Abstract
Systems biology and infectious disease (host-pathogen-environment) research and development is becoming increasingly dependent on integrating data from diverse and dynamic sources. Maintaining integrated resources over long periods of time presents distinct challenges. This paper describes experiences and lessons learned from integrating data in two five-year projects focused on pathosystems biology: the Pathosystems Resource Integration Center (PATRIC, http://patric.vbi.vt.edu/), with a goal of developing bioinformatics resources for the research and countermeasures development communities based on genomics data, and the Resource Center for Biodefense Proteomics Research (RCBPR, http://www.proteomicsresource.org/), with a goal of developing resources based on the experiment data such as microarray and proteomics data from diverse sources and technologies. Some challenges include integrating genomic sequence and experiment data, data synchronization, data quality control, and usability engineering. We present examples of a variety of data integration problems drawn from our experiences with PATRIC and RBPRC, as well as open research questions related to long term sustainability, and describe the next steps to meeting these challenges. Novel contributions of this work include (1) an approach for addressing discrepancies between experiment results and interpreted results and (2) expanding the range of data integration techniques to include usability engineering at the presentation level.
Introduction
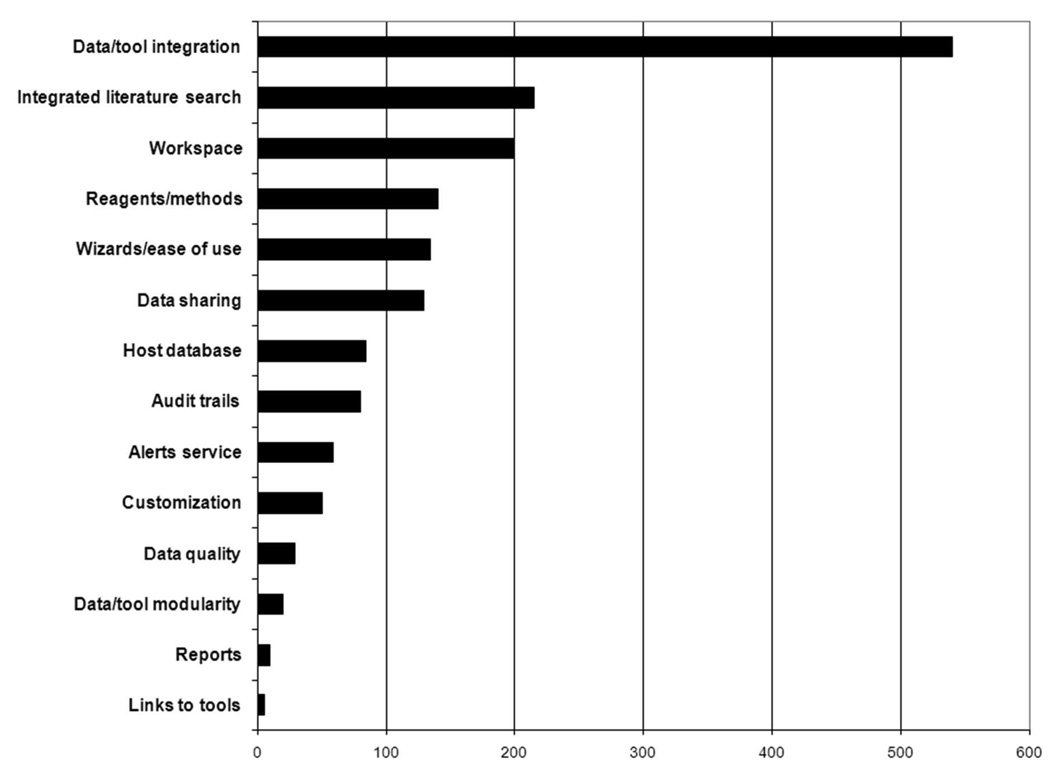
Data integration is a pervasive problem in bioinformatics research. In 2003, a National Science Foundation Blue-Ribbon Advisory Panel on Cyberinfrastructure identified data, information, and knowledge management services as fundamental elements of cyberinfrastructure [1]. Some of these services entail storing and providing access to large numbers of data sets, such as those provided by the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/) and the European Bioinformatics Institute (EBI, http://www.ebi.ac.uk/). Another key technical feature required in e-science cyberinfrastructure is the ability to integrate data from diverse sources and of varying types. In-depth interviews with 17 diverse infectious disease researchers, conducted by our Cyberinfrastructure Group and a consulting team from the Accelrys Corporation (http://accelrys.com/), showed that data and tool integration was the single most important function sought by researchers in emerging infectious disease research portals (see Figure 1). We expect the data integration we see in today’s bioinformatics climate to become significantly more challenging as the volume, rate of production, and complexity of data types increases.
Figure 1.
Seventeen infectious disease researchers were asked to allocate 100 resource units to 14 proposed features of an infectious disease research portal according to the relative value of each feature. Data and tool integration was more than twice as valued as the closest of the other features.
Data integration difficulties stem, in part, from several, often co-existing, specific sub-problems such as how to store, access, and analyze multiple types of data; how to work with multiple databases simultaneously; how to integrate structured data, such as that found in relational databases, with unstructured data, such as literature; and how to make data accessible and usable to life sciences researchers. Although these problems occur throughout bioinformatics, we focus in this paper on the particular challenges and lessons learned in two five-year projects dedicated to supporting the work of infectious disease researchers developing data for target discovery programs and seeking to develop new vaccines, diagnostics, and therapeutics:
The PathoSystems Resource Integration Center (PATRIC, http://patric.vbi.vt.edu/) is one of several bioinformatics research centers funded by the National Institutes of Health through the National Institute of Allergy and Infectious Diseases (NIAID). PATRIC provides genomic information related to organisms that NIAID has identified as significant to biodefense and emerging or re-emerging human diseases. PATRIC’s database provides access to nucleic acid features, protein features and attributes, metabolic pathways, host-pathogen interactions, and links to published literature. During its first phase, from 2004 to 2009, PATRIC provided information on eight organisms; following renewal in 2009, PATRIC will provide information on all bacteria identified by NIAID as significant to biodefense and emerging or re-emerging human diseases. The expanded scope of the project underlies the need for the multi-level data integration approach described here.
The Resource Center for Biodefense Proteomics Research (RCBPR, http://www.proteomicsresource.org/) is a NIAID-funded project providing data and resources on proteomics experiments related to infectious disease research. The project supports seven proteomics research centers and the scientific community through a collaboration between Social & Scientific Systems, Inc., Georgetown University Medical Center, and our CyberInfrastructure Group. The RCBPR database hosts several experiment types, including mass spectrometry, microarray, protein interaction, and clones. The RCBPR completed in 2009 and data was transferred to Pathogen Portal (http://www.pathogenportal.org) which is now under development by the PATRIC team.
