Abstract
The heart adapts to an increased workload through the activation of a hypertrophic response within the cardiac ventricles. This response is characterized by both an increase in the size of the individual cardiomyocytes and an induction of a panel of genes normally expressed in the embryonic and neonatal ventricle, such as atrial natriuretic peptide (ANP). ANP and brain natriuretic peptide (BNP) exert their biological actions through activation of the natriuretic peptide receptor-1 (Npr1). The current study examined mice lacking Npr1 (Npr1−/−) activity and investigated the effects of the absence of Npr1 signaling during cardiac development on embryo viability, cardiac structure and gene and protein expression. Npr1−/−embryos were collected at embryonic day (ED) 12.5, 15.5 and neonatal day 1 (ND 1). Npr1−/−embryos occurred at the expected Mendelian frequency at ED 12.5, but knockout numbers were significantly decreased at ED 15.5 and ND 1. There was no indication of cardiac structural abnormalities in surviving embryos. However, Npr1−/−embryos exhibited cardiac enlargement (without fibrosis) from ED 15.5 as well as significantly increased ANP mRNA and protein expression compared to wild-type (WT) mice, but no concomitant increase in expression of the hypertrophy-related transcription factors, Mef2A, Mef2C, GATA-4, GATA-6 or serum response factor (SRF). However, there was a significant decrease in Connexin-43 (Cx43) gene and protein expression at mid-gestation in Npr1−/−embryos. Our findings suggest that the mechanism by which natriuretic peptide signaling influences cardiac development in Npr1−/− mice is distinct from that seen during the development of pathological cardiac hypertrophy and fibrosis. The decreased viability of Npr1−/−embryos may result from a combination of cardiomegaly and dysregulated Cx43 protein affecting cardiac contractility.
Keywords: Akt1, Atrial natriuretic peptide, Calcineurin A, Cardiac hypertrophy, Connexin 43, Gene expression, Heart development, Npr1, Transcription factor
1. Introduction
Atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP) regulate blood pressure by vasodilation, by increasing vascular permeability resulting in a net movement of fluid out of the vascular compartment and by modulating the rate of diuresis and natriuresis through the kidneys [1,2]. During development the natriuretic peptide system is functional by mid-gestation, and is capable of responding to volume stimuli, regulating blood pressure and salt and water balance in the developing embryo [3,4]. Circulating levels of ANP have been shown to be higher in the fetus than in adults, and fetal ventricles express higher levels of both ANP and BNP than adult ventricles [5,6]. Both ANP and BNP are expressed within the myocardium from ED 8.0 and are two of the first markers of heart formation [7,8]. After birth, ANP is down regulated in the ventricles, and in the adult expression in the atria is 1000-fold higher than in the ventricles. Re-activated ventricular ANP expression occurs in response to cardiac stress and serves as the most highly conserved marker of hypertrophy [9]. In association with the reappearance of ANP expression in the adult ventricle, cardiac contractile protein genes, normally only active during cardiac development, are also re-expressed leading to the identification of the embryonic gene program characteristic of pathological ventricular hypertrophy [10,11].
Transgenic mouse models lacking the natriuretic peptide receptor (NPR)-A gene (Npr1−/−) have been widely used to illustrate the essential role of natriuretic peptide signaling in suppressing the development of cardiac hypertrophy and fibrosis on cardiac function and survival [12–16]. These studies have focused on the effect of disrupted natriuretic peptide signalling in adult animals only. Further investigations utilizing these and other animal models have characterized the molecular mechanisms by which natriuretic peptide signaling mediates the hypertrophic response in a variety of pathological states [16–19].
Furthermore, fetal models of cardiovascular pathophysiology suggest the natriuretic peptide system in the fetal heart and circulation may regulate both physiological fluid balance and cellular development within cardiomyocytes [20]. Both immune and non-immune fetal hydrops are associated with higher than normal fetal concentrations of circulating ANP [21], while fetal cardiac distress is associated with increased BNP expression reminiscent of heart failure in the adult [22]. Despite this, few studies have investigated natriuretic peptide signaling during the process of heart development itself.
A large number of studies have identified the critical role of natriuretic peptide signaling in the development of pathological hypertrophy in adults. In this study we tested the hypothesis that ablation of Npr1 signaling during cardiac development would result in the early establishment of a hypertrophic phenotype. The aim of this study was to determine if gene and protein expression during cardiac development in Npr1−/− mice was characteristic of pathological or physiological cardiac hypertrophy.
2. Materials and methods
2.1. Generation and genotyping of mice
Npr1−/− mice and WT controls were generated as previously described [12]. Experiments were performed according to the protocols approved by the Animal Ethics Committee of the University of Otago.
Male and female Npr1−/− and WT embryos from timed matings between Npr1+/− dams and Npr1−/− sires and WT dams and sires were collected at ED 12.5 and 15.5 and neonatal pups collected at ND 1. Npr1−/− dams were not used for breeding in this study due to reduced conception rates, poor maternal behaviour and increased rates of infanticide in the few successful pregnancies observed. Npr1−/− embryos and ND 1 pups from these matings were not used in the current study. Embryos were collected after sacrificing the pregnant dams by cervical dislocation following Halothane BP™ (Nicolas Piramal I Ltd, London) anesthesia. Embryos were rapidly dissected out of the uterine horns and separated from the placenta in ice-cold, RNase-free, phosphate buffered saline solution. Embryos were digitally photographed against a grid and ruler to assess body dimensions. The embryonic left hind limb was removed for DNA extraction and the embryos immersion-fixed in 10% neutral buffered formalin. ND 1 pups were sacrificed by rapid decapitation, tails removed for DNA extraction and whole bodies immersion-fixed as above. Genomic DNA was extracted via a standard phenol–chloroform extraction protocol from embryonic left hind limbs or tail-tips of ND 1 mice and genotyped as previously described [12,23].
Systolic and diastolic blood pressures in conscious dams were measured by a non-invasive, computerized tail–cuff system (ADInstruments, Dunedin, New Zealand) as previously described [23]. The mice were familiarized to the procedure by being placed in the restrainer and the tail cuff system for a period of 7 days, after which baseline mean arterial pressure (MAP) measurements were made for each animal (mean of at least ten recordings), and repeated at 10 and 15 days post coitum (dpc) or until sacrifice.
2.2. Gender determination of collected embryos
To determine the gender of embryos a PCR-based assay was established utilizing a variant of the structural maintenance of chromosomes (SMC) gene located on the X-chromosome (accession number AC083816), which has a Y-chromosome homolog containing a 30-bp intronic deletion. The region of the gene surrounding the deletion site was amplified, using the following primers (forward 5′ CCGCTGCCAAATTCTTTGG and reverse 5′CTCAAAAGCCAAAAGCTTCA). Amplification of the X-chromosomal SMC variant produced a 330-bp fragment and the Y-chromosomal variant, produced a 300-bp fragment visualized by gel electrophoresis.
2.3. Histological characterization of Npr1−/− and WT embryos
Before being sectioned, embryos and ND 1 pups were either transferred to 10% neutral-buffered formalin containing 10% sucrose as a cryoprotectant for 24 h and then embedded in OCT medium (Miles; Elkhart, IN, n=6 per gender and genotype at each gestational age), or paraffin embedded (n=6 per gender and genotype at each gestational age). An equal number of embryos and neonates per group were embedded such that the heart could be sectioned in either the transverse or sagittal plane. The use of two anatomical planes ensured that cardiac morphology could be thoroughly examined, especially the locations of the great vessels and the structures of the interventricular and interatrial septa. Histological analysis was performed on 20-µm-thick serial cryostat sections and 3-µm-thick serial paraffin sections (Leica Microsystems), stained with either hematoxylin and eosin (H&E) or Masson's trichrome and examined under bright-field illumination (Olympus BX50). Composite photo images of the entire heart structure were compiled using Photoshop Elements 4.0 (Adobe Systems Inc.). Heart size for each embryo, as well as individual chamber dimensions, ventricular septal length and width, and free wall thickness in each chamber were determined on transverse sections under light microscopy with an eyepiece graticule (Olympus Model WH10×3) using a modified Cavalieri method [24]. ED 12.5 and 15.5 hearts were viewed at a total magnification of 100×, while ND 1 hearts were viewed at a total magnification of 40×. Additionally, the number of nuclei per photographic field was recorded within representative regions of the free wall of the mid left ventricle (LV) at 400× magnification. In all cases the mean of five independent sections per specimen was used for statistical analysis.
