalk region and the protein Rpl12 (L11p) at the stalk base are neither visible in the control nor in the 60S subunit map of our complex.
Biochemical characterization of 60S subunit–ligand interactions
To seek supporting evidence for the position of MBP-Nmd3 on the 60S subunit, helices appearing to make contact with the mass assigned to MBP-Nmd3 in the cryo-EM map were probed for altered sensitivity to RNaseV1, a nuclease specific for double-stranded RNA. In these assays, 60S subunits were incubated alone, with MBP-Nmd3, or with GST-Nmd3 and treated with RNaseV1. The GST-Nmd3 reactions were used to control for MBP-specific effects. After RNaseV1 treatment, the rRNA was extracted, and reverse transcription was performed using radio-labeled primers. Primer extension reactions were compared with a DNA sequencing ladder to identify the positions of cleavages.
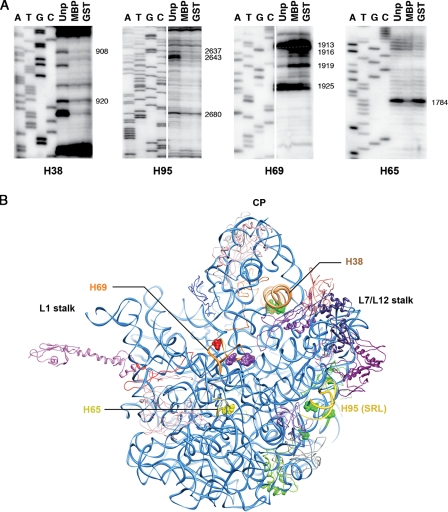
Protection by both MBP- and GST-Nmd3 against RNaseV1 was observed for H38 at four positions (Fig. 3 A). Two of these positions (bases 1045 and 1054) correspond to E. coli 23S bases 908 and 920. MBP- and GST-Nmd3 binding also protected three positions in H95 against cleavage. These positions (3003, 3009, and 3047) correspond to E. coli 23S bases 2637, 2643, and 2680 (Fig. 3 A). A strong enhancement of RNaseV1 cleavage was observed with both Nmd3 fusion proteins (i.e., GST- and MBP-Nmd3) at three positions in H69, nt 2253, 2256, and 2259 (corresponding to E. coli 23S nt 1913, 1916, and 1919). Furthermore, a GST-Nmd3–specific protection was seen in H69 at position 2265 (corresponding to E. coli 23S position 1925), and an MBP-Nmd3–specific RNaseV1 protection was observed at position 2142 of H65 (corresponding to E. coli 23S nt 1784). Primer extension analysis did not reveal significant changes in other regions of 25S, 5.8S, or 5S rRNA (unpublished data).
Figure 3.
rRNA protection: MBP-Nmd3 interaction with helices H38, H65, H69, and H95 of 25S. (A) 60S subunits were incubated with no protein, MBP-Nmd3, or GST-Nmd3 and treated with RNaseV1. The rRNA was extracted, and primer extension reactions were performed to identify regions of altered sensitivity to RNaseV1. Sequencing reaction lanes are marked by the dideoxynucleotide present in the mixture. Primers used were H38, AJO1061; H65, AJO501; H69, AJO1060; and H95, AJO1135. Numbers indicate positions (E. coli numbering) of nucleotides showing major alteration in sensitivity to RNaseV1. Unp, unprotected (no Nmd3); MBP, MBP-Nmd3; GST, GST-Nmd3. (B) An interface view of the 50S subunit of the 70S E. coli crystal structure (PDB ID 2AW4) showing the position of the helices concerned. Nucleotides marked in green are protected from RNaseV1 by Nmd3; nucleotides in purple show enhancement of cleavage upon interaction with Nmd3; the nucleotide in red shows protection from RNaseV1 cleavage by GST; and the nucleotide in yellow is protected from RNaseV1 cleavage by MBP.