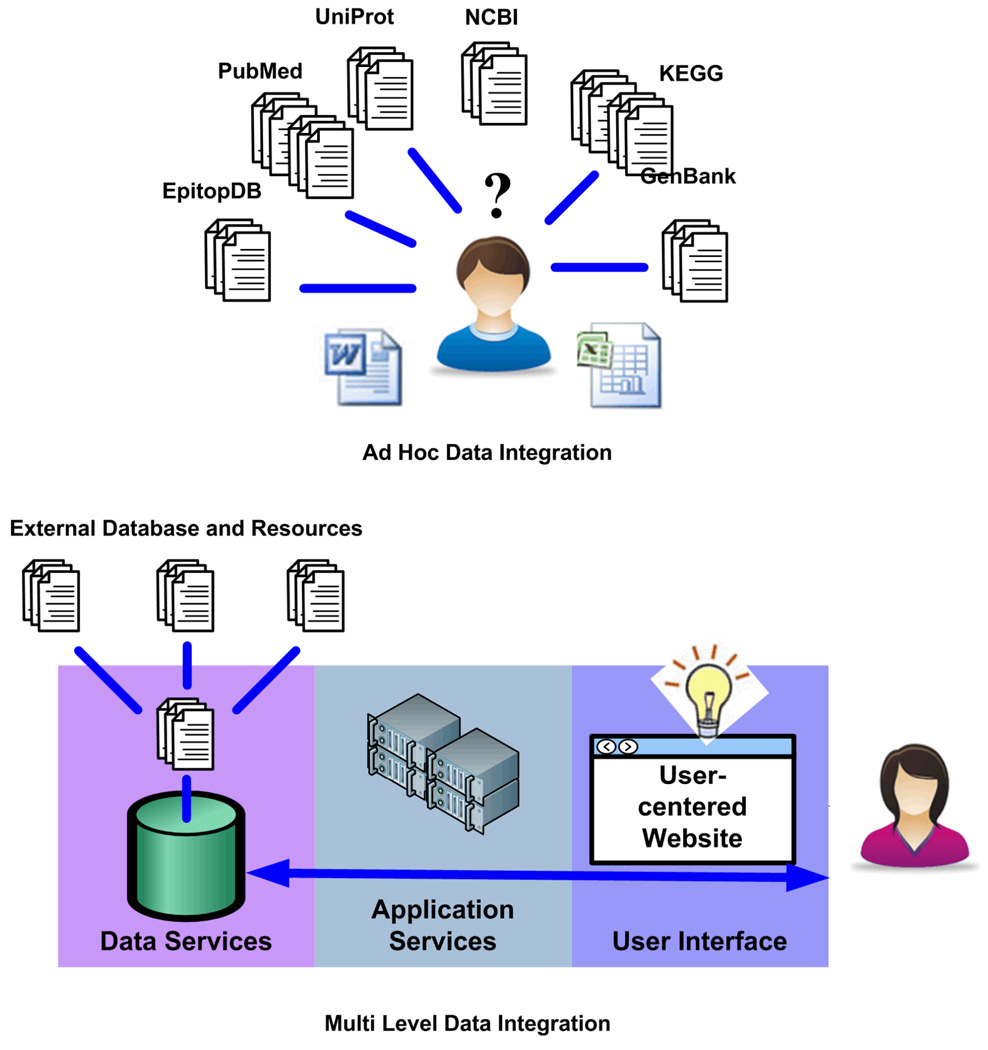
The PATRIC and RCBPR are integrated Web resources for infectious disease researchers. When integrated data services and applications are unavailable, infectious disease researchers are required to perform their own integration of diverse data sources, sometimes resorting to ad hoc tools such as spreadsheets and word processing documents to provide a common foundation for data integration. This approach is not only time consuming, but shifts the onus of data integration to the researcher (i.e., application / Web resource user), thus impeding on their cognitive resources and hence their ability to gain insight. The end result is often an overwhelmed or confused user (Figure 2, left). We instead propose a data integration approach that strives to create integrated Web resources, such as PATRIC and RCBPR, by addressing data integration at three distinct software architectural levels: the data services level, the application services level, and the user interface level (see Figure 2, right). From our experiences, data integration at any one of these levels is necessary, but not sufficient to create a bona fide Web resource exhibiting high utility and value. By addressing data integration at all three software architecture levels, we lay the foundation for Web resources with high scalability, reliability, maintainability, and usability.
Figure 2.
Lacking integrated data services, infectious disease researchers must devise their own integration methods, sometimes resorting to ad hoc tools such as spreadsheets and text documents (left). By providing data integration at the data and application services levels along with user interfaces designed to support interaction with the broad range of integrated data now available, infectious disease researchers are relieved of data integration challenges (right).
Table 1 summarizes the challenges and solutions across the three levels. The remainder of this paper provides further explanation and details about our experiences and lessons learned integrating data at each of the levels. Specifically, Subsection 1 of Results and Discussion details several challenges encountered and solutions developed when integrating data at the database and application services levels of the systems architecture. Subsection 2 focuses on the user interface and need for intuitive access to data, with particular emphasis on information retrieval in increasingly integrated Web resources. The challenges and solutions to both the database/application levels and the user interface level of systems architecture are outlined in Table 1. Subsection 3 summarizes the lessons learned from our experiences in data integration across these two projects that may be of use to other data integration projects. The conclusion section ends the paper with a brief discussion of future work.
Table 1.
Data integration challenges and solutions encountered when integrating genomic and post-genomic experiment data related to infectious disease research.
| Challenges | Solutions |
|---|---|
| Tolerance for identification of gene or proteins |
Maintaining multiple interpretations of data |
| Ensuring completeness, validity, accuracy and currency of data |
Formalizing data staging and review process; implementing multiple data quality controls |
| Managing multiple identifier schemes | Short term, use mapping service, but long term use reduced number of identifier schemes and equivalence mappings in Semantic Web |
| Data synchronization | Use access on demand APIs with canonical data structures when possible and bulk load only when APIs cannot meet requirements |
| Literature integration | Use APIs to query literature repositories; provide end users with "smart query" capabilities |
| Maximizing the use of limited post- genomic experimental evidence for gene and protein predictions. |
Treat experiments using orthologs as indirect evidence for gene and protein predictions. |
| Usability | Employ usability engineering to complement and inform software development; include support for integrated information, data and tools, progressive filtering, and context sensitivity |
Results and Discussion
1. Data Integration in Databases and Application Servers
The overall goals of both PATRIC and RCBPR are to support the discovery of targets for use in the development of vaccines, diagnostics, and therapeutics. To that end, infectious disease researchers need access to a wide array of data, including supporting experimental evidence for assertions and annotations. This is a common requirement in life science research and a variety of approaches have been applied [2–17], yet challenges remain [6]. Our experience with PATRIC and RCBPR revealed five broad categories of data integration challenges:
Using sequence similarity as a proxy for sequence identity
Ensuring completeness, validity, accuracy, and currency of data across separate databases
Managing and mapping between multiple identifier schemes
Synchronizing data across separate databases
Integrating literature and structured data
Using indirect experiment evidence for gene and protein predictions
Each of these challenges is described in detail in the remainder of this subsection.
1.1. Similarity Thresholds and Data Integration
Sequence similarity searching is a fundamental bioinformatics technique that allows researchers to search for sequences that match one another, possibly with slight variations. Unlike other disciplines, bioinformatics must temper its concept of identity to allow for some variation. For example, if a leucine in a protein were substituted with an isoleucine, the resulting protein could, in many cases, be considered the “same” (functionally) as the original. This is an example of using similarity as a proxy for (functional) identity. Although toleration for variation is required in bioinformatics data integration, there is no universally accepted threshold that can be used in all circumstances to distinguish an identity match from a mismatch.