2.4. RNA isolation and quantitative real-time PCR analysis
Total RNA was isolated from whole ED 12.5 and 15.5 embryos and excised hearts (atria and ventricles combined) of ND 1 as described previously (n=6 per gender and genotype at each gestational age) [23]. Briefly tissues were homogenized in a Retsch MM301 tissue mill at 30 Hz for 10 min in 800 µl pre-chilled TRIzol™ (Invitrogen, Carlsbad, CA). Chloroform (160 µl) was added and samples were centrifuged at 12,000 g for 15 min at room temperature. The RNA-containing supernatant was purified by RNeasy Midi and Mini columns according to manufacturer's instructions (Qiagen, Hilden, Germany).
Quantitative real-time PCR (RT-PCR) analysis was performed on cDNA generated from 2.5 µg of total RNA as previously described [23]. Oligonucleotide primer sequences and PCR annealing temperatures are given in Table 1. PCR reactions were performed in a total volume of 30 µl containing 1 µl cDNA, 0.4 mM primers, 1× PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, 10 nM Sybr Green 1 (Roche, IN) and 1U TAQ-Ti DNA polymerase (Fisher Biotec, Australia). The PCR reaction conditions were optimized for each gene of interest, and PCR products were confirmed by sequencing on an ABI 3100-Avant Genetic Analyzer (Foster City, CA). Levels of mRNA expression were evaluated by quantitative RT-PCR in either a Rotor-Gene RG-3000 real-time PCR machine (Corbett Research, Australia) or a Light Cycler 480 (Roche Diagnostics, IN) as previously described [23]. For each assay a hotstart at 96 °C was performed for 2 min followed by 30 cycles of denaturation at 94 °C for 30 s, annealing for 35 s at the gene-specific annealing temperature (Table 1) and extension at 72 °C for 30 s, after which a melt curve was performed. Each sample was assayed in duplicate and gene expression levels were quantified against a standard curve and expressed as amount of message per µg of total RNA.
Table 1.
Primer sequences and annealing temperatures for RT-PCR assays.
| mRNA | Primers | Annealing temperature |
|---|---|---|
| Angiotensin converting enzyme | fwd TGCCTCCCAAGGAATTAGAA | 59 °C |
| rev CCATGCCCATAGCAATTCTT | ||
| Angiotensinogen | fwd AAGACCTCCCCTGTGAATGA | 65 °C |
| rev CCTGCCTCATTCAGCATCTT | ||
| Angiotensin type 1 receptor | fwd GGAAACAGCTTGGTGGTGAT | 63 °C |
| rev ACATAGGTGATTGCCGAAGG | ||
| ANP | fwd GAACCTGCTAGACCACCT | 56 °C |
| rev CCTAGTCCACTCTGGGCT | ||
| BNP | fwd AAGCTGCTGGAGCTGATAAGA | 56 °C |
| rev GTTACAGCCCAAACGACTGAC | ||
| Connexin 43 | fwd AACAGTCTGCCTTTCGCTGT | 58 °C |
| rev GGGCACAGACACGAATATGA | ||
| GATA-4 | fwd AATGCCTGTGGCCTCTATCA | 58 °C |
| rev CTGGTTTGAATCCCCTCCTT | ||
| GATA-6 | fwd AAGATGAATGGCCTCAGCAG | 56 °C |
| rev CATATAGAGCCCGCAAGCAT | ||
| Mef2A | fwd ACTCAAGGGCCTCTCCAAAT | 54 °C |
| rev CTTGATGGGGGAATGACAAC | ||
| Mef2C | fwd ATTTGGGAACTGAGCTGTGC | 56 °C |
| rev CGCTCATCCATTATCCTCGT | ||
| Serum response factor | Fwd GAGATTCTGGGATTGCTCCA | 58 °C |
| rev CAACAACCCTAGAGGGCAAA |
Sequences are listed 5′ to 3′. Forward primers are designated by fwd and reverse primers by rev.
2.5. Immunohistochemistry
Npr1−/− and WT mice were collected at ED 12.5 and 15.5 and ND1 (n=6 per gender and genotype at each gestational age), fixed in 10% formalin overnight at 4 °C followed by ethanol dehydration and paraffin embedding. Immunohistochemistry was performed in duplicate using indirect immunoperoxidase staining (Envision®+ Dual Link System-HRP, DakoCytomation, CA), on 3 µm serial sections on poly-l-lysine coated slides.
Primary antibodies used for immunohistochemistry included, GATA-4, Mef2, SRF from Santa Cruz Biotechnology Inc. (Santa Cruz, CA) and Cx43 (Zymed laboratories, San Francisco, CA). ANP antiserum was a generous gift from Associate Professor Tim Yandle (Endolab, University of Otago-Christchurch). ANP antiserum was raised in New Zealand white rabbits injected and boosted at monthly intervals with rat ANP (3–28) conjugated to bovine thyroglobulin followed by boosting with human ANP (1–28) conjugated to bovine thyroglobulin. The ANP antiserum exhibited high cross-reactivity to human, rat and mouse ANP (>95%), but <5% cross-reactivity to other natriuretic peptides.
Briefly, immunohistochemistry involved deparaffinized sections being heat-treated for 2 min in citrate buffer (pH 6.0), quenching of endogenous peroxidase activity with 3% hydrogen peroxidase in distilled water (15 min at room temperature) and non-specific binding of IgG suppressed with 1% normal goat serum in phosphate buffered saline (15 min at room temperature). The sections were incubated with the primary antisera diluted to working concentrations of 1:300 for ANP, Cx43, Mef2 and GATA-4 and 1:500 for SRF at room temperature for 1 h then visualized using liquid DAB+ (DakoCytomation, CA). Sections were counter-stained with alum hematoxylin. Negative controls were obtained by omission of the primary antibody.
Representative antibody staining within the left ventricular free wall and left atria was photographed for semi-quantitative densitometry of DAB+ staining using light microscopy under bright-field illumination (Olympus BX50 microscope) and Leica digital camera, DFC420. Intensity of DAB+ staining was determined using Meta-Morph™ image analysis software (v6.2.6, Molecular Devices), as previously described [25,26].
2.6. Statistical analysis
All results are expressed as means±SEM. The effects of genotype (Npr1−/− vs. WT), gender and gestational age were tested by three-way factorial ANOVA. Subsequent comparisons between genotypes at specific ages were performed using independent t-tests. Bivariate relationships between expression levels for all genes and proteins were tested with Pearson's correlation. All statistical analyses were performed using SPSS version 11 (SPSS Inc., Chicago, IL). A p value of <0.05 was considered significant.
3. Results
3.1. Colony genotype distribution
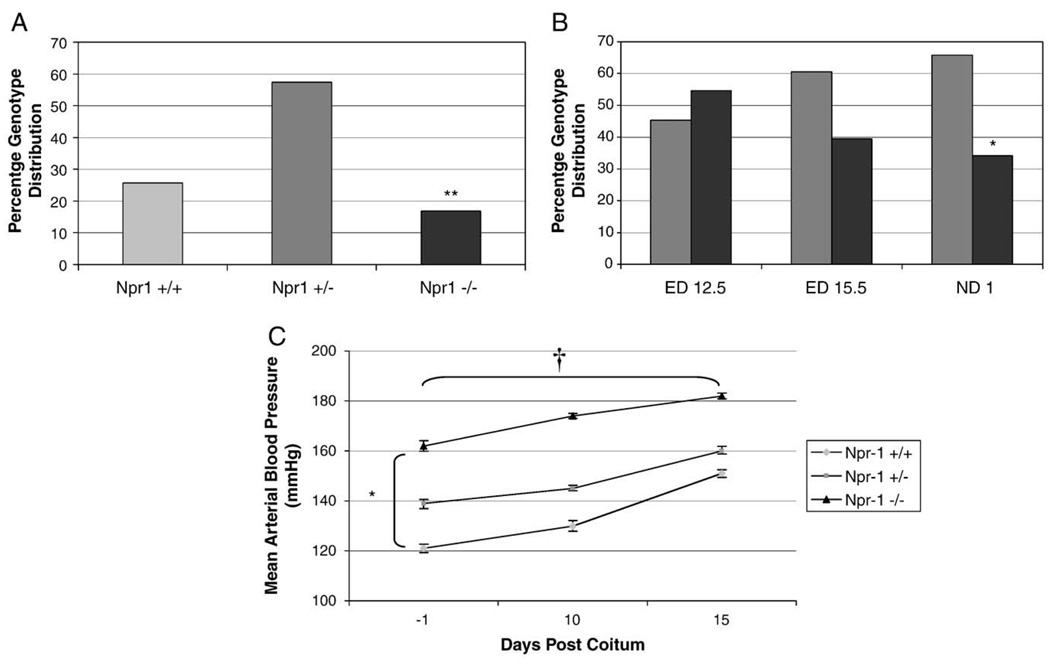
The genotype distributions at weaning (approximately 4 weeks after birth), identified that within this colony genotype distributions were non-Mendelian, with significantly fewer than expected Npr1−/− mice surviving to 4 weeks of age (Fig. 1A, 17% Npr1−/−, 58% Npr1+/− and 25% WT mice respectively, n = 250, p = 0.009).
