Abstract
The adaptor protein Nmd3 is required for Crm1-dependent export of large ribosomal subunits from the nucleus. In this issue, Sengupta et al. (2010. J. Cell Biol. doi:10.1083/jcb.201001124) identify a binding site for yeast Nmd3 on 60S ribosomal subunits using cryoelectron microscopy and suggest a conformational model for its release in the cytoplasm. The study provides the first detailed structural description of a ribosome biogenesis factor in complex with the large subunit.
In eukaryotic cells, the production of ribosomes depends on a highly dynamic multistep process that takes place sequentially: first in the nucleolus, then in the nucleoplasm, and finally in the cytoplasm. During this process, ribosomal RNA (rRNA) is transcribed, modified, processed, and assembled with ribosomal proteins into ribosomal subunits with the concomitant participation of ∼200 trans-acting factors that ensure the correct maturation of ribosomes (Staley and Woolford, 2009). Early on, in the nucleolus, the maturation pathways of the large (60S) and small (40S) ribosomal subunits diverge, and each is assembled and exported separately from the nucleus (Staley and Woolford, 2009). Although many steps of the process and factors involved are known, most mechanistic details of ribosome biogenesis and the binding and action of trans-acting factors are still elusive. In this issue, Sengupta et al. use cryo-EM to provide structural insights into a binding site for the nuclear export adaptor Nmd3 on the large ribosomal subunit. This work represents an important step in filling the gap in our understanding of how preribosome structure and the positioning of trans-acting factors therein direct ribosome maturation.
Nmd3 is a highly conserved adaptor protein that is required for the export of 60S ribosomal subunits from yeast to humans (Ho and Johnson, 1999; Thomas and Kutay, 2003; Trotta et al., 2003). Although predominantly cytoplasmic, Nmd3p shuttles between the nucleus and the cytoplasm and facilitates 60S subunit export by providing a nuclear export signal that recruits the export receptor Crm1 in the nucleus (Ho et al., 2000; Gadal et al., 2001). Nmd3 binds in vivo to free 60S subunits but not 40S subunits or 80S ribosomes, indicating that it is released before subunit joining and translation initiation (Ho and Johnson, 1999; Ho et al., 2000). Recent studies have demonstrated that the GTPase Lsg1 is required for the removal of Nmd3 from 60S subunits as part of the final maturation events in the cytoplasm (Hedges et al., 2005; Lo and Johnson, 2009). Moreover, additional data provided evidence that association of the ribosomal protein Rpl10L to cytoplasmic 60S subunits is a prerequisite of Nmd3 release (Hedges et al., 2005; West et al., 2005).
Although the molecular events of Nmd3 binding to 60S subunits have been studied extensively, its actual site of interaction on 60S subunits is still unknown. In the past, several studies have speculated on a potential binding site for Nmd3 based on its functional and physical interaction with Rpl10L (Karl et al., 1999; Gadal et al., 2001; Hedges et al., 2005; West et al., 2005). Because Rpl10L is positioned close to the intersubunit joining face, this region has also been suggested as a potential binding site for Nmd3 (Eisinger et al., 1997; Spahn et al., 2001). In their study, Sengupta et al. (2010) provide the first direct evidence for this idea. By performing cryo-EM on purified mature 60S subunits with and without a recombinantly expressed maltose-binding protein (MBP)–Nmd3 fusion protein, the authors identify the site of MBP-Nmd3 binding. By comparing cryo-EM maps obtained by single particle reconstruction of mature control 60S subunits to those that contained MBP-Nmd3, they are able to identify an additional density on Nmd3-bound subunits covering the intersubunit region that is close in mass to their recombinantly expressed Nmd3 fusion protein. In addition, the authors observe conformational changes in the Nmd3-bound 60S subunits relative to the control 60S subunits in several surrounding regions, including in the GTPase-associated center, in the region around the central protuberance and peptidyl-transferase center, and at the base of the L1 stalk; in these areas, the authors observe a conformational switch toward Nmd3 in Nmd3-bound subunits, suggesting that binding of Nmd3 leads to a tighter conformation and thus less accessibility across the intersubunit region.
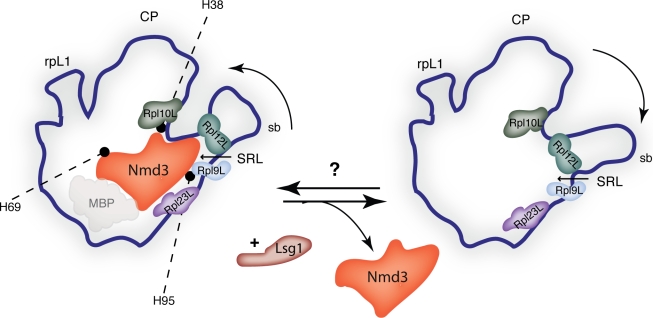
To confirm as well as tentatively narrow down the observed binding region by biochemical means, the authors go on to probe for altered sensitivity of 60S subunits to RNaseV1 in the presence and absence of MBP-Nmd3 as well as a second fusion protein, GST-Nmd3. Based on observed protection by either both or only one fusion protein, they suggest that Nmd3 binds to helices 38, 69, and 95 of the 25S rRNA and propose a binding site for Nmd3 closer to the sarcin–ricin loop and the central protuberance than the base of the L1 stalk (Fig. 1). To determine further details between MBP-Nmd3 and the large subunit, Sengupta et al. (2010) align their cryo-EM map with a quasiatomic model of the 60S subunit (Spahn et al., 2001). The overlay not only supports the results obtained in their RNA protection assay but also identifies the nearest large ribosomal subunit proteins: Rpl23, Rpl9, Rpl12, and Rpl10 (Fig. 1). Interestingly, although the interface containing Rpl10L was pulled toward Nmd3 upon binding of the latter, the authors did not observe a direct interaction between Ndm3 and Rpl10L. However, based on the morphological features of the Nmd3-bound RNA helices (H38 and H95) in the presence and absence of Nmd3 and their proximity to the binding site of Rpl10L, they propose a model for Nmd3 release, suggesting that the relaxation of these helices upon initial Rpl10L binding and GTP hydrolysis by Lsg1 may accommodate correct binding of Rpl10L while at the same time facilitating the release of Nmd3.