Consider the following example: A mass spectrometry experiment was conducted on Brucella melitensis biovar Abortus 2308. At the time, a complete genome sequence and annotations of Brucella melitensis biovar Abortus 2308 were not available. Therefore, peptides were identified by comparing Brucella melitensis biovar Abortus 2308 mass spectra to the proteins from other Brucella species that had been sequenced and annotated such as Brucella abortus bv. 1 str. 9–941, Brucella melitensis 16M, Brucella suis 1330, and Brucella ovis. In order to preserve the integrity of the experiment, results must be presented on the RCBPR website exactly as they were published by the experimenters. However, when integrating with genomic data in PATRIC, these experiment results could not be integrated as if they were evidence for the existence of proteins in Brucella abortus bv. 1 str. 9–941, Brucella melitensis, Brucella suis, or Brucella ovis because the experiment was not conducted on those strains, but instead conducted exclusively on Brucella melitensis biovar Abortus 2308. The dilemma was resolved as follows: first, the proteomics results were reported as generated in the RCBPR database, and second, those results were mapped to proteins for Brucella melitensis biovar Abortus 2308 in PATRIC. In addition, results were also mapped to proteins in other Brucella strains in PATRIC using ortholog groups and presented as indirect evidence.
This solution has its drawbacks. A researcher browsing Brucella melitensis biovar Abortus 2308 data in PATRIC would find experimental evidence for the existence of specific proteins in Brucella melitensis biovar Abortus 2308. However, if the researcher reviews the same experiment results on the RCBPR website, it may give an impression that the conclusions were derived based on the experiment performed on other Brucella strains. If researchers in this position reviewed the experiment metadata or consulted with the experiment publication they would discover the experiment was in fact performed on Brucella melitensis biovar Abortus 2308, but this is not immediately apparent in the application.
1.2. Ensuring Completeness, Validity, Accuracy, and Currency of Data
Any data repository that presents inaccurate data risks undermining researchers’ confidence and trust in the repository as a reliable source of data; however, researchers will have varying tolerance for error depending on how they are using the data. Addressing this challenge entails a balancing of competing demands. For example, a researcher studying a transmembrane protein as a potential drug target requires accurate information about protein structure, and an error in a single domain could invalidate conclusions drawn about that protein as a potential target. That same type of error, although unwelcome, might be tolerable for a bioinformatics researcher developing a new ortholog predication algorithm.
Another consideration facing curators and repository developers is the inherent limits on the quality of data. Errors introduced during the experiment itself may not be detected without careful expert evaluation. Other errors are more easily detected but there may be no generally accepted means of correcting them. For example, if a data value is missing, should this be reflected as a null value in the database or should another value be used in its place, such as the mean or median of that attribute’s values, or should an arbitrarily chosen value, such as 0, be selected? An acceptable solution for a statistician may be unacceptable for a vaccine researcher. Limited budgets and production schedules also impose limits on how much remediation is possible with data quality problems. The solutions to this challenge include using a formalized process for data staging and review and employing multiple types of data quality checks.
Data staging is routinely employed when loading data to the RCBPR database. First, data is loaded into a separate, temporary database schema where it is reviewed before integrating it with existing production data. Since the data is staged in a separate database, any errors that occur, either in the data itself or with the data loading process, will not adversely affect the live, publicly released data. Moreover, recovering from errors in a staging schema is easier than from a production schema because there are fewer dependencies with other data. Once obvious errors are corrected, multiple types of data quality checks are applied in the staging schema, such as checking for missing values, checking ranges of values, verifying referential integrity, and applying arbitrary checks, such as minimum length of a peptide. These simple quality control checks help quickly identify bugs in data loading programs and errors in data files.
The solution proposed here is necessarily limited. A formal data staging and review process will help identify potential problems with data quality, but determining the appropriate resolution to those problems will be highly specific to the domain and use cases for the Web resource.
1.3. Managing Multiple Identifier Schemes
Bioinformaticians routinely work with multiple identifier schemes and map from one scheme to another. For example, a researcher using NCBI accession numbers may need to find corresponding data using UniProtKB accession numbers. Several cross referencing services have been developed [18–20]. Methods that work well with curated entities in large public repositories, such as Entrez Protein (http://www.ncbi.nlm.nih.gov/sites/entrez?db=protein/) and UniProt (http://www.uniprot.org/), are not always sufficient when working with experimental data that is not yet fully curated and may include proteins that are not yet available in these databases. For example, the results of a mass spectrometry experiment may provide evidence for the existence of what was thought to be a hypothetical gene product. That hypothetical entity may not have an identifier assigned in any of the major genomic repositories, so conventional identifier mapping systems, such as the Protein Identifier Cross Reference service (PICR, http://www.ebi.ac.uk/Tools/picr/) [18], are not suitable options for this mapping problem. In such cases, a local RCBPR or PATRIC identifier can be assigned and mapped to other identifier schemes as they become available.
When mappings between identifier schemes are not available, similarity-based matching may be used. One must determine an appropriate similarity cutoff and track metadata about sequences to avoid potentially mapping to an incorrect strain or organism (see Subsection 2).
1.4. Data Synchronization
Data integration methods must take into account the fact that data and especially its interpretation may change over time, and that at any given point in time, two or more databases that are, from a users’ perspective, perceived to contain the most up-to-date data, indeed may not. This problem of data synchronization introduces challenges above and beyond those faced when working with relatively static data.
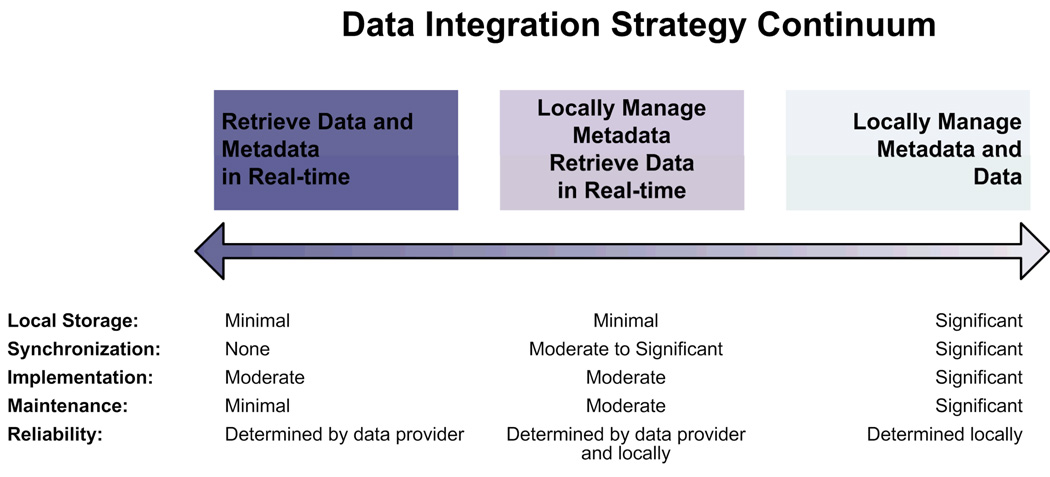
We present a continuum of data synchronization schemes (Figure 3) that range from real-time data access with no synchronization demands to a “copy and store” approach that requires software developers to completely manage synchronization across all data sources. Figure 3 presents three strategies within this continuum and summarizes the storage, synchronization, implementation, maintenance, and reliability demands of each strategy.
Figure 3.
Data synchronization strategies lie in a continuum that at a high level range from “access on demand” (left end), providing real-time access to data stored and managed by other data providers, to “copy and store” (right end), placing data management responsibilities solely on the primary Web resource. The tradeoffs associated with implementation, maintainability, and reliability allow software developers to adapt an approach to meet their specific needs.