Fig. 1.
Npr1 genotype distribution at weaning (A) and during gestation (B) and maternal blood pressure changes during gestation (C). Black, dark grey and light grey bars represent Npr1−/−Npr1+/− and WT groups respectively. Significant differences between the genotypes are indicated by *p<0.05, **p<0.01 and †p<0.001. Maternal blood pressures were calculated from n=8 per group, except for Npr1−/− where n = 4.
Litter sizes were comparable between WT and Npr1+/− dams collected at ED 12.5 (Npr1+/− 5.1±0.8 vs. WT 6.3±1.1), at ED 15.5 (Npr1+/− 5.9±0.5 vs. WT 6.2±0.7) and at ND 1 (Npr1+/− 5.3±0.5 vs. WT 5.6±0.6, n=332 from 59 litters). The effect of genotype on gestational survival, genotype distribution during embryogenesis was studied in embryos collected from Npr1+/−/Npr1−/− timed matings (Fig. 1B). At ED 12.5 the numbers of Npr1−/− embryos and Npr1+/− embryos were approximately equivalent, as expected from heterozygote-knockout matings (45% Npr1+/− and 55% Npr1−/−). By ED 15.5 the proportion of Npr1−/− embryos had declined to 39%. At ND 1 the number of Npr1−/− mice was further reduced, with only 34% of the collected embryos identified as Npr1−/−, instead of the expected 50% (n=130, p<0.05).
3.2. Maternal blood pressure during pregnancy
Blood pressure was monitored throughout pregnancy in Npr1−/−, Npr1+/− and WT dams. At baseline, mean arterial pressure (MAP, Fig. 1C) was significantly different between genotypes, with the average MAP of WT dams being 121 mm Hg compared to either Npr1+/− or Npr1−/− (139 and 162 mm Hg, respectively, p<0.05). The MAP in all three genotypes increased significantly with duration of pregnancy (p<0.001), but there were no differences in the extent to which blood pressure increased between genotypes. The observed initial difference in MAP due to genotype was maintained and remained significant throughout pregnancy (p<0.001).
3.3. Embryonic Npr1−/− and WT heart structure
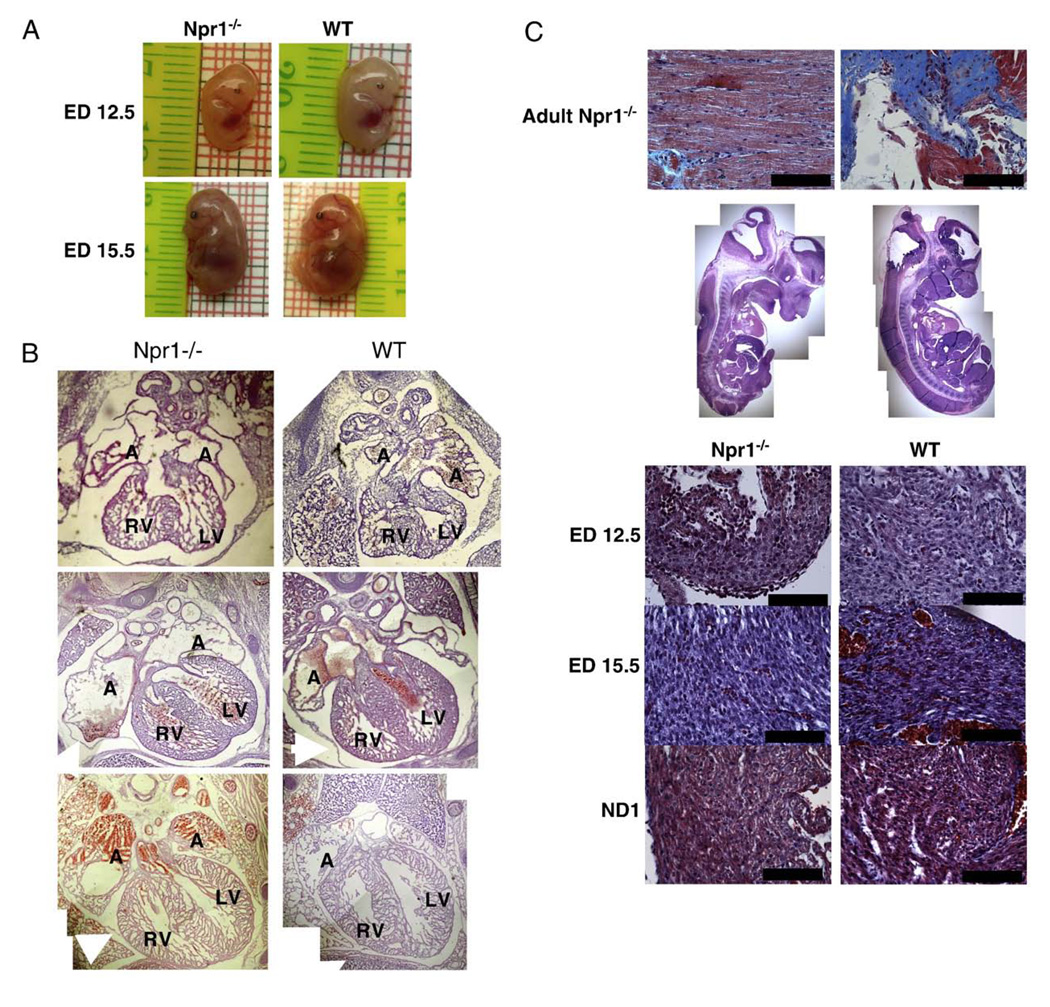
There were no physical abnormalities evident between Npr1−/− and WT embryos at any age (Fig. 2A and B.) No cardiac anatomical abnormalities were apparent in H&E stained serial sections of the thoracic regions of Npr1−/− compared with WT mice at any of the gestational ages studied (Fig. 2B). Masson's trichrome staining did not identify aberrant collagen deposition within the hearts of Npr1−/− mice at any of the gestational ages studied (Fig. 2C).
Fig. 2.
Representative digital photographs of male mice at EDs 12.5 and 15.5 (A), comparative composite H&E stained hearts of Npr1−/− and WT mice at ED 12.5 in the transverse plane, ED 15.5 and ND 1 hearts in the coronal plane (B), and Masson’s trichrome stained paraffin sections from the left ventricle (C). The top row images are comparative sections from a 6-month-old Npr1−/− mouse, on the left a section through the LV wall and on the right a positively stained cardiac valve. Row 2 displays composite photomerges of H&E stained sections from ED 12.5 Npr1−/− and WT mice. Rows 3–5 are representative Masson’s trichrome stained sections from ED 12.5, ED 15.5 and ND 1 mice respectively. Structures labeled: A=right atria; RV=right ventricle; LV=left ventricle; white arrow indicates the pericardium. Scale bar represents 100 µm.
Both ED 12.5 and 15.5 Npr1−/− mice tended to be lighter than WT embryos (Table 2), however, the difference was not significant. There was no overall difference in body weight at ND 1 between genotypes. As an alternative measure embryonic crown-rump lengths (in mm) were calculated from the digital photos taken at embryo collection (ED 12.5 Npr1−/− 9.33±0.39 vs. WT 9.7±0.12, ED 15.5 Npr1−/− 14.39±0.22 vs. WT 14.42±0.22), with no significant difference in embryo length at either gestational age (p=0.078 and 0.937 respectively).
Table 2.