H38 is part of 25S rRNA domain II, which accounts for most of the solvent-side surface of the large subunit. However, the tip of this helix (A-site finger), adjacent to the CP, protrudes toward the subunit interface side and participates in the formation of the intersubunit bridge B1a. In contrast, H65 and H69 belong to domain IV, which accounts for most of the intersubunit surface of the large subunit. H69 is positioned at the center of the large subunit interface and participates in the formation of two essential intersubunit bridges, B2a and B2b (Yusupov et al., 2001). H65 is also exposed to the subunit surface on the intersubunit side. H95 (SRL; rRNA domain VI) is situated below the P-protein stalk base region (Ban et al., 2000), and part of it is exposed to the solvent (Figs. 2 B and 3 B). Based on the protection assay results, a tentative identification of the positions for MBP and Nmd3 in the density can be made. Our results enable us to suggest that the SRL/CP proximal part of the differential mass observed in the cryo-EM structure accounts for Nmd3, whereas the distal part close to H65 likely represents the MBP portion of the fusion protein (see next section).
Identification of Nmd3–60S subunit interactions
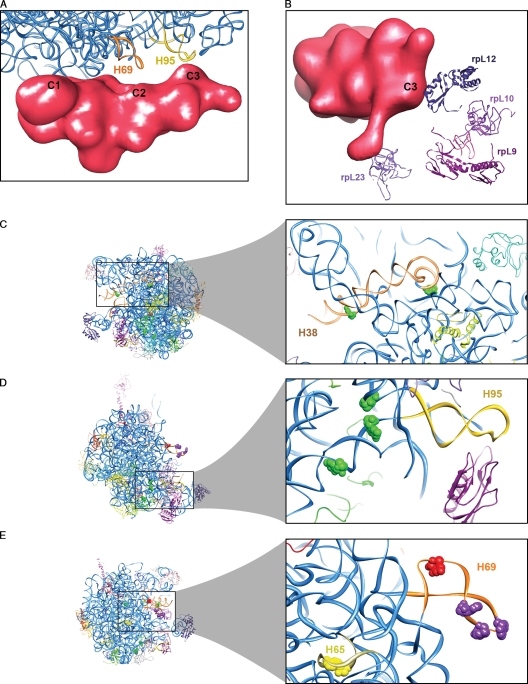
Overall, the MBP-Nmd3 density appears as a complex, extended mass stretching from the Rpl1 stalk base side of the 60S subunit to the P-protein stalk base (Fig. 2, C and D). Connections between the MBP-Nmd3 complex and the large subunit are clearly visible in three places (marked as C1, C2, and C3 in Figs. 2 D and 4 A). To determine the molecular details of interaction between the large subunit and the MBP-Nmd3 density, the quasiatomic model of the 60S subunit previously derived by cryo-EM and homology modeling (Spahn et al., 2001) was used (PDB ID 1S1I). This model was aligned, as a rigid body, to the 60S subunit within the map of the complex. The analysis of the ligand–subunit interactions is consistent with the biochemical results: the anchoring region (C2) of the center of the MBP-Nmd3 mass on the ribosome was determined as being formed at the H69 region of 25S rRNA (Fig. 4 A). Additionally, contact C1 with the part of rRNA domain IV that contains H65 and contact C3 with the H95/SRL region were identified. Based on MBP and Nmd3 molecular masses and our biochemical data, we expect the MBP-Nmd3 density in the cryo-EM map to be roughly split evenly between the two, with the Nmd3 part being closest to H69/SRL. We suggest Nmd3 to be directly interacting with H95 and H38 of the 60S subunit at the C3 point of contact (Fig. 2, C and D; Fig. 3 A; and Fig. 4 A). The C2 point of contact involves the central part of the MBP-Nmd3 density (Figs. 2 D and 4 A). This region of the density is most likely at the borderline between Nmd3 and MBP. This interpretation is supported by the fact that in Fig. 3 A, we observe multiple H69 sites of altered RNaseV1 sensitivity for both MBP- and GST-Nmd3 (suggesting Nmd3 interaction) along with a protection specific for the GST-Nmd3 control (suggesting the GST protein tag to be interacting). The C1 point of contact is at the end of the combined MBP-Nmd3 density closest to Rpl1 and is therefore expected to represent solely the interaction between MBP and the 60S subunit. Indeed, we observe an MBP-specific protection from RNaseV1 cleavage in H65 strongly supporting this interpretation.
Figure 4.