When databases depend on a copy and store strategy for data synchronization, all data and metadata must be locally managed and stored, and data from external sources must be regularly refreshed to remain synchronized with the source system. In some cases, bulk data load programs take advantage of standardized data interchange formats (e.g., MGED [21] and GFF3 format [22]); however, significant effort is still required to develop reliable data loading programs that can gracefully degrade and recover from failure. Moreover, there are issues with the need for local storage: bandwidth and compute resources to execute bulk loads to keep local copies of data up to date; maintenance of a local database for storing the data and responding to queries; and the potential of not having the most up-to-date data between updates. In spite of these limitations, the high level of performance, reliability, and control over the data itself can often offset the disadvantages.
On the other end of the spectrum is the “access on demand” strategy which uses application programming interfaces (APIs), created by data providers, to allow external developers to programmatically access data in public repositories in real-time. Examples of APIs for life sciences include NCBI ’ s E-Utils (http://www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html/), EBI’s UniProt Java API (http://www.ebi.ac.uk/uniprot/remotingAPI/), and the Protein Data Bank (PDB) API for retrieving data using standard Web protocols (Figure 4). Instead of bulk loading large amounts of data in anticipation of user needs, developers can create applications that query a data source and retrieve just the information users need at any specific point in time or location within the Web resource. For example, when a PATRIC user navigates to the Brucella literature page, the PubMed API is used to retrieve, in real-time, the most recent Brucella publications. This approach has several advantages, the most obvious being that there is no overhead of copying and persistently storing data as described above. However, even robust APIs may not always scale well to, for example, the number of concurrent users or the size of result sets. In some cases, the data provider may not have the resources to support the networking and storage infrastructure required to meet community needs for on demand access.
Figure 4.
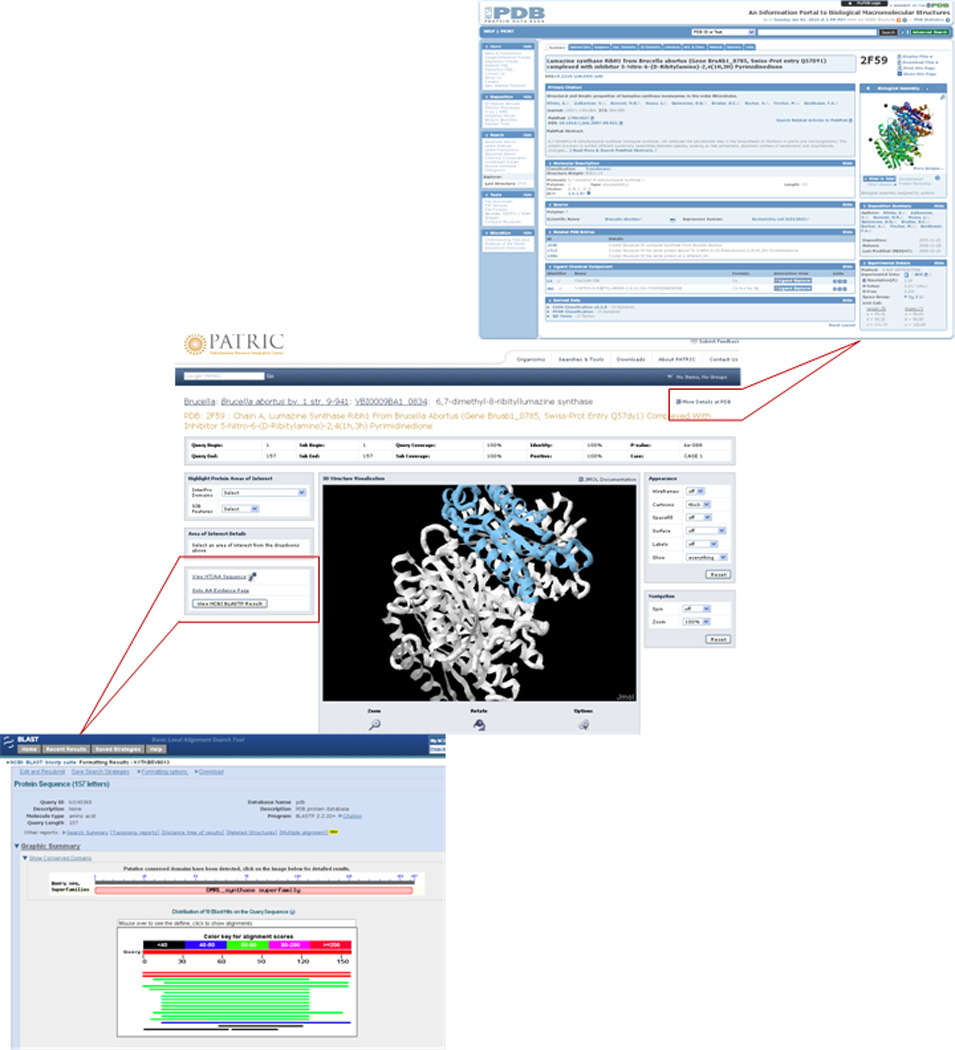
Protein structure data and BLASTP services are made available via API from PDB and NCBI, respectively. Structural data does not have to be stored locally to be integrated at a fine grained level of detail.
Database application developers must also take into account, and be able to mitigate, the risks of using an external data source. For example, if the external database is down or the API services are unavailable, the application using that API needs to degrade gracefully and avoid losing functionality. Moreover, data providers may place limits on the amount of data extracted in a single query or the total amount of data extracted by a single user within a given time period. With less control over the data source, developers may need to expend more effort on designing programs that gracefully degrade when an API is unavailable and effectively recover when the data sources become available again.
Experience with both store and copy and access on demand strategies leaves us with a distinct preference for the access on demand strategy unless the limitations of a particular API adversely impact required functionality. APIs allow for more rapid deployment and access to the latest data at minimal cost. Of course, in some cases, a hybrid strategy that lies in the middle of the continuum may be most appropriate for the specific application.