Embryo and neonatal wet weights at collection.
| WT | Npr1−/− | p-value | |
|---|---|---|---|
| ED 12.5 | |||
| Males | 88.0±8 | 85.4±12 | 0.860 |
| Females | 88.5±12 | 70.9±17 | 0.421 |
| ED 15.5 | |||
| Males | 472.7±31 | 418.9±15 | 0.091 |
| Females | 387.8±9 | 432.2±15 | 0.060 |
| ND 1 | |||
| Males | 1447.5±63 | 1373.4±42 | 0.321 |
| Females | 1292.8±58 | 1419.8±36 | 0.067 |
All weights are recorded in mg and presented as mean±SEM.
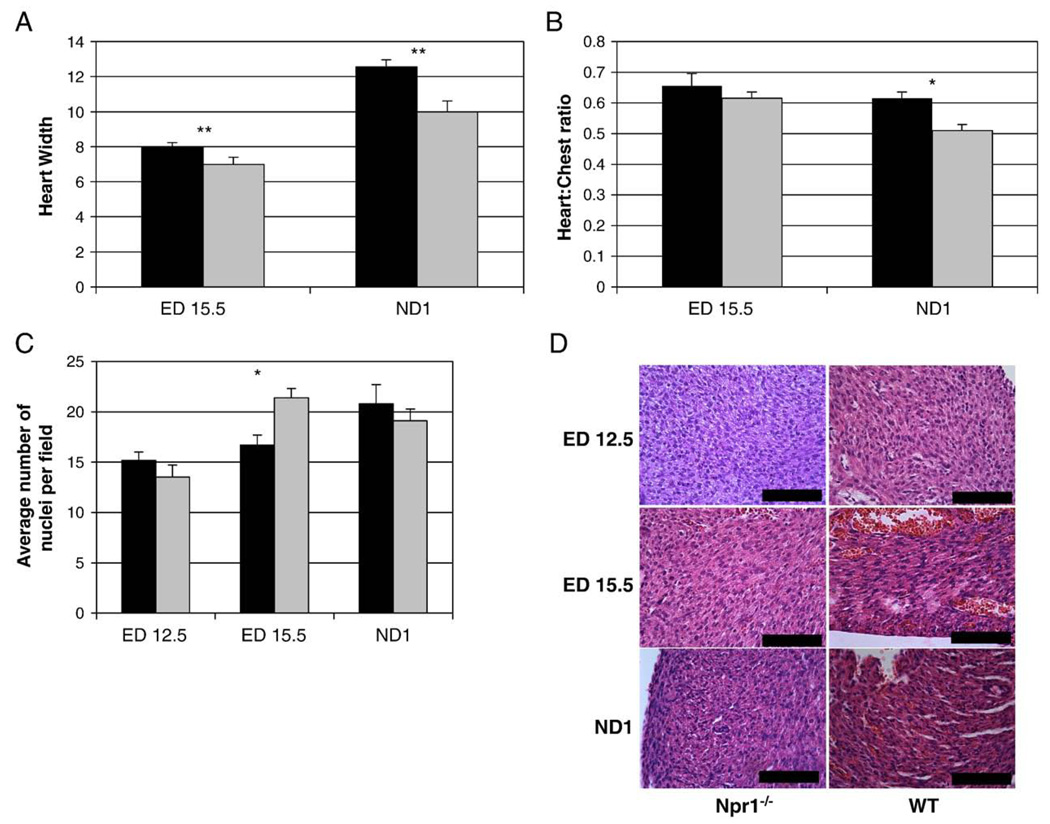
Heart dimensions were compared between Npr1−/− and WT mice from ED 15.5 (at ED 12.5 heart dimensions were not quantifiable). An increase in heart size was apparent from ED 15.5 in Npr1−/− mice compared to WT (p=0.003, Fig. 3A), while male and female Npr1−/− hearts at ND 1 were 22% and 29% larger respectively than WT littermates (p=0.0008). In addition, a significant increase in the ratio between heart size and chest size, an indication of cardiomegaly, was observed in ND 1 Npr1−/− mice compared to WT (p<0.05, Fig. 3B).
Fig. 3.
Calculated heart size at ED 15.5 and ND 1 (A) and calculated heart size to chest size ratio at ED 15.5 and ND 1 in Npr1−/− mice compared to WT (B), left ventricular free-wall myocyte nuclei count (C) in Npr1−/− mice compared to WT and paraffin embedded H&E stained sections from the left ventricular free-wall (D). Black and light grey bars represent Npr1−/− and WT groups respectively. The y axis for both A and B are in arbitrary units determined from and eye piece graticule as described in Methods. Significant differences between the genotypes are indicated by *p<0.05 and **p<0.01. Scale bar represents 100 µm.
In Npr1−/− mice the increased heart size was due to significantly increased interventricular septal thickness (3.16±0.03 vs. 2.36±0.01 graticule units, p=0.0002) and a trend towards increased left ventricular free wall thickness (2.22±0.07 vs. 1.82±0.02 graticule units, p=0.070) compared to WT mice (2.22±0.07 vs. 1.82±0.02 graticule units, p=0.070). There were no significant differences in left ventricular chamber volume (7.97±0.13 vs. 7.01±0.21 units, p=0.611) or thickness of right ventricular free wall (1.84±0.01 vs. 1.73±0.01 units, p=0.179) between genotypes. Furthermore while there was no significant difference in the number of cardiomyocyte nuclei within the left ventricular free wall at either ED 12.5 and ND 1, ED 15.5 Npr1−/− mice had significantly fewer nuclei than WT mice (17 nuclei vs. 21 nuclei, p=0.0019, Fig. 3C), indicating cardiac enlargement resulted from hypertrophy rather than hyperplasia.
3.4. Natriuretic peptide expression during development
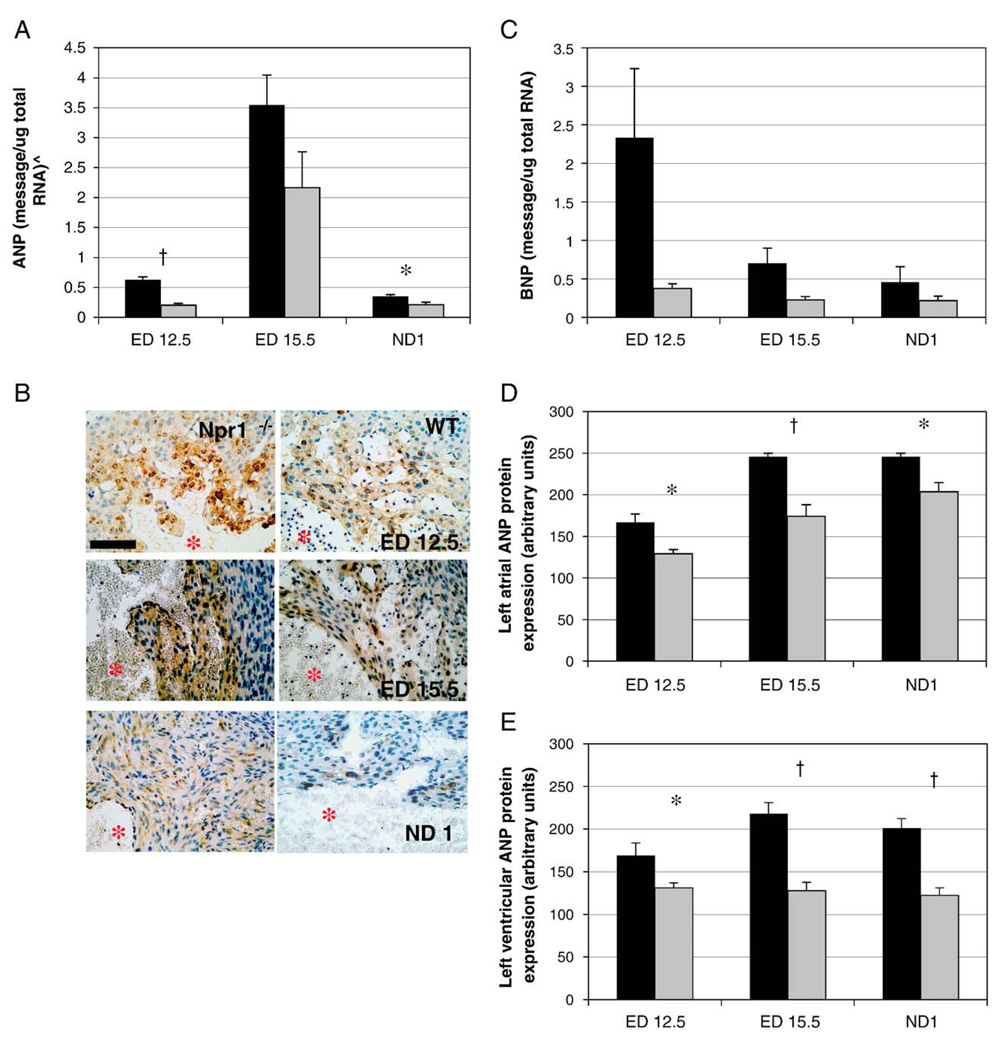
As expected, quantitative real-time PCR demonstrated higher mRNA levels of both the cardiac natriuretic peptides, ANP and BNP, in Npr1−/− mice compared to WT at all three gestational ages. In particular ANP message expression was significantly increased in Npr1−/− mice at ED 12.5 (whole embryos) and ND 1 (isolated hearts, Fig. 4A) when compared to WT. While not significant, BNP gene expression levels tended to be higher in Npr1−/− mice compared to WT at all three gestational ages (Fig. 4B).