MBP-Nmd3 interaction with the 60S subunit. (A and B) Close-up view of the quasiatomic structure of the 60S subunit (PDB ID 1S1I; Spahn et al., 2001), with the MBP-Nmd3 density showing connections with the rRNA helices (A) and nearby proteins (B). (C–E) Close-up views of the rRNA helices and the nucleotides showing altered protection patterns upon MBP-Nmd3 binding in the 50S subunit of the E. coli 70S ribosome crystal structure: H38 (C), H95 (D), and H65 and H69 (E). Nucleotides are colored as in Fig. 3 B.
Cryo-EM maps at the resolution achieved in this study cannot confirm the identity of interacting nucleotides suggested by the protection assay. However, to participate in the interaction with the MBP-Nmd3 mass, these nucleotides are required to be exposed on the subunit interface. Examination of the 50S subunit (PDB ID 2AW4) within the E. coli 70S ribosome crystal structure (Schuwirth et al., 2005) elucidated that, indeed, all of the nucleotides identified in the protection assay were situated at the intersubunit side of the rRNA helices concerned (Fig. 3 B). However, except for the base 1919 (E. coli number) on H69, none of the nucleotides were totally exposed on the surface (Fig. 4, C–E). The fact that they are accessible to the binding by MBP-Nmd3 must be attributed to a difference in the conformation of the relevant helices caused by subunit association. In fact, H38 and H69 are disordered in the crystal structure of the isolated 50S subunit (Ban et al., 2000), suggesting that these helices are initially in a variety of different conformations and stabilized only upon subunit joining. In addition, the observed change in large subunit conformation upon MBP-Nmd3 binding indicates that an induced-fit mechanism might be at work.
A thread of density at the P-protein stalk base side of the MBP-Nmd3 density is visible (Fig. 2 C, asterisk). It is likely that this part of the density does not belong to MBP-Nmd3 but rather represents a conformational change in this region of the ribosome caused by ligand binding. Apparently, the nearest neighbors of MBP-Nmd3 among the large subunit proteins are Rpl23 (L14 family), Rpl9 (L6), Rpl12 (L11), and Rpl10 (L10e; Fig. 4 B).
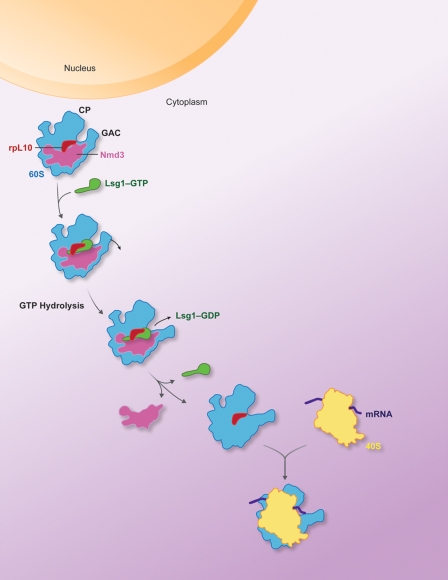
The cryo-EM map presented in this study is of Nmd3 (fused with MBP) in complex with a mature 60S subunit. However, during ribosome assembly in eukaryotes, Nmd3 initially binds to pre-60S particles in the nucleus to direct their export to the cytoplasm (Ho et al., 2000b; Gadal et al., 2001). After export, the pre-60S particle undergoes a series of maturation steps involving the release of trans-acting factors and the assembly of certain ribosomal proteins (Panse and Johnson, 2010) and culminates in the release of Nmd3 (Lo et al., 2010). The release of Nmd3 depends on the presence of ribosomal protein Rpl10 and the activity of the GTPase Lsg1 (Hedges et al., 2006). Thus, at the time of Nmd3 release, the subunit is presumably mature. The complex that we have reconstituted, which contains Rpl10 (unpublished data), may represent a late intermediate of 60S maturation, after Rpl10 loading but before Nmd3 release. We have also previously suggested that both Nmd3 and Lsg1 can bind to mature subunits as well as nascent subunits (Ho et al., 2000a). Considering that Nmd3 appears to be the last factor released from the nascent subunit, its binding to mature subunits could simply be a reversal of this step, possibly as a means of inhibiting 60S function under certain conditions, for example during stress response.