1.5. Literature Integration
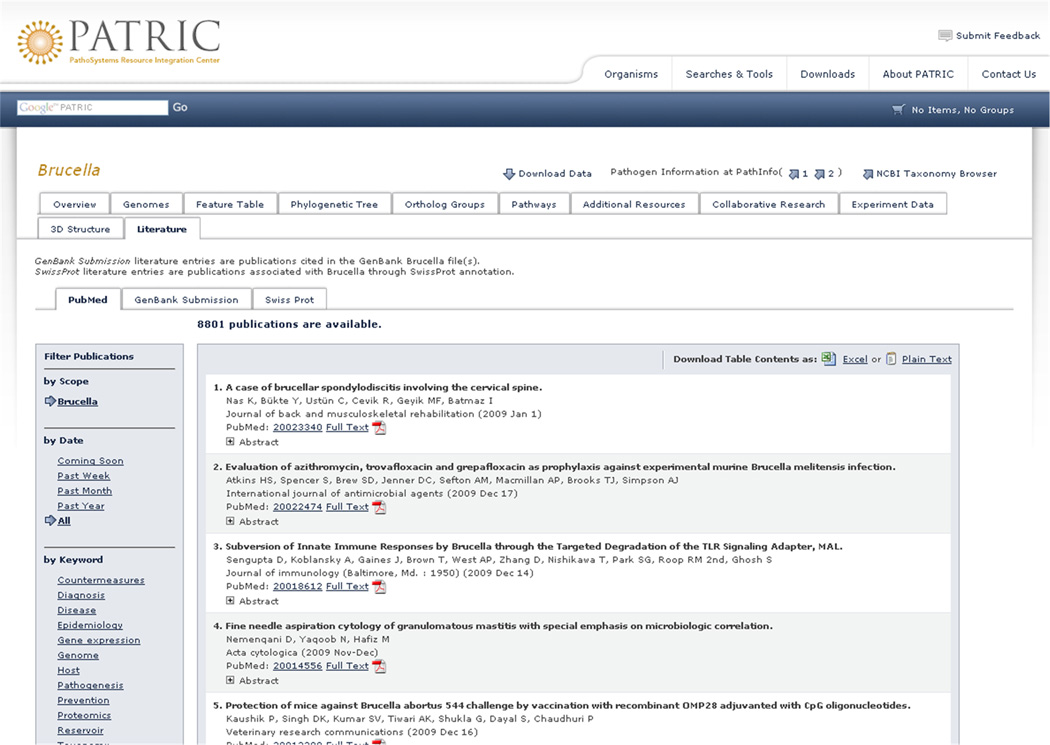
Based on users’ input (Figure 1), one of the top three requirements of an infectious disease research portal is literature integration. In both structured data integration and literature integration, we have issues of data synchronization, copy and store versus access on demand strategies, and control over infrastructure. Fortunately, literature searching APIs from NCBI [23] are readily available, appear to scale well, and do not impose limits on the amount of data queried or retrieved. Figure 5 shows the use of the PubMed API on the PATRIC website, allowing users to not only browse the most recent and relevant publications in real-time, but also filter (left side of Figure 5) by date and keyword in real-time.
Figure 5.
Literature is integrated into PATRIC using APIs providing access on demand. This strategy ensures that PATRIC users have access to a complete and up-to-date literature repository for life science research.
Developers have three broad options for literature integration. One is to implement the copy and store strategy with crawlers, such as wget (http://www.gnu.org/software/wget/). Minimal data quality checks are required with literature and copies can be refreshed as frequently as needed, limited by the bandwidth and compute resource allotted to refreshes. This approach duplicates the core functionality of sites such as MedLine, PubMed, and Google Scholar, albeit on a presumably much smaller scale. Another alternative is a modified copy and store strategy in which only metadata is downloaded and stored (Figure 3, center of continuum). This strategy is appropriate for applications, such as CiteULike (http://www.citeulike.org/), in which individuals and collaborators wish to track literature relevant to their work. It reduces storage requirements compared to the first option while allowing developers more control over access to metadata and how it is related to other locally stored data. This method continues to duplicate some of the functionality of PubMed, Medline, and Google Scholar. The third strategy is to use literature APIs for access on demand. This approach takes advantage of the literature search infrastructure provided by literature repositories and minimizes the amount of code required to add search capabilities into an application.
In addition to designing literature for efficient retrieval with sufficient levels of precision and recall, Web resource designers must address the need to navigate potentially large result sets. Both semantically aware tools [24–26] and usability design principals have been applied to address this challenge [27].
1.6. Post-Genomic Experiment Data Integration
Gene and protein prediction algorithms, while essential bioinformatics tools, cannot provide sufficient evidence for the existence and function of genes or proteins. Evidence from microarray and mass spectrometry experiments, however, complement these predictive tools and thus are valuable to vaccine, diagnostics, and therapeutics researchers. The rate of genome sequencing and subsequent predictive analysis exceeds the rate at which post-genomic experiments are preformed. The PATRIC application compensates for this gap by integrating experimental evidence based on orthologous groups of genes and proteins.
2. Usability Engineering
Data integration requirements are not isolated to the data or applications services layers. One of the contributions of the work descried here is to extend data integration techniques to include usability engineering. Specifically, usability engineering was used to address three sub-problems in data integration: the need for flexible user task flows; efficient navigation of large, integrated data sets; and supporting serendipitous discovery in intenerated data sets.
Users’ expectations of web-based systems are on the rise. The traditional “hunt and peck” approach, where users have to click through several links and wait for pages to load, only to have to back up and click down another path, is no longer acceptable to savvy web users. This is especially true with intensive data integration Web resources, where the volume and diversity of data demands a more sophisticated approach to user interface (UI) design. As such, attracting and retaining users by creating a compelling and truly useful web resource for integrated data services was one of our biggest challenges.
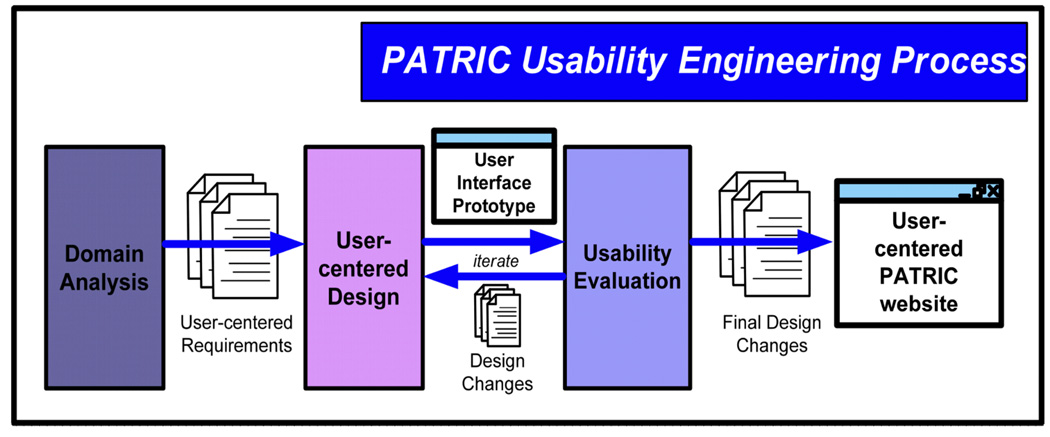
For the PATRIC website, we addressed these concerns by employing a well-established approach to developing user interfaces that identifies and addresses the most important data integration issues from a user’s perspective. Specifically, we applied a usability engineering approach to developing the PATRIC website, soliciting users’ needs and opinions through all stages of the software development lifecycle. Usability engineering is a cost-effective, user-centered process that ensures a high level of effectiveness, efficiency, and safety in complex interactive user interfaces [28]. High usability – ease of use and usefulness – is ensured via the usability engineering process, which revolves around early and continual representative user participation. Figure 6 shows the three major usability engineering activities, which are briefly described following the figure.
Figure 6.
We applied a usability engineering process to develop the PATRIC website. Using this approach, data integration implications on the user interface were identified and addressed early and iteratively throughout the website development lifecycle.
As shown in Figure 6, activities in the PATRIC usability engineering process include domain analysis, user–centered design, and various kinds of usability evaluations of the user interface. Domain analysis activities generate vital information used through the usability engineering process by determining answers to two critical questions: “Who are the users?” and “what specific tasks will they perform?” User-centered design activities employ artifacts of users’ current workflow and de facto standards in the domain (e.g., life sciences) to iteratively evolve a UI design from hand sketches, to static representations (e.g., PowerPoint slides), to working Web prototypes. This approach allows users to provide feedback on the most critical UI design elements early in the process without getting distracted by the polishing UI elements (e.g., color, font shape and size, icons, etc.). To evaluate user interfaces, we employ both expert evaluation (identifying PATRIC usability problems by comparing its UI to established usability design guidelines [29]) and user-based evaluations (assessing the PATRIC UI by having representative users perform benchmark tasks and observing the users’ performance while collecting quantitative and qualitative data to identify problems and redesign recommendations [30]).