Fig. 4.
Gestational expression of ANP mRNA (A), BNP mRNA (B), left ventricular ANP protein staining (C), and quantitation of staining for left atrial ANP protein (D) and left ventricular ANP protein (E)in Npr1−/−mice compared to WT control mice. ^Quantitated gene levels are expressed as picograms of message per microgram of total RNA (pg/µg total RNA), except for ND 1 where ANP expression gene expression is expressed as nanograms of message per microgram of total RNA (ng/µg total RNA). Scale bar represents 100 µm and red asterisks denote left ventricular lumen. Protein expression represents the accumulation of ANP protein measured by immunohistochemistry using MetaMorph™ image analysis software. Black and grey bars represent Npr1−/− and WT groups respectively. Significant differences between the genotypes are indicated by *p<0.05 and †p<0.001.
Immunohistochemical analysis showed aggregations of ANP protein throughout the atria and within the trabecular region of the cardiac ventricles of Npr1−/− and WT mice during gestation (Fig. 4C). ANP protein staining was significantly greater in both the left atria (Fig. 4D) and LV (Fig. 4E) of Npr1−/− compared to WT mice at all three gestational time points. As expected ANP protein expression within the LV was significantly greater in Npr1−/− mice than WT mice (193.6±7 in Npr1−/− vs. 124.8±5 in WT, p<0.001) and was maximally expressed at ED 15.5. There was no significant gender influence on ANP protein expression in either the atria or ventricles (p=0.570 and 0.859, respectively). In WT mice, levels of ANP expression within the left atria increased significantly with gestational age (p<0.001), however, in Npr1−/− mice left atrial ANP protein levels appeared to peak at ED 15.5.
3.5. Transcription factor expression during development
Gene and protein expression levels of transcription factors implicated in cardiac development; GATA-4, GATA-6, Mef2A, Mef2C and SRF, were investigated in Npr1−/− mice compared to WT and are shown in Table 3. GATA-4 mRNA and Mef2 protein levels were significantly reduced in Npr1−/− mice at ND1, but neither gene or protein expression of the other transcription factors was altered.
Table 3.
Levels of cardiac gene and protein expression in Npr1−/− mice compared to WT during cardiac development.
| ED 12.5 |
ED 15.5 |
ND 1 |
|||||
|---|---|---|---|---|---|---|---|
| WT | Npr1−/− | WT | Npr1−/− | WT | Npr1−/− | ||
| GATA | GATA-4 mRNA | 0.000135±0.00002 | 0.0000987±0.00001 | 0.000501±0.00007 | 0.000646±0.00006 | 0.00979±0.002 | 0.0046±0.0007* |
| GATA-6 mRNA | 0.00438±0.0005 | 0.00329±0.0004 | 0.00352±0.002 | 0.00171±0.0002 | 0.145±0.02 | 0.0168±0.02 | |
| GATA-4 protein | 169.9±8 | 170.3±8 | 178.7±9 | 183.2±10 | 189.1±10 | 181.8±13 | |
| Mef2 | MEF2A mRNA | 0.0229±0.002 | 0.0193±0.003 | 0.0439±0.006 | 0.0573±0.007 | 0.100±0.01 | 0.117±0.01 |
| MEF2C mRNA | 0.00832±0.0007 | 0.00501±0.0006 | 0.00424±0.0007 | 0.0067±0.0007 | 0.000223±0.00004 | 0.000235±0.00004 | |
| MEF2^ protein | 147.9±11 | 176.5±12 | 157.2±10 | 140.9±12 | 163.0±24 | 102.2±22* | |
| SRF | mRNA | 0.0000543±0.00003 | 0.00206±0.002 | 0.000302±0.0001 | 0.000471±0.0002 | 0.0328±0.01 | 0.0164±0.009 |
| protein | 208.3±10 | 209.6±7 | 171.7±7 | 171.6±9 | 184.1±9 | 187.4±10 | |
All data presented is calculated mean±SEM, where males and females were combined. mRNA levels are absolute amounts of gene expression in pg message/µg of total RNA. Protein expression represents the accumulation of specific protein measured by immunohistochemical densitometry using MetaMorph™ image analysis software and is expressed as arbitrary intensity units.
p<0.05 Npr1−/− compared to age matched WT.
Mef2 antibody was a universal antibody detecting Mef2A, Mef2C and Mef2D proteins.
Gestational age significantly influenced gene expression levels (p<0.001 for all genes except BNP where p=0.008). All transcription factor gene expression was significantly correlated to gestational age (Pearson's correlation coefficients of r=−0.422, 0.561, 0.665, 0.725 and 0.614, p<0.01 for Mef2A, Mef2C, GATA-4, GATA-6 and SRF respectively).
3.6. Renin–angiotensin system gene expression during development
Gene expression levels of angiotensinogen, the angiotensin II type I receptor (AT1R) and the angiotensin converting enzyme (ACE) were investigated. There was no significant difference in expression levels between genotypes at any stage of development (Table 4).
Table 4.
Levels of renin–angiotensin system and pro-hypertrophic gene expression in Npr1−/− mice compared to WT during cardiac development.
| ED 12.5 |
ED 15.5 |
ND 1 |
||||
|---|---|---|---|---|---|---|
| WT | Npr1−/− | WT | Npr1−/− | WT | Npr1−/− | |
| Angiotensinogen | 0.00122±0.0005 | 0.00046±0.0001 | 0.00421±0.003 | 0.00225±0.001 | 0.00161±0.0007 | 0.00056±0.0003 |
| ACE | 0.00012±0.0001 | 0.00011±0.0001 | 0.00050±0.0002 | 0.00060±0.0002 | 0.00018±0.0001 | 0.00004±0.00003 |
| AT1R | 0.00034±0.00004 | 0.00026±0.0001 | 0.0612±0.01 | 0.06265±0.008 | 0.0351±0.0003 | 0.0347±0.0004 |
| Akt1 | 0.859±0.1 | 0.443±0.01* | 0.492±0.02 | 0.750±0.09 | 0.0007±0.0002 | 0.004±0.002 |
| Calcineurin A | 0.207±0.05 | 0.098±0.03 | 0.0015±0.0008 | 0.0107±0.005 | 0.042±0.02 | 0.165±0.05* |
All data presented are calculated mean±SEM, where males and females were combined. mRNA levels are absolute amounts of gene expression in pg message/µg of total RNA.
3.7. Hypertrophy-related gene expression during development
Expression levels of markers of physiologically derived cardiac hypertrophy, Akt1, and pathological cardiac hypertrophy, Calcineurin A, were investigated (Table 4). While both genes were differentially expressed with gestational age, Akt1 expression was significantly reduced in Npr1−/− mice only at ED12.5 compared to WT (p=0.024), while Calcineurin A expression was significantly increased in the hearts of ND 1 Npr1−/− mice (p=0.031).