With the binding of Nmd3, the interface of the 60S subunit containing Rpl10 is apparently pulled toward the ligand. The resulting conformational change of the 60S subunit may reflect strain induced by Nmd3 binding. Although there is no direct interaction between the isolated mass and the 60S subunit at the Rpl10 region visible at this resolution of the cryo-EM map, the morphological features at the Rpl10-binding site and H38 (A-site finger) regions in the 60S subunit appear different in the complex as compared with the control 60S subunit map (Fig. 2 A). However, the resolution of the maps does not allow us to model this protein accurately inside the density. We speculate that the conformation of the Rpl10-binding site changes upon GTP hydrolysis on Lsg1, allowing the 60S subunit to relax into the more open position. This relaxation may facilitate proper accommodation of Rpl10 and the coordinated release of Nmd3 (Fig. 5).
Figure 5.
Schematic representation of the proposed mechanism for Nmd3 release. The Nmd3-bound 60S subunit in the cytoplasm presents a different conformation compared with the unbound 60S subunit, characterized as the result of a closing motion, as if gripping the ligand Nmd3. The usual multiple binding sites of translational GTPase factors, EF1A, eEF2, eRF3, are in the GAC region, as identified in eukaryotic and prokaryotic systems. In the present 60S complex, this region is partially blocked by the ligand. Thus, the cleft region between CP and GAC, close to the protein Rpl10 region, is a likely binding region for Lsg1. Conformational changes in Lsg1 as well as in the Rpl10-binding cleft, which accompany GTP hydrolysis, allow the 60S subunit to adopt a relaxed conformation and thus facilitate Nmd3 release. Next, the unbound 60S subunit is ready for association with the 40S initiation complex.
Materials and methods
Protein purification
GST-(TEV)-Nmd3.
2 liters of strain BJ5464 (MATα ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1) with pAJ235 (GAL1::GST-Nmd3; Ho et al., 2000a) was grown in selective medium containing 1% raffinose to OD600 of ∼0.3. Expression was induced with 1% galactose for 6 h. Cells were harvested, washed, and resuspended in two volumes of lysis buffer: 20 mM Tris, pH 7.5, 500 mM KCl, 0.5% Triton X-100, 10% glycerol, 1 mM DTT, and 1 mM EDTA plus protease inhibitors (PIs; 1 mM PMSF, 1 µM leupeptin, and 1 µM pepstatin). Cells were disrupted by vortexing with glass beads. The crude extract was clarified by centrifugation two times for 10 min at 15,000 g at 4°C. Clarified extract was incubated for 2 h at 4°C with 750 µl of glutathione beads. The beads were washed with 20 mM Tris, pH 7.5, 500 mM KCl, and 1 mM EDTA plus PIs, and protein was eluted with 50 mM glutathione in 50 mM Tris, pH 8. Concentration was determined by Bradford assay to be 8.5 µM.
MBP-(TEV)-HIS6-Nmd3.
4 liters of BJ5464 with pAJ1381 (GPD::MBP-HIS-TEV-NMD3 LEU2 2 micron; this study) was grown to OD600 of 0.6 in selective medium containing 2% glucose. Cells were harvested, washed, and resuspended in two volumes of extract buffer: 50 mM Tris, pH 8, 450 mM NaCl, 100 mM KCl, and 10% glycerol plus PIs. Cells were disrupted and clarified as described for GST-(TEV)-Nmd3. Imidazole and NP-40 were added to 10 mM and 0.01%, respectively. 1.5 ml of Ni–nitrilotriacetic acid beads was added, and the sample was rocked for 2 h at 4°C. The beads were washed three times with extract buffer, and the protein was eluted with 1.5 ml of extract buffer supplemented with 250 mM imidazole. The eluate was then incubated with 1.5 ml of amylose resin for 2 h at 4°C. The beads were washed with wash buffer: 20 mM Tris, pH 7.5, 50 mM NaCl, and 10% glycerol plus PIs. Proteins were eluted with wash buffer supplemented with 50 mM maltose. Concentration was determined by Bradford assay to be 0.75 µM.
Ribosomes.