The details of our strategy and experiences applying a usability engineering approach to PATRIC over the course of five years is beyond the scope of this paper. However, of interest are some of the basic design philosophies that we developed during our work in response to specific data integration challenges identified through early engagement with representative users. Prior work in data integration and usability has focused more on data modeling design tool usability and less on the usability of the application’s user interface [31]. Below, we present the challenges we identified that are rooted in data integration but have specific implications on user interface design. We also present the respective design philosophies and provide descriptive examples of UI design solutions.
2.1. Supporting Flexible User Task Flows
We learned early in the usability engineering process that PATRIC would need to support a wide variety of user classes, each with diverse types of user goals. This is very different than most traditional websites where users’ goals are very focused and prescripted. For example, on the online auction site eBay, users are either attempting to sell a product or bid on/purchase a product (with buyers needing to browse and search first in order to buy). Contrast this to PATRIC where our set of users goals can be defined (in part) by users background/expertise, where we identified many user classes representative of PATRIC users (e.g., microbiologist, structural biologist, immunologist, molecular biologist, drug/vaccine/diagnostics researcher/developer, epidemiologist, educators, and so on), each with their own set of specific end goals in mind and approaches to browsing, collecting, and analyzing PATRIC data. As a result, it is nearly impossible to know a priori the types of paths users will take through the PATRIC site in pursuit of their goal(s). Compounding the problem was the fact that PATRIC supported many different types of data and tools to generate/filter data. Given that there is no way of knowing exactly which sets of data types and tools would be useful for any specific user class or individual, we were faced with the challenge of creating a UI that allows users to easily access the data and tools they need without navigating in an antiquated “stove-piped” fashion. That is, we did not want to force users to have to navigate down a single prescribed path to find one type of data, then back up to the home page only to drill down a second prescribed path in order to find the second piece of data or tool, etc.
We have implemented an integrated approach to navigation through the PATRIC website. Our integrated approach allows users to seamlessly access, for example, organism summary information, tools, searches, and specific detailed data without having to go to different pages for each. While it is impossible to provide all possible data and tools on a single page, it is possible to provide numerous links to relevant data and tools throughout the website, resulting in fewer web pages that are “dead ends,” and more pages that offer obvious navigation paths to other related data and useful/common/preferred tools. As an example, PATRIC pages that present organism overview information (e.g., Brucella summary information) also provide numerous links to specific data types as well as specific searches and tools, resulting in an entry point for Brucella researchers that supports flexible navigation to many different areas of the site that may be of interest.
2.2. Finding the Needle in the Haystack
A challenge inherent in any large data integration effort is allowing users to wade through the vast amounts of data to find those small pieces of data that are relevant to their specific goals. The two broad categories of approaches to this problem prevalent on the Web today are search and browse. Google (http://www.google.com/) has changed the way in which users find relevant information by providing a simple text box for searching, while other sites such as Amazon. com (http://www.amazon.com/) and Home Depot (http://www.homedepot.com/) rely on hierarchical navigation schemes to support product browsing. While more advanced approaches to finding data are typically seen in these prominent commercial websites, many have not been adopted by life science websites. As such, most life science websites have antiquated or basic search and browse mechanisms that are cumbersome or inefficient to use. Complicating the matter is that even the best commercial approaches to browsing may not apply to life science with rich data integration due to the fact that the nature of the relationships between the data elements is not as straightforward as those in other domains.
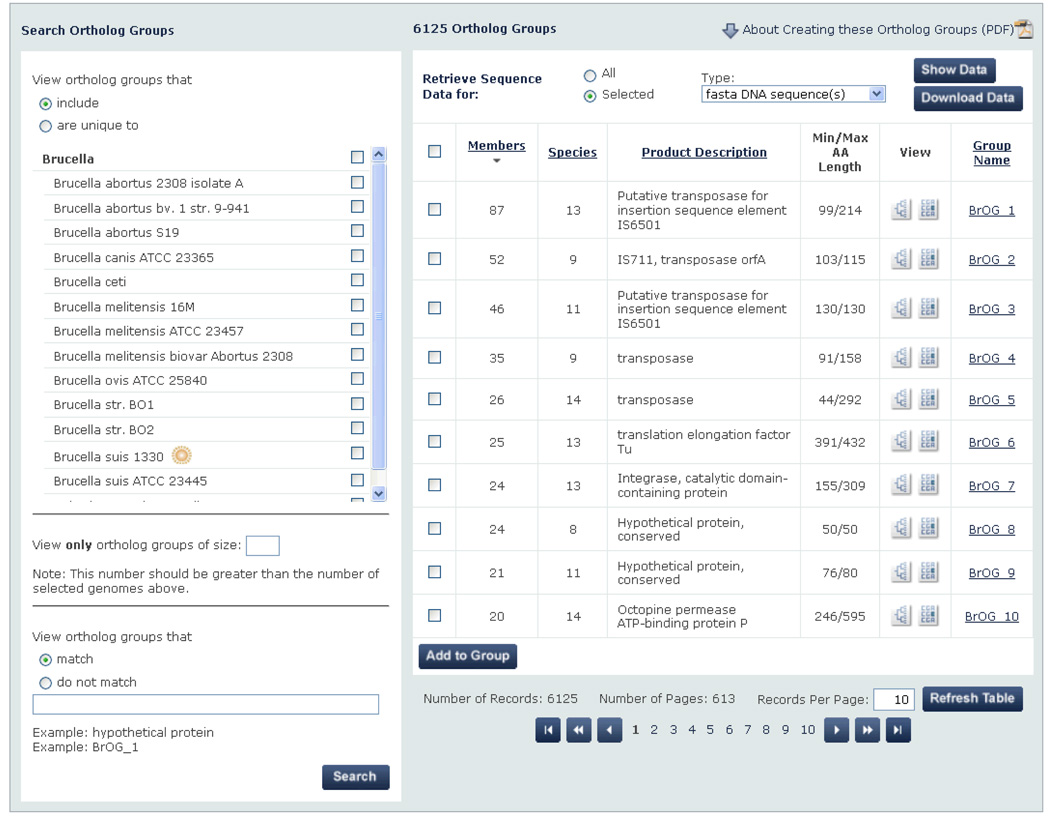
To address this challenge, we implemented progressive filtering where appropriate throughout the PATRIC website. Progressive filtering supports different levels of filtering and drill down capabilities within a single page, allowing users to easily remove information clutter and progressively create a smaller set of meaningful data. For example, as shown in Figure 7, users can winnow down a set of 6125 ortholog groups using the set of filters present in the left hand side of the page. When the filter is applied, the list of ortholog groups refreshes to display the resulting smaller set.
Figure 7.
Through embedded progressive filtering, users can narrow this list of 6125 ortholog groups (right hand side) using a variety of filtering criteria (left hand side – not all shown due to space constraints) integrated in a single workspace. This was one type of progressive filtering approach used in PATRIC to allow users to find the “needle in the haystack.”