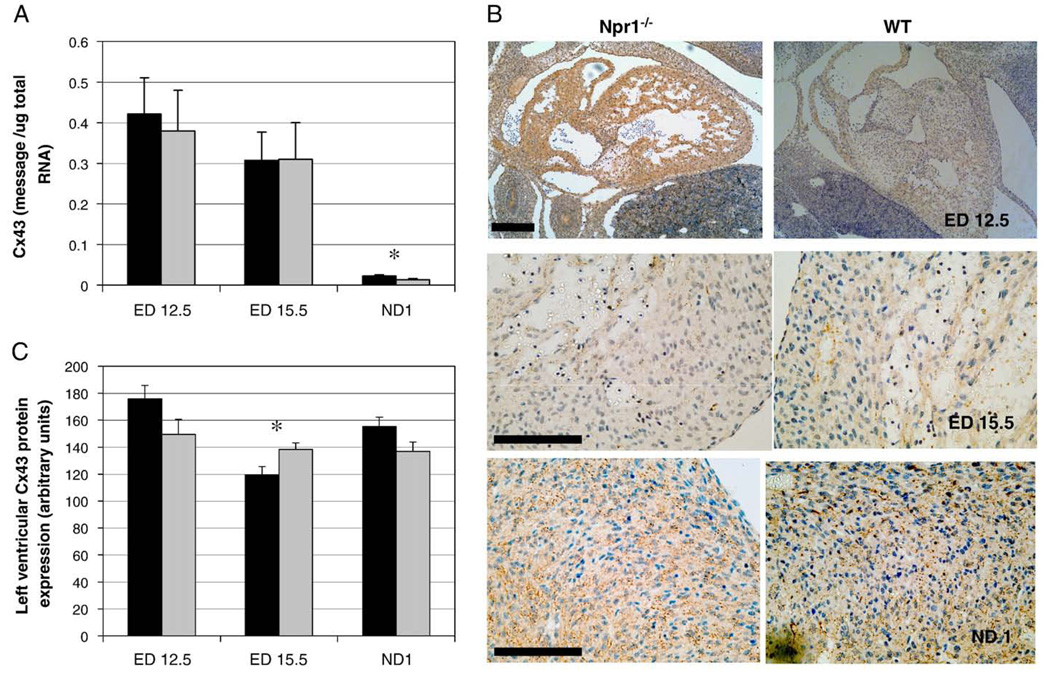
3.8. Connexin 43 gene and protein expression in Npr1−/− mice during development
Cx43 gene and protein expression decreased significantly with gestational age irrespective of Npr1 genotype (Fig. 5A). However at ND 1 mRNA levels were significantly higher in Npr1−/− hearts compared to WT (p=0.044, Fig. 5A). Additionally, developmental age, gender and Npr-1 genotype in combination influenced Cx43 gene expression (p=0.019).
Fig. 5.
Gestational expression of Cx43 mRNA (A) and protein (B, C) in Npr1−/− and WT mice. (B) Representative micrographs of Cx43 protein expression in whole hearts of ED 12.5 and left ventricles of ED 15.5 and ND 1 mice. (C) Quantitation of Cx43 protein expression in the left ventricle of Npr1−/− and WT mice. Quantitated gene levels are expressed as picograms of message per microgram of total RNA (pg/µg total RNA). Protein expression represents the accumulation of Cx43 protein measured by immunohistochemistry using MetaMorph™ image analysis software. Black and grey bars represent Npr1−/− and WT groups, respectively. Significant differences between the genotypes are indicated by *p<0.05. Scale bar represents 100 µm.
Cx43 protein is detected in the adult myocardium along the myocyte margins, with Cx43 positive protein regions forming aggregations of gap junctions. During cardiac development Cx43 protein was expressed within both the myocardium of the cardiac atria and ventricles in both Npr1−/− and WT mice (Fig. 5B), but excluded from the regions of the cardiac cushions and cardiac valves (Fig. 5B). Maximum Cx43 protein expression was evident at ED 12.5 in both Npr1−/− and WT mice. Cx43 hemi-channel aggregations at the myocardial cell membrane were also detected in the myocardium from ED 12.5; however they were more diffuse in localization compared to the high density of aggregations evident in the hearts of ND 1 and adult mice.
Cx43 protein expression was significantly different between genotypes dependent on gestational age (p=0.014, Fig. 5C), with levels of Cx 43 protein significantly reduced in the ventricles of Npr1−/− mice at ED 15.5 (p=0.029).
4. Discussion
In the current study, Npr-1 gene ablation was shown to reduce gestational survival and contribute to left ventricular hypertrophy (LVH) in the developing hearts of Npr1−/− mice. Despite the presence of cardiomyocyte hypertrophy, there was no significant change in gene expression of hypertrophy-related transcription factors. There was, however, a decrease in Cx43 protein expression at mid-gestation. The absence of any apparent gross developmental abnormalities in the whole embryos or apparent defects of cardiac structure, suggests that the decreased viability of Npr1−/−embryos may result from a combination of cardiomegaly, increased calcineurin A expression and decreased Cx43 protein affecting cardiac contractility. This is the first study to examine cardiac structure, gene expression and protein expression changes in Npr1−/− mice during embryonic development.
Npr-1 gene ablation is characterized by incremental increases in blood pressure relative to the number of disrupted alleles [13], as well as the development of cardiac hypertrophy and fibrosis in post-natal life [12]. The findings from this study suggest that Npr-1 activity is important in embryonic survival as significantly fewer than expected Npr1−/− embryos survived to birth and subsequently to weaning.
Reduced post-natal survival has been previously reported in an alternative strain of Npr-1 null mice, where the total number of null mice at 3 weeks of age was only 70% of that expected [27]. It was reported that a large proportion of the gestational deaths were attributable to fetal hydrops. Within our colony, the proportion of Npr1−/− mice surviving at 4 weeks of age was comparable at 68%. Furthermore we identified that the decline in the number of Npr1−/− mice occurred across a continuum from ED 12.5 until birth, indicating that Npr1−/− embryos were implanted at a normal rate but a degree of embryonic lethality was evident at mid/late gestation. Fetal hydrops was not observed in our colony, nor was there evidence of cardiac structural abnormalities among the surviving embryos at collection to account for loss of developmental viability. However, significant left ventricular hypertrophy was apparent from ED 15.5 onwards in Npr1−/− mice.
Maternal hypertension has been shown to damage and prematurely age the placental vasculature [28] and is also associated with inter-uterine growth retardation, reduced birth weight and increased neonatal death [29,30]. In surrogate studies using spontaneously hypertensive rats Di Nicolantonio et al. have shown that the genetic identity of the fetus determines the growth rate of both the fetus and the placenta independent of maternal phenotype [31]. In this study maternal blood pressure was monitored throughout pregnancy. While there was a significant genotype influence on blood pressure, the degree of blood pressure elevation as pregnancy progressed was not significantly different between Npr1+/−, Npr1−/− and WT dams. Nor were there significant differences in embryo body weight between genotypes at any time point investigated. There was no indication maternal hypertension contributed to the gestational death or resulted in reduced birth weight of Npr1−/− mice during development.
Histological examination identified an increase in heart dimensions and subsequent heart size to chest size ratio due to thickening of the left heart structures, namely the LV free-wall and IVS, in Npr1−/− mice from ED 15.5. Previously Kuhn et al. have shown Npr-1 null mice exhibit cardiomyocyte enlargement at both 4 and 12 months of age [15]. Leu et al. have shown that during the first four post-natal days the volume of cardiomyocytes remains constant despite a concomitant increase in cardiac mass, indicating that normal cardiac growth during this developmental phase is due to cell division [32]. In the current study the decreased nuclei number suggests the increase in heart mass of Npr1−/− mice during gestation resulted from enlargement of the myocardial cells (hypertrophy), rather than through increased cell numbers (hyperplasia) in the left ventricle.