3 liters of BJ5464 (for wild-type 60S) and AJY2757 (MATα ade2 leu2 ura3 his3 trp1 rpl25Δ::HIS3 containing pAJ909: Rpl25-13xmyc URA3 CEN; this work; for 60S-Rpl25-myc) were grown in yeast extract–peptone–dextrose to OD600 of 0.8. Cells were washed with binding buffer (50 mM KCl, 20 mM Tris, pH 7.4, 10 mM MgCl2, and 6 mM β-mercaptoethanol plus PIs), resuspended in 12 ml of binding buffer, lysed by vortexing with glass beads, and clarified twice as described for GST-(TEV)-Nmd3. The crude extract was layered over 2.5-ml 1 M sucrose cushions in binding buffer and centrifuged for 2 h at 32,000 rpm in an SW40 rotor (Beckman Coulter). The pellet was resuspended by stirring in 2.5 ml of binding buffer. Insoluble material was removed by centrifugation, and the soluble fraction was layered onto four 250-µl 1 M sucrose cushions in binding buffer and centrifuged for 1 h at 80,000 rpm in a TLA 100.3 rotor (Beckman Coulter). The pellets were resuspended in 1 ml of dissociation buffer: 20 mM Tris, pH 7.4, 0.5 M KCl, 8 mM MgCl2, and 6 mM b-mercaptoethanol plus PIs incubated 5 h at 4°C to dissociate ribosomes. The sample was then centrifuged through 5–20% sucrose gradients in dissociation buffer for 11 h at 23,000 rpm in an SW28 rotor (Beckman Coulter). Fractions containing the 60S and 40S peaks were pooled separately and concentrated using an Amicon Ultra 100K (Millipore), and buffer was changed to binding buffer. Final concentrations were [60S] = 0.14 µM, [40S] = 0.23 µM, and [60S-Rpl25-myc] = 0.46 µM.
Binding of MBP-Nmd3 and GST-Nmd3 to 60S and 80S
Cushion assay.
60S was incubated with or without 40S in binding buffer and sedimented through a 7–47% sucrose gradient. 60S and 80S ribosomes were recovered, MBP-Nmd3 was added, and binding was allowed to proceed for 1 h at 4°C. The reaction was layered on top of a 700-µl 60% sucrose cushion in binding buffer and centrifuged 60 min at 60,000 rpm in a TLA 100.3 rotor. The supernatant was carefully removed and precipitated with trichloroacetic acid. All samples were resuspended in equal volumes of SDS-PAGE sample buffer, separated by SDS-PAGE, and stained with Coomassie blue.
Immunoprecipitation assay.
60S-Rpl25-myc subunits were incubated with increasing amounts of MBP-Nmd3 in binding buffer at 4°C for 1 h. Subunits were immunoprecipitated by the addition of 9e10 monoclonal anti-myc antibody and protein A beads. Bound proteins were separated by SDS-PAGE and stained with Coomassie blue.
RNaseV1 protection assays
2.1 pmol of 60S was incubated with and without 3.75 pmol MBP-Nmd3 or 25.5 pmol GST-Nmd3 in 50 µl final volume of binding buffer for 1 h at 4°C with rocking. The reactions were treated with 0.05 U RNaseV1 (Applied Biosystems) for 20 min at 16°C. Samples were extracted with acid phenol/CHCl3, precipitated with ethanol, and resuspended in 12.5 µl H2O. Primer extension using 32P-labeled primers was performed as described previously (Beltrame and Tollervey, 1995), and products were separated on a 7-M urea polyacrylamide gel. Sequencing reactions were performed with Sequenase version 2.0 (USB) using pJD211.LEU as the template. Oligonucleotides used were AJO501, 5′-ACTGGGCAGAAATCACAT-3′; AJO1060, 5′-GTAGATAGGGACAGTGGGAA-3′; AJO1061, 5′-GTTCTGCTTACCAAAAATGG-3′; and AJO1135, 5′-AGAGCCATAATCCAGCGG-3′.