2.3. Supporting Serendipitous Discovery
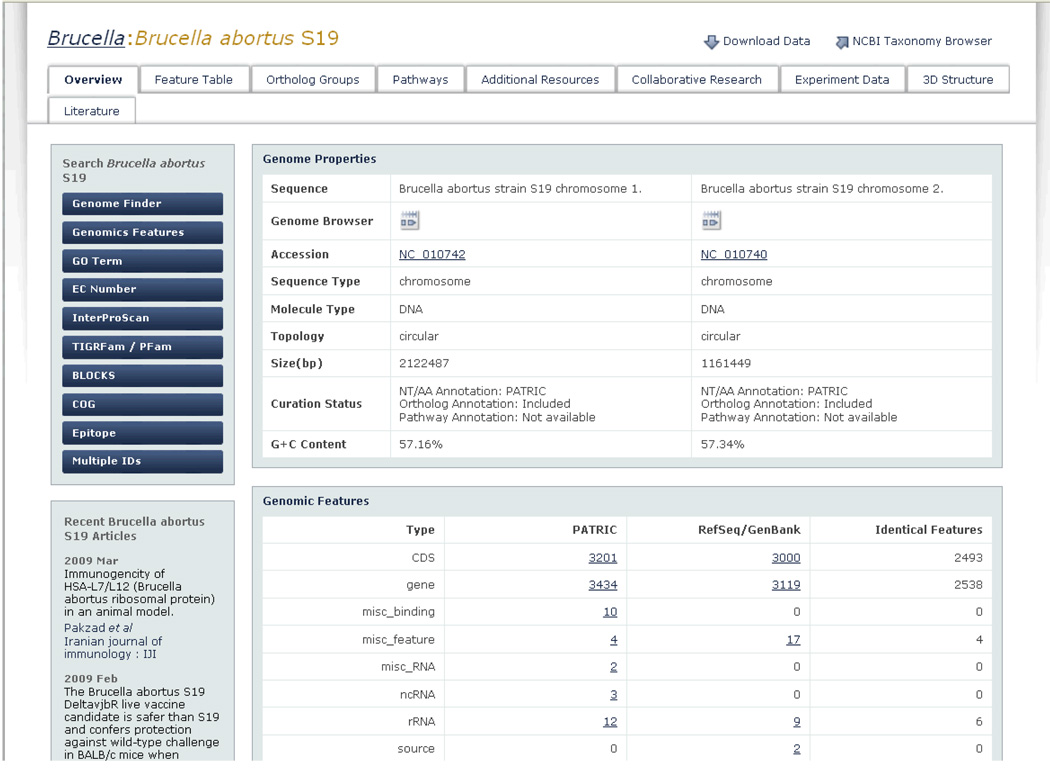
Users of websites or applications that store large volumes of diverse data are often focused on a specific task: employing pre-conceived notions of what type of data they are looking for and approaches to finding that data. These notions and approaches are learned over time and are often honed to optimize the efforts needed to accomplish a particular research task or goal. One of the interesting user-centered requirements we identified during our usability engineering efforts was that focused users want direct and intuitive access to data and tools to suit their needs. However, they are also interested in related information and tools that may provide additional insight into their problem or alternatively enhance a user’s interest so that an alternate path may be taken, sometimes resulting in a serendipitous discovery. Here we define “serendipitous discovery” as some newfound knowledge or association between data that is unplanned or unexpected. As we designed the PATRIC UI, we wanted to embrace this concept by providing a rich and meaningful workspace with context-sensitive access to related data and tools. For example, when considering a Brucella abortus S19 researcher looking for a specific protein or orthologous groups of proteins, instead of simply creating pages that provide details on proteins and ortholog groups, we also created Brucella abortus S19 workspace entry pages with direct access to information on Brucella abortus S19 related pathways, experiment data, and literature. While a particular researcher may not choose this path, the presence of these context-sensitive visual cues allows users to at least be aware that this information exists.
To meet this goal, we employed the notion of context sensitivity, allocating screen real estate to other data and tools related specifically to the current context. This context-sensitive approach provides options (controls, filters, tools, access to data, and so on) that are appropriate to the user’s current scope. Figure 8 depicts a specific example where the current context is the specific genome Brucella abortus S19. While the page’s main goal is to provide overview information on Brucella abortus S19, we also provide context-sensitive search tools specific to Brucella abortus S19 (on the left side of the screen), as well as access to other data (e.g., 3D structures) specific to Brucella abortus S19. This same design philosophy is used for all genome overview pages throughout PATRIC, with access to context-sensitive tools and data geared to each respective genome. In this manner, we provide consistent design (e.g., search tools on the left, access to related data on top), but with context-sensitive scoping of data.
Figure 8.
To support serendipitous discovery and relationships between data, PATRIC provides context-sensitive access to multiple tools and data types. Through the use of visual cues (e.g., visually distinct tabs and buttons) users are given the opportunity to view relevant data that they may not have thought to seek out (e.g., experiment data).
3. Lessons Learned
Accommodate multiple constraints on data and derived information
Experiment data should reflect the actual data generated in experiments as well as reflect the information contained in the data. The data produced in a Brucella melitensis biovar Abortus 2308 experiment indicated peptides and proteins belonging to other strains of Brucella were found, although experimenters did not include other strains in the experiment. Both the actual data produced and the experimenters’ interpretation of the data should be available to researchers using the PATRIC and RCBPR applications. Similarly, when potential discrepancies arise, as much information as possible should be released to allow researchers using the application to reason about the results.
Implement formal quality control procedures
Bioinformatics data can reflect complex biological phenomenon and that often implies large volumes of data and complex data structures. Errors can and do occur. Use formal quality control procedures including staging schemas to isolate new data sets and quality control checks to detect broad classes of errors, such as missing data, referential integrity violations, and values outside expected ranges.
Standardize identifier mapping procedures
Multiple identifier schemes and the need to map between them will likely continue. Standardize rules and procedures for mapping between identifier schemes and re-use them across applications. This improves consistency and lowers the cost of maintaining mapping services.
Use on demand data access when possible
Retrieving data as needed can reduce data storage and management overhead; reduce extraction, transformation, and load complexity; and reduce costs. The scalability, reliability, and availability of APIs limit the use of access on demand data.
Employ a usability engineering process
Applying a usability engineering process not only produces more practical user interfaces, but also can identify potential end-user challenges inherent in sites with large, diverse data integration. Identifying these challenges early in the website development process ensures that they can be addressed early, which in turn reduces website development costs. A convenient aspect of usability engineering is that the process is very flexible, allowing website development teams to scope usability engineering efforts to best meet the project needs. Even the most modest usability engineering efforts are always better than none.
Survey the commercial webspace to inform design
Professional website developers in high-profile commercial sites have already examined user interface challenges associated with data integration. By examining these sites, designers and developers can get ideas on how others are providing intuitive user access to complex information spaces.
Conclusions
Data integration is a common requirement in infectious disease research. The PATRIC and RCBPR projects indicate that practical, useful data integration is possible, but there are challenges. These challenges can be met with a combination of design principles, data quality control measures, and usability engineering practices. Although we have made progress with respect to some challenges, others remain. In the future we will address the need for canonical data structures for describing important bioinformatic entities, e.g., genes, proteins, annotations, and provenance. Much of the work in this area will be done by community collaboration and we anticipate adopting community standards for describing complex data structures as they become available. We also anticipate improving the amount, quality, and accessibility of metadata. Semantic Web representations and reasoning methods hold the promise of more automated analysis and will be incorporated into future work [32]; Semantic Web approaches have already been adopted in a number of life sciences applications [4, 13, 33–39]. For subsequent releases of the PATRIC website, we will be soliciting user feedback through a number of new mechanisms, including bi-annual users workshops (to support user-based evaluation), as well as online surveys to gather feedback from online users as they experience usability problems or think of website enhancements.