Several studies using both experimental manipulation and genetic models have explored the gene expression changes associated with pathological cardiac hypertrophy [14,33]. These studies and our data strongly suggest that ANP-mediated Npr-1 signaling has a direct inhibitory effect on cardiac hypertrophy. Horsthuis et al. have shown that distinct regulatory sequences and divergent regulatory pathways control fetal ANP expression and the reactivation of ventricular ANP expression during cardiac disease [34]. Despite the increased levels of natriuretic peptide gene and protein expression, there was no detected increase in gene expression of the hypertrophy-related, cardiac transcription factors; Mef2A, Mef2C, GATA-4, GATA-6 and SRF or genes of the renin–angiotensin system. This contrasts with adult Npr1−/− mice, in which we previously reported cardiac hypertrophy was correlated to increased expression of both the GATA-4 and Mef2C genes in aged animals [23]. Interestingly our observation of unaltered RAS gene expression in embryos contrasts with a study by Li et al. [35] where cardiac hypertrophy and fibrosis in adult Npr1−/− mice was ascribed to augmented cardiac AT1R signaling. This further illustrates differential role of natriuretic peptide signaling in the developing and mature cardiovascular systems. In the absence of up-regulation of traditional transcriptional markers of pathological cardiac hypertrophy we investigated the expression of Atk1, a down-stream effector of the phosphoinositol-3 kinase p110α subunit (PI3K-P110α), recently implicated as key component of physiological cardiac hypertrophy [36–38]. Contrary to our expectations significant down-regulation of expression was observed at ED 12.5. Interestingly, Calcineurin A expression increased in Npr1−/− mice from ED 15.5 (not significant) culminating in significantly elevated expression at ND 1 once cardiomyocyte hypertrophy is observed. These result complement our previous data where calcium/calmodulin signaling was implicated in the early establishment of cardiac hypertrophy in young Npr1−/− mice [23].
Cx43, like the natriuretic peptides, is a marker of myocardial cell development during embryogenesis [7]. In the heart, Cx protein-derived gap junctions provide the pathway for intercellular current flow, enabling coordinated action potential propagation and contraction. In adult ventricular muscle, cell-to-cell coupling is formed predominantly by Cx43 channels. Deranged expression and organization of Cx43 gap junctions in the ventricle have been demonstrated in a variety of cardiovascular pathologies, including hypertrophy [39]. Cardiomyocyte-restricted deletion of Cx43 has been shown to result in increased gestational attrition of null mice from ED 13.5 through to post-natal day 6 in the absence of major cardiac structural malformations [40]. Dysregulation of Cx43 activity has been associated with impaired electrical action of the heart, namely an increased prevalence of tachyarrhythmias [41,42], such impaired cardiac conduction might account for the observed gestation deaths in Npr1−/− mice, particularly the sporadic rate of deaths occurring along a developmental continuum.
This study has demonstrated that during embryonic development of Npr1−/− mice, there is progressive LVH from ED 15.5, but no concomitant up-regulation of hypertrophy-associated, transcription factor genes or components of the renin–angiotensin system. However increased ANP expression during embryonic development was associated with reduced expression of both Cx43 and SRF genes in Npr1−/− mice. While the morphological cardiac enlargement in embryonic and adult Npr1−/− mice is similar, the different molecular characteristics of LVH evident in Npr1−/− mice from ED 15.5 compared to adult Npr1−/− mice indicates that the role of natriuretic peptide signaling in cardiac development is distinct from that traditionally attributed to either pathological or physiological cardiac remodeling.
Acknowledgements
This work was supported by the National Heart Foundation of New Zealand (grant 1056). The authors would like to thank Howard Potter (Molecular Pathology, Canterbury Health Labs) for work that involved sequencing PCR products, Rachel Purcell (Children's Cancer Research Group, University of Otago-Christchurch) for help with Immunohistochemistry techniques, Professor Mike Dragunow (Department of Pharmacology and Clinical Pharmacology, University of Auckland) for MetaMorph data analysis and Dr. Anna Pilbrow (Christchurch Cardioendocrine Research Group, University of Otago) for statistical advice.
References
- 1.Holmes SJ, Espiner EA, Richards AM, Yandle TG, Frampton C. Renal, endocrine and haemodynamic effects of human brain natriuretic peptide in normal men. J. Clin. Endocrinol. Metab. 1993;76:91–96. doi: 10.1210/jcem.76.1.8380606. [DOI] [PubMed] [Google Scholar]
- 2.Espiner EA, Richards AM, Yandle TG, Nicholls MG. Natriuretic hormones. Endocrinol. Metab. Clin. N. Am. 1995;24:481–509. [PubMed] [Google Scholar]
- 3.Lang CC, Choy AMJ, Turner K, Tobin R, Coutie W, Struthers AD. The effect of intravenous saline loading on plasma levels of brain natriuretic peptide in man. J. Hypertens. 1993;11:737–741. doi: 10.1097/00004872-199307000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Jaekle RK, Sheikh AU, Berry DD, Washburn L, Rose JC. Hemodynamic and hormonal responses to atrial distension in the ovine fetus. Am. J. Obstet. Gynecol. 1995;173:694–701. doi: 10.1016/0002-9378(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 5.Cameron VA, Aitken GD, Ellmers LJ, Kennedy MA, Espiner EA. The sites of gene expression of atrial, brain, and C-type natriuretic peptides in mouse fetal development: temporal changes in embryos and placenta. Endocrinology. 1996;137:817–824. doi: 10.1210/endo.137.3.8603590. [DOI] [PubMed] [Google Scholar]
- 6.Kingdom JC, Jardine AG, Doyle J, Connell JM, Gilmore DH, Whittle MJ. Atrial natriuretic peptide in the fetus. BMJ (Clinical research ed. 1989;298:1221–1222. doi: 10.1136/bmj.298.6682.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, Bao ZZ, Palmer S, Biben C, Harvey RP, Moorman AF. Chamber formation and morphogenesis in the developing mammalian heart. Dev. Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- 8.Zeller R, Bloch KD, Williams BS, Arceci RJ, Seidman CE. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev. 1987;1:693–698. doi: 10.1101/gad.1.7.693. [DOI] [PubMed] [Google Scholar]
- 9.Day ML, Schwartz D, Wiegand RC, Stockman PT, Brunnert SR, Tolunay HE, Currie MG, Standaert DG, Needleman P. Ventricular atriopeptin: unmasking of mRNA and peptide synthesis by hypertrophy or dexamethasone. Hypertension. 1987;9:485–491. doi: 10.1161/01.hyp.9.5.485. [DOI] [PubMed] [Google Scholar]
- 10.Lattion AL, Michel JB, Arnauld E, Corvol P, Soubrier F. Myocardial recruitment during ANF mRNA increase with volume overload in the rat. Am. J. Physiol. 1986;251:H890–H896. doi: 10.1152/ajpheart.1986.251.5.H890. [DOI] [PubMed] [Google Scholar]
- 11.Takayanagi R, Imada T, Inagami T. Synthesis and presence of atrial natriuretic factor in rat ventricle. Biochem. Biophys. Res. Commun. 1987;142:483–488. doi: 10.1016/0006-291x(87)90300-7. [DOI] [PubMed] [Google Scholar]