Cryo-EM and image reconstruction
Purified 60S subunits at 32 nM in binding buffer were applied to cryo-EM grids at 4°C according to a standard procedure (Wagenknecht et al., 1988) but using the Vitrobot (FEI). The images of control 60S ribosomal subunit were recorded at −180°C on a field emission gun electron microscope (Tecnai F20; FEI) at 200 kV on SO163 film (Kodak) and a magnification of 50,000 according to standard low-dose procedures and were digitized with a step size of 14 µm on an Imaging Scanner (Carl Zeiss, Inc.). The final pixel size corresponded to 2.82 Å on the object scale. Micrographs for the MBP-Nmd3–bound 60S subunit were recorded at −180°C on SO163 film on a Tecnai F20 field emission gun electron microscope at 200 kV (numerical aperture of objective lenses was 70 µm, and a magnification of 50,760) and digitized with a step size of 7 µm on the Imaging Scanner. Image processing was performed with the final pixel size corresponding to 2.76 Å on the object scale.
The image processing using SPIDER (Frank et al., 1996) included a 3D projection alignment procedure with correction of the contrast transfer function and enhancement of the high-resolution Fourier amplitudes based on x-ray solution scattering data. When using all particles, the density in the map corresponding to the ligand (MBP-Nmd3) was low compared with that of the ribosome, indicating lower than 100% occupancy. Therefore, we used a supervised classification method (Valle et al., 2003) to identify a subpopulation with high occupancy of the ligand. For the final reconstruction according to supervised classification, 19,317 ribosome particles (∼60% of the total number of particles selected for the complex) were used to obtain a refined cryo-EM map of the MBP-Nmd3–bound 60S subunit (16.3 Å). The final resolution of the constant transfer function–corrected volume was estimated by the Fourier shell correlation criterion (Fig. S1) with a cutoff value of 0.5 (Malhotra et al., 1998). The falloff of the Fourier amplitudes toward higher spatial frequencies was corrected as previously described (Gabashvili et al., 2000), using x-ray solution scattering data for the E. coli 70S ribosome.
Isolation of the ligand mass from the Nmd3-MBP–bound 60S subunit map was performed according to a two-step segmentation procedure. In brief, the 60S subunit was first masked out from the map of the complex. Next, selection of the mass of interest from the remaining masses was performed using a clustering procedure (http://www.wadsworth.org/spider_doc/spider/docs/techs/isolate.html). The software O (Jones et al., 1991) was used for manually docking the 60S quasiatomic model (Spahn et al., 2001) as a rigid piece into the cryo-EM 60S map. Amira (Visage Imaging, Inc.) and PyMOL (DeLano Scientific) were used for graphic visualizations.
Online supplemental material
Fig. S1 shows the Fourier shell correlation curve for the cryo-EM reconstruction of the MBP-Nmd3–60S subunit complex. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201001124/DC1.
Acknowledgments
We thank Richard Gursky and Xing Meng for their assistance with the EM and both Greg Allen and Lila Iino-Rubenstein with assistance in the preparation of figures.
This work was supported by National Institutes of Health (NIH) grant GM 53655 to A.W. Johnson and by the Howard Hughes Medical Institute and NIH grants R37 GM 29169 and R01 GM 55440 to J. Frank.