Acknowledgements
PATRIC is funded through NIAID contract number HHSN266200400035C awarded to Dr. Bruno Sobral; RCBPR is funded through NIAID contract number HHSN266200400061C awarded to SSS with a subcontract to Dr. Bruno Sobral. Thanks to Lisa Gunderman for assisting with proofreading and formatting the manuscript.
REFERENCES
- 1.Atkins D, Droegemeier K, et al. National Science Foundation. 2003 [Google Scholar]
- 2.Blake J, Bult C. Biomedical Ontologies. 2006;39:314. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Bodenreider O. Data Integration in the Life Sciences. 2008:1. [Google Scholar]
- 4.Cheung K-H, Yip KY, Smith A, deKnikker R, Masiar A, Gerstein M. Bioinformatics. 2005;21:i85. doi: 10.1093/bioinformatics/bti1026. [DOI] [PubMed] [Google Scholar]
- 5.Daehee, Jennifer, Deena, Andrea, Alistair, Stephen, Pedro, Andrew, Hamid Proceedings of the National Academy of Science. 2005;102 [Google Scholar]
- 6.Goble C, Stevens R. J Biomed Inform. 2008 doi: 10.1016/j.jbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 7.Hwang D, Smith JJ, Leslie DM, Weston AD, Rust AG, Ramsey S, de Atauri P, Siegel AF, Bolouri H, Aitchison JD, Hood L. Proc Natl Acad Sci U S A. 2005;102:17302. doi: 10.1073/pnas.0508649102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joyce AR, Palsson BO. Nat Rev Mol Cell Biol. 2006;7:198. doi: 10.1038/nrm1857. [DOI] [PubMed] [Google Scholar]
- 9.Lenzerini M. PODS '02: Proceedings of the twenty-first ACM SIGMOD-SIGACT-SIGART symposium on Principles of database systems; ACM Press; 2002. p. 233. [Google Scholar]
- 10.Louie B, Mork P, Martin-Sanchez F, Halevy A, Tarczy-Hornoch P. J Biomed Inform. 2007;40:5. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Orchard S, Hermjakob H, Julian RK, Jr, Runte K, Sherman D, Wojcik J, Zhu W, Apweiler R. Proteomics. 2004;4:490. doi: 10.1002/pmic.200300694. [DOI] [PubMed] [Google Scholar]
- 12.Parker D, Hsiao R, Xing Y, Resch A, Lee C. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 2008;5:432. doi: 10.1109/TCBB.2007.1073. [DOI] [PubMed] [Google Scholar]
- 13.Post LJG, Roos M, Marshall MS, van Driel R, Breit TM. Bioinformatics. 2007;23:3080. doi: 10.1093/bioinformatics/btm461. [DOI] [PubMed] [Google Scholar]
- 14.Sahoo Satya S, Bodenreider O, Zeng K, S A. 16th International World Wide Web Conference (WWW2007); Banff, Canada. 2007. [Google Scholar]
- 15.Shulaev V. Brief Bioinform. 2006;7:128. doi: 10.1093/bib/bbl012. [DOI] [PubMed] [Google Scholar]
- 16.Wang K. Proceedings, American Medical Informatics Association Fall Symposium; 2006. p. 779. [Google Scholar]
- 17.Xiang Z, Tian Y, He Y. Genome Biol. 2007;8:R150. doi: 10.1186/gb-2007-8-7-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cote R, Jones P, Martens L, Kerrien S, Reisinger F, Lin Q, Leinonen R, Apweiler R, Hermjakob H. BMC Bioinformatics. 2007;8 doi: 10.1186/1471-2105-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith M, Kunin V, Goldovsky L, Enright AJ, Ouzounis CA. Bioinformatics. 2005;21:3429. doi: 10.1093/bioinformatics/bti548. [DOI] [PubMed] [Google Scholar]
- 20.Iragne F, Barre A, Goffard N, de Daruvar A. Bioinformatics. 2004;20:2331. doi: 10.1093/bioinformatics/bth241. [DOI] [PubMed] [Google Scholar]
- 21.Whetzel P, Parkinson H, Causton H, Fan L, Fostel J, Fragoso G, Game L, Heiskanen M, Morrison N, Rocca-Serra P, Sansone S-A, Taylor C, White J, Stoeckert C. Bioinformatics. 2006;22:866. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 22.S. O. Project.
- 23.National Center for Biotechnology Information. 2009 [Google Scholar]
- 24.Doms A, Schroeder M. Nucl. Acids Res. 2005;33:W783. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanteru B, Shaik J, Yeasin M. BMC Genomics. 2008;9:S10. doi: 10.1186/1471-2164-9-S1-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muller H-M, Kenny EE, Sternberg PW. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolchini D, Finkelstein A, Perrone V, Nagl S. Bioinformatics. 2009;25:406. doi: 10.1093/bioinformatics/btn633. [DOI] [PubMed] [Google Scholar]
- 28.Hix D, Hartson HR. Developing User Interfaces: Ensuring Usability through Product and Process. New York, New York: John Wiley, Inc.; 1993. [Google Scholar]
- 29.Nielsen J. Usability Engineering. Boston, MA: Academic Press Professional; 1993. [Google Scholar]
- 30.Hix D, Hartson H. Developing User Interfaces: Ensuring Usability through Product and Process. New York: John Wiley and Sons, Inc.; 1993. [Google Scholar]
- 31.Dominovic C. IADIS European Conference Data Mining 2008; Amsterdam, The Netherlands. 2008. [Google Scholar]
- 32.Cannata N, Schröder M, Marangoni R, Romano P. BMC bioinformatics. 2008;9 Suppl 4 doi: 10.1186/1471-2105-9-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith AK, Cheung KH, Yip KY, Schultz M, Gerstein MK. BMC Bioinformatics. 2007;8 Suppl 3:S5. doi: 10.1186/1471-2105-8-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith A, Cheung K, Krauthammer M, Schultz M, Gerstein M. Bioinformatics. 2007;23:3073. doi: 10.1093/bioinformatics/btm452. [DOI] [PubMed] [Google Scholar]
- 35.Kohler J, Philippi S, Lange M. Bioinformatics. 2003;19:2420. doi: 10.1093/bioinformatics/btg340. [DOI] [PubMed] [Google Scholar]
- 36.Lam H, Marenco L, Clark T, Gao Y, Kinoshita J, Shepherd G, Miller P, Wu E, Wong G, Liu N, Crasto C, Morse T, Stephens S, Cheung K-H. BMC Bioinformatics. 2007;8:S4. doi: 10.1186/1471-2105-8-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belleau1 F, Nolin M, Tourigny N, Rigault P, Morissette J. WWW 2007. Canada: Banff; 2007. [Google Scholar]
- 38.Cases I, Pisano DG, Andres E, Carro A, Fernandez JM, Gomez-Lopez G, Rodriguez JM, Vera JF, Valencia A, Rojas AM. Nucl. Acids Res. 2007;35:W16. doi: 10.1093/nar/gkm280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freifeld CC, Mandl KD, Reis BY, Brownstein JS. J Am Med Inform Assoc. 2008;15:150. doi: 10.1197/jamia.M2544. [DOI] [PMC free article] [PubMed] [Google Scholar]