Figure 1.
The 60S ribosome subunit with and without Nmd3. A schematic of the 60S subunit is shown from the intersubunit side, in the classical crown view. Nmd3 bound to a mature 60S subunit (left). The opaque area below bound Nmd3 denotes an MBP-binding area as described in Sengupta et al. (2010). The nearest identified large ribosomal proteins (Rpl9L, Rpl10L, Rpl12L, and Rpl23L) are shown, as well as the RNA helices bound by Nmd3, as determined by RNase protection assay in Sengupta et al. (2010). Release of Nmd3 is mediated by the GTPase Lsg1 (Hedges et al., 2005) and leads to a change in orientation of the stalk base (sb; right). The mechanism of Nmd3 recruitment to 60S subunits is unknown. CP, central protuberance; SRL, sarcin–ricin loop.
This study by Sengupta et al. (2010) represents the first direct demonstration of the position of Nmd3 on the intersubunit side of the 60S particle, which is consistent with its lack of association with 80S ribosomes, and adds one item to the still very short list of preribosomal assembly factors whose position within ribosomal particles has been identified. Although we have to keep in mind that the authors are looking at the binding of Nmd3 to mature 60S subunits that already contain Rpl10L, it is very likely that the identified binding site for Nmd3 is the same in pre-60S subunits. It remains to be seen whether different conformational changes take place upon Nmd3 binding to nucleolar/nuclear pre-60S subunits.
There are several interesting questions regarding Nmd3 that remain. What is its role at the subunit joining interface: is it perhaps monitoring correct folding and/or assembly at this crucial site, or is it simply a placeholder for Rpl10L? What is Nmd3’s function on mature 60S subunits? And finally, if not Rpl10L, who or what recruits Nmd3 to pre-60S subunits? Although this study represents an important step forward in understanding the integration of structural and functional mechanisms during ribosome maturation, more such data are needed. Several different methods are currently being used to identify the positioning of different trans-acting factors within ribosomal subunits, and in the future, we should be able to combine that information with our current knowledge of ribosome maturation to gain a truly comprehensive picture of the dynamic organization of the ribosome biogenesis pathway (Tang et al., 2008; Ulbrich et al., 2009; Granneman et al., 2010).
Acknowledgments
M. Oeffinger is funded by a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (NSERC-386315-2010).
References
- Eisinger D.P., Dick F.A., Trumpower B.L. 1997. Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol. Cell. Biol. 17:5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Strauss D., Kessl J., Trumpower B., Tollervey D., Hurt E. 2001. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 21:3405–3415 10.1128/MCB.21.10.3405-3415.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S., Petfalski E., Swiatkowska A., Tollervey D. 2010. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J. 10.1038/emboj.2010.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J., West M., Johnson A.W. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24:567–579 10.1038/sj.emboj.7600547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Johnson A.W. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.H., Kallstrom G., Johnson A.W. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057–1066 10.1083/jcb.151.5.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl T., Onder K., Kodzius R., Pichová A., Wimmer H., Th r A., Hundsberger H., Löffler M., Klade T., Beyer A., et al. 1999. GRC5 and NMD3 function in translational control of gene expression and interact genetically. Curr. Genet. 34:419–429 10.1007/s002940050416 [DOI] [PubMed] [Google Scholar]
- Lo K.Y., Johnson A.W. 2009. Reengineering ribosome export. Mol. Biol. Cell. 20:1545–1554 10.1091/mbc.E08-10-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J., Bussiere C., Pallesen J., West M., Johnson A.W., Frank J. 2010. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J. Cell Biol. 189:1079–1086 10.1083/jcb.201001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 107:373–386 10.1016/S0092-8674(01)00539-6 [DOI] [PubMed] [Google Scholar]
- Staley J.P., Woolford J.L., Jr 2009. Assembly of ribosomes and spliceosomes: complex ribonucleoprotein machines. Curr. Opin. Cell Biol. 21:109–118 10.1016/j.ceb.2009.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Sahasranaman A., Jakovljevic J., Schleifman E., Woolford J.L., Jr 2008. Interactions among Ytm1, Erb1, and Nop7 required for assembly of the Nop7-subcomplex in yeast preribosomes. Mol. Biol. Cell. 19:2844–2856 10.1091/mbc.E07-12-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas F., Kutay U. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116:2409–2419 10.1242/jcs.00464 [DOI] [PubMed] [Google Scholar]
- Trotta C.R., Lund E., Kahan L., Johnson A.W., Dahlberg J.E. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22:2841–2851 10.1093/emboj/cdg249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich C., Diepholz M., Bassler J., Kressler D., Pertschy B., Galani K., Böttcher B., Hurt E. 2009. Mechanochemical removal of ribosome biogenesis factors from nascent 60S ribosomal subunits. Cell. 138:911–922 10.1016/j.cell.2009.06.045 [DOI] [PubMed] [Google Scholar]
- West M., Hedges J.B., Chen A., Johnson A.W. 2005. Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol. Cell. Biol. 25:3802–3813 10.1128/MCB.25.9.3802-3813.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]