Footnotes
Abbreviations used in this paper:
- CP
- central protuberance
- GAC
- GTPase-associated center
- MBP
- maltose-binding protein
- PI
- protease inhibitor
- rRNA
- ribosomal RNA
- SRL
- sarcin–ricin loop
References
- Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 289:905–920 10.1126/science.289.5481.905 [DOI] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 14:4350–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B., Katahira J., Kowalinski E., Bange G., Yao W., Sekimoto T., Baumgärtel V., Boese G., Bassler J., Wild K., et al. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 27:767–779 10.1016/j.molcel.2007.06.034 [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190–199 10.1006/jsbi.1996.0030 [DOI] [PubMed] [Google Scholar]
- Fried H., Kutay U. 2003. Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60:1659–1688 10.1007/s00018-003-3070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Senger B., Saveanu C., Fasiolo F. 2003. Ribosome assembly in eukaryotes. Gene. 313:17–42 10.1016/S0378-1119(03)00629-2 [DOI] [PubMed] [Google Scholar]
- Gabashvili I.S., Agrawal R.K., Spahn C.M., Grassucci R.A., Svergun D.I., Frank J., Penczek P. 2000. Solution structure of the E. coli 70S ribosome at 11.5 A resolution. Cell. 100:537–549 10.1016/S0092-8674(00)80690-X [DOI] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405–3415 10.1128/MCB.21.10.3405-3415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage A.E., Tollervey D. 2004. A surfeit of factors: why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 1:10–15 [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A.W. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567–579 10.1038/sj.emboj.7600547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J., Chen Y.I., West M., Bussiere C., Johnson A.W. 2006. Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J. Biol. Chem. 281:36579–36587 10.1074/jbc.M606798200 [DOI] [PubMed] [Google Scholar]
- Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 65:2334–2359 10.1007/s00018-008-8027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Johnson A.W. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom G., Johnson A.W. 2000a. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 6:1625–1634 10.1017/S1355838200001291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom G., Johnson A.W. 2000b. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057–1066 10.1083/jcb.151.5.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung N.J., Lo K.Y., Patel S.S., Helmke K., Johnson A.W. 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 19:735–744 10.1091/mbc.E07-09-0968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.W. 2009. Ribosomal export. Nuclear Transport. Kehlenbach R.H., editor Landes Bioscience, Austin, TX: 69–84 [Google Scholar]
- Jones T.A., Zou J.Y., Cowan S.W., Kjeldgaard M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47:110–119 10.1107/S0108767390010224 [DOI] [PubMed] [Google Scholar]
- Klein D.J., Moore P.B., Steitz T.A. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340:141–177 10.1016/j.jmb.2004.03.076 [DOI] [PubMed] [Google Scholar]
- Köhler A., Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8:761–773 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Lo K.Y., Li Z., Bussiere C., Bresson S., Marcotte E.M., Johnson A.W. 2010. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol. Cell. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A., Penczek P., Agrawal R.K., Gabashvili I.S., Grassucci R.A., Jünemann R., Burkhardt N., Nierhaus K.H., Frank J. 1998. Escherichia coli 70 S ribosome at 15 A resolution by cryo-electron microscopy: localization of fMet-tRNAfMet and fitting of L1 protein. J. Mol. Biol. 280:103–116 10.1006/jmbi.1998.1859 [DOI] [PubMed] [Google Scholar]
- Panse V.G., Johnson A.W. 2010. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35:260–266 10.1016/j.tibs.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Paschal B.M. 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 6:187–198 10.1111/j.1600-0854.2005.00270.x [DOI] [PubMed] [Google Scholar]
- Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. 2005. Structures of the bacterial ribosome at 3.5 A resolution. Science. 310:827–834 10.1126/science.1117230 [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 107:373–386 10.1016/S0092-8674(01)00539-6 [DOI] [PubMed] [Google Scholar]
- Thomas F., Kutay U. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116:2409–2419 10.1242/jcs.00464 [DOI] [PubMed] [Google Scholar]
- Tran E.J., Bolger T.A., Wente S.R. 2007. SnapShot: nuclear transport. Cell. 131:420 10.1016/j.cell.2007.10.015 [DOI] [PubMed] [Google Scholar]
- Trotta C.R., Lund E., Kahan L., Johnson A.W., Dahlberg J.E. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22:2841–2851 10.1093/emboj/cdg249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. 2003. Locking and unlocking of ribosomal motions. Cell. 114:123–134 10.1016/S0092-8674(03)00476-8 [DOI] [PubMed] [Google Scholar]
- Wagenknecht T., Grassucci R., Frank J. 1988. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol. 199:137–147 10.1016/0022-2836(88)90384-1 [DOI] [PubMed] [Google Scholar]
- West M., Hedges J.B., Chen A., Johnson A.W. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 25:3802–3813 10.1128/MCB.25.9.3802-3813.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W., Roser D., Köhler A., Bradatsch B., Bassler J., Hurt E. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell. 26:51–62 10.1016/j.molcel.2007.02.018 [DOI] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science. 292:883–896 10.1126/science.1060089 [DOI] [PubMed] [Google Scholar]
- Zemp I., Kutay U. 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581:2783–2793 10.1016/j.febslet.2007.05.013 [DOI] [PubMed] [Google Scholar]