- 12.Oliver PM, Fox JE, Kim R, Rockman HA, Kim HS, Reddick RL, Pandey KN, Milgram SL, Smithies O, Maeda N, Hypertension cardiac hypertrophy. and sudden death in mice lacking natriuretic peptide receptor A. Proc. Natl. Acad. Sci. U. S. A. 1997;94:14730–14735. doi: 10.1073/pnas.94.26.14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver PM, John SW, Purdy KE, Kim R, Maeda N, Goy MF, Smithies O. Natriuretic peptide receptor 1 expression influences blood pressures of mice in a dose-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2547–2551. doi: 10.1073/pnas.95.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knowles JW, Esposito G, Mao L, Hagaman JR, Fox JE, Smithies O, Rockman HA, Maeda N. Pressure-independent enhancement of cardiac hypertrophy in natriuretic peptide receptor A-deficient mice. J. Clin. Invest. 2001;107:975–984. doi: 10.1172/JCI11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn M, Holtwick R, Baba HA, Perriard JC, Schmitz W, Ehler E. Progressive cardiac hypertrophy and dysfunction in atrial natriuretic peptide receptor (GC-A) deficient mice. Heart. 2002;87:368–374. doi: 10.1136/heart.87.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellmers LJ, Knowles JW, Kim HS, Smithies O, Maeda N, Cameron VA. Ventricular expression of natriuretic peptides in Npr1(−) mice with cardiac hypertrophy and fibrosis. Am. J. Physiol. Heart Circ. Physiol. 2002;283:H707–H714. doi: 10.1152/ajpheart.00677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez MJ, Garbers DL, Kuhn M. The guanylyl cyclase-deficient mouse defines differential pathways of natriuretic peptide signaling. J. Biol. Chem. 1997;272:23064–23068. doi: 10.1074/jbc.272.37.23064. [DOI] [PubMed] [Google Scholar]
- 18.Molkentin JD, Dorn II IG. Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu. Rev. Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 19.Vellaichamy E, Sommana NK, Pandey KN. Reduced cGMP signaling activates NF-kappaB in hypertrophied hearts of mice lacking natriuretic peptide receptor-A. Biochem. Biophys. Res. Commun. 2005;327:106–111. doi: 10.1016/j.bbrc.2004.11.153. [DOI] [PubMed] [Google Scholar]
- 20.Walther T, Stepan H, Faber R. Dual natriuretic peptide response to volume load in the fetal circulation. Cardiovasc. Res. 2001;49:817–819. doi: 10.1016/s0008-6363(00)00303-5. [DOI] [PubMed] [Google Scholar]
- 21.Nimrod C, Keane P, Harder J, Davies D, Kondo C, Takahashi Y, Wong T, Maloney J, Nicholson S. Atrial natriuretic peptide production in association with nonimmune fetal hydrops. Am. J. Obstet. Gynecol. 1988;159:625–628. doi: 10.1016/s0002-9378(88)80022-x. [DOI] [PubMed] [Google Scholar]
- 22.Itoh H, Sagawa N, Hasegawa M, Okagaki A, Inamori K, Ihara Y, Mori T, Suga S, Mukoyama M, Shirakami G, et al. Brain natriuretic peptide levels in the umbilical venous plasma are elevated in fetal distress. Biol. Neonate. 1993;64:18–25. doi: 10.1159/000243966. [DOI] [PubMed] [Google Scholar]
- 23.Ellmers LJ, Scott NJ, Piuhola J, Maeda N, Smithies O, Frampton CM, Richards AM, Cameron VA. Npr1-regulated gene pathways contributing to cardiac hypertrophy and fibrosis. J. Mol. Endocrinol. 2007;38:245–257. doi: 10.1677/jme.1.02138. [DOI] [PubMed] [Google Scholar]
- 24.Gundersen HJ, Jensen EB. The efficiency of systematic sampling in stereology and its prediction. J. Microsc. 1987;147:229–263. doi: 10.1111/j.1365-2818.1987.tb02837.x. [DOI] [PubMed] [Google Scholar]
- 25.Dragunow M, Cameron R, Narayan P, O'Carroll S. Image-based high-throughput quantification of cellular fat accumulation. J. Biomol. Screen. 2007;12:999–1005. doi: 10.1177/1087057107306502. [DOI] [PubMed] [Google Scholar]
- 26.Graham ES, Ball N, Scotter EL, Narayan P, Dragunow M, Glass M. Induction of Krox-24 by endogenous cannabinoid type 1 receptors in Neuro2A cells is mediated by the MEK-ERK MAPK pathway and is suppressed by the phosphatidylinositol 3-kinase pathway. J. Biol. Chem. 2006;281:29085–29095. doi: 10.1074/jbc.M602516200. [DOI] [PubMed] [Google Scholar]
- 27.Lopez MJ, Wong SK, Kishimoto I, Dubois S, Mach V, Friesen J, Garbers DL, Beuve A. Salt-resistant hypertension in mice lacking the guanylyl cyclase-A receptor for atrial natriuretic peptide. Nature. 1995;378:65–68. doi: 10.1038/378065a0. [DOI] [PubMed] [Google Scholar]
- 28.Soma H, Yoshida K, Mukaida T, Tabuchi Y. Morphologic changes in the hypertensive placenta. Contrib. Gynecol. Obstet. 1982;9:58–75. [PubMed] [Google Scholar]
- 29.Martyn CN, Barker DJ, Jespersen S, Greenwald S, Osmond C, Berry C. Growth in utero, adult blood pressure, and arterial compliance. Br. Heart J. 1995;73:116–121. doi: 10.1136/hrt.73.2.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiong X, Mayes D, Demianczuk N, Olson DM, Davidge ST, Newburn-Cook C, Saunders LD. Impact of pregnancy-induced hypertension on fetal growth. Am. J. Obstet. Gynecol. 1999;180:207–213. doi: 10.1016/s0002-9378(99)70176-6. [DOI] [PubMed] [Google Scholar]
- 31.Di Nicolantonio R, Koutsis K, Wlodek ME. Fetal versus maternal determinants of the reduced fetal and placental growth in spontaneously hypertensive rats. J. Hypertens. 2000;18:45–50. doi: 10.1097/00004872-200018010-00007. [DOI] [PubMed] [Google Scholar]
- 32.Leu M, Ehler E, Perriard JC. Characterisation of postnatal growth of the murine heart. Anat. Embryol. (Berl) 2001;204:217–224. doi: 10.1007/s004290100206. [DOI] [PubMed] [Google Scholar]
- 33.Kishimoto I, Rossi K, Garbers DL. A genetic model provides evidence that the receptor for atrial natriuretic peptide (guanylyl cyclase-A) inhibits cardiac ventricular myocyte hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2703–2706. doi: 10.1073/pnas.051625598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Horsthuis T, Houweling AC, Habets PE, de Lange FJ, el Azzouzi H, Clout DE, Moorman AF, Christoffels VM. Distinct regulation of developmental and heart disease-induced atrial natriuretic factor expression by two separate distal sequences. Circ. Res. 2008;102:849–859. doi: 10.1161/CIRCRESAHA.107.170571. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Kishimoto I, Saito Y, Harada M, Kuwahara K, Izumi T, Takahashi N, Kawakami R, Tanimoto K, Nakagawa Y, Nakanishi M, Adachi Y, Garbers DL, Fukamizu A, Nakao K. Guanylyl cyclase-A inhibits angiotensin II type 1A receptor-mediated cardiac remodeling, an endogenous protective mechanism in the heart. Circulation. 2002;106:1722–1728. doi: 10.1161/01.cir.0000029923.57048.61. [DOI] [PubMed] [Google Scholar]
- 36.McMullen JR, Jennings GL. Differences between pathological and physiological cardiac hypertrophy: novel therapeutic strategies to treat heart failure. Clin. Exp. Pharmacol. Physiol. 2007;34:255–262. doi: 10.1111/j.1440-1681.2007.04585.x. [DOI] [PubMed] [Google Scholar]
- 37.Porrello ER, Bell JR, Schertzer JD, Curl CL, McMullen JR, Mellor KM, Ritchie RH, Lynch GS, Harrap SB, Thomas WG, Delbridge LM. Heritable pathologic cardiac hypertrophy in adulthood is preceded by neonatal cardiac growth restriction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009;296:R672–R680. doi: 10.1152/ajpregu.90919.2008. [DOI] [PubMed] [Google Scholar]
- 38.Rigor DL, Bodyak N, Bae S, Choi JH, Zhang L, Ter-Ovanesyan D, He Z, McMullen JR, Shioi T, Izumo S, King GL, Kang PM. Phosphoinositide 3-kinase Akt signaling pathway interacts with protein kinase Cbeta2 in the regulation of physiologic developmental hypertrophy and heart function. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H566–H572. doi: 10.1152/ajpheart.00562.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Severs NJ, Coppen SR, Dupont E, Yeh HI, Ko YS, Matsushita T. Gap junction alterations in human cardiac disease. Cardiovasc. Res. 2004;62:368–377. doi: 10.1016/j.cardiores.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Eckardt D, Kirchhoff S, Kim JS, Degen J, Theis M, Ott T, Wiesmann F, Doevendans PA, Lamers WH, de Bakker JM, van Rijen HV, Schneider MD, Willecke K. Cardiomyocyte-restricted deletion of connexin43 during mouse development. J. Mol. Cell. Cardiol. 2006;41:963–971. doi: 10.1016/j.yjmcc.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 41.Beauchamp P, Choby C, Desplantez T, de Peyer K, Green K, Yamada KA, Weingart R, Saffitz JE, Kleber AG. Electrical propagation in synthetic ventricular myocyte strands from germline connexin43 knockout mice. Circ. Res. 2004;95:170–178. doi: 10.1161/01.RES.0000134923.05174.2f. [DOI] [PubMed] [Google Scholar]
- 42.Danik SB, Liu F, Zhang J, Suk HJ, Morley GE, Fishman GI, Gutstein DE. Modulation of cardiac gap junction expression and arrhythmic susceptibility. Circ. Res. 2004;95:1035–1041. doi: 10.1161/01.RES.0000148664.33695.2a. [DOI] [PMC free article] [PubMed] [Google Scholar]