Summary
Activation of the c-JUN N-terminal kinase (JNK) pathway is implicated in a number of important physiological processes, from embryonic morphogenesis to cell survival and apoptosis. JNK stimulatory phosphatase 1 (JSP1) is a member of the dual specificity phosphatase subfamily of protein tyrosine phosphatases (PTPs). In contrast to other dual specificity phosphatases, which catalyze inactivation of mitogen-activated protein kinases, expression of JSP1 activates JNK-mediated signaling. JSP1 (and its relative DUSP15) are unique among members of the PTP family in that they contain a potential myristoylation site at the N-terminus (MGNGMXK). In this study, we investigated whether JSP1 was myristoylated and examined the functional consequences of myristoylation. Using mass spectrometry, we showed that wild type JSP1, but not a JSP1 mutant in which glycine 2 was mutated to alanine (JSP1-G2A), was myristoylated in cells. Abrogation of myristoylation did not impair the intrinsic phosphatase activity of JSP1, but changed the subcellular localization of the enzyme. Compared to wild type, the ability of non-myristoylated JSP1 to induce JNK activation and phosphorylation of the transcription factor c-JUN was attenuated. Upon expression of wild type JSP1, a subpopulation of cells, with highest levels of the phosphatase, was induced to float off the dish and undergo apoptosis. In contrast, cells expressing similar levels of JSP1-G2A remained attached, further highlighting that the myristoylation mutant was functionally compromised.
Keywords: Apoptosis, JNK, JSP1, myristoylation, phosphatase
Introduction
Mitogen-activated protein kinases (MAPK) signaling pathways are critical regulators of cellular responses to environmental stimuli, such as growth signals and stress, that modulate cell behavior, such as proliferation, differentiation, or cell death [1–4]. All MAPK pathways consist of a central three-tiered core signaling module in which MAPK kinase kinases (MAPKKK or MEKK) phosphorylate MAPK kinases (MAPKK or MEK) on Ser/Thr residues with concomitant activation. MAPKKs are dual specificity kinases that, upon activation, phosphorylate both the Tyr and Thr residue of the conserved TXY motif in the activation loop of MAPKs, resulting in MAPK activation. Activated MAPKs phosphorylate specific Ser and Thr residues in target substrates, which include effector protein kinases, like MAPK-activated protein kinases (MAPKAP-Ks) and transcription factors, such as Activator Protein-1 (AP-1) [1–3].
Four major subgroups of MAPKs have been delineated in mammals, i.e. extracellular signal-regulated kinases (ERK1/2), Jun N-terminal kinases (JNK1/2/3), p38 proteins (p38α/β/γ/δ), and ERK5, which are activated by distinct sets of stimuli [1–4]. Of particular importance to this study is the JNK family of MAPKs, which are predominantly activated by proinflammatory cytokines and a variety of environmental stresses [2, 4]. The JNK family comprises three distinct genes, JNK1-3, with further structural diversity due to alternative mRNA splicing. The Jnk1 and Jnk2 genes are expressed ubiquitously, whereas expression of Jnk3 is largely restricted to brain, heart, and testis [4, 5]. JNK is phosphorylated and activated by the MAPKKs MKK4 and MKK7 [6, 7], with MKK7 primarily responding to cytokines, whereas MKK4 is preferentially activated by environmental stress [5, 8]. Depending on the stimulus and cellular context, the JNK pathway has been implicated in both apoptosis and cell survival [9].
The duration and extent of MAPK activation depends not only upon the activity of kinases, but also the protein phosphatases that dephosphorylate the Tyr and Ser/Thr residues in substrate proteins that are part of the MAPK signaling modules. Although protein phosphatases have long been viewed as negative regulators that terminate MAPK signaling, it is now evident that they play an important role in determining the magnitude and duration of MAPK activation, which determines the cellular response [10, 11]. Moreover, protein phosphatases can also regulate MAPK signaling positively. For example, the prototypic member of the protein tyrosine phosphatase (PTP) superfamily, PTP1B, acts as a positive mediator of the ErbB2-induced signaling pathways that trigger mammary tumorigenesis and metastasis [12, 13], and the tyrosine phosphatase SHP-2 is necessary for activation of ERK in response to a number of growth factors, including insulin growth factor-1, platelet-derived growth factor, and epidermal growth factor [14, 15].
Various protein phosphatases have been implicated in the regulation of MAPK signaling, including the subfamily of PTPs known as dual-specificity phosphatases (DSPs) [16, 17]. DSPs form a structurally and functionally heterogeneous subgroup of the PTP superfamily, and share little sequence similarity beyond the conserved active-site signature motif HCX5R. Although they use the same catalytic mechanism as the classical PTPs, the catalytic cleft of DSPs is shallower, which allows accommodation of both phosphorylated Ser/Thr and Tyr residues [18]. Several DSPs have been established as MAPK phosphatases (MKPs) that dephosphorylate the Tyr and Thr residues in the activation loop of MAPKs and thereby attenuate signaling [17, 19]. In addition, there is a group of low molecular weight DSPs that lack the regulatory N-terminal Cdc25 homology domain found in the MKPs [18]. One member of this subgroup is the dual-specificity phosphatase 22 (DUSP22), which was first identified by this lab as JNK-stimulatory phosphatase 1 (JSP1) [20]. Subsequently, it was also reported as JNK pathway-associated phosphatase (JKAP), which is a splice isoform of JSP1 [21], Low-molecular weight DSP2 (LMW-DSP2) [22], and VHR-related MKPX (VHX) [23]. JSP1 is expressed in multiple tissues [20, 23], although expression of the murine splice isoform JKAP was shown to be testis- and liver-specific [21]. JSP1 preferentially dephosphorylates Tyr residues in assays in vitro [20] and was shown to stimulate JNK activation specifically, thus acting as a positive regulator of JNK signaling [20, 21]. However, other reports have implicated JSP1 in the negative regulation of MAPK function [22–25].
JSP1 contains a putative myristoylation consensus sequence at its N-terminus (Met-Gly-X3-Asn-Lys-). In the present study, we aimed to confirm JSP1 myristoylation by mass spectrometry, as well as to analyze its possible role in regulating JSP1 functional activity. We demonstrated that JSP1 was myristoylated at its N-terminus, which was not necessary for the intrinsic phosphatase activity of the enzyme. However, myristoylation determined the subcellular localization of JSP1, and was required for JSP1-induced activation of the JNK signaling pathway. When overexpressed, wild type JSP1, but not a myristoylation mutant, induced apoptosis in cells, further highlighting the importance of myristoylation for JSP1 functional activity.
Results
JSP1 was myristoylated in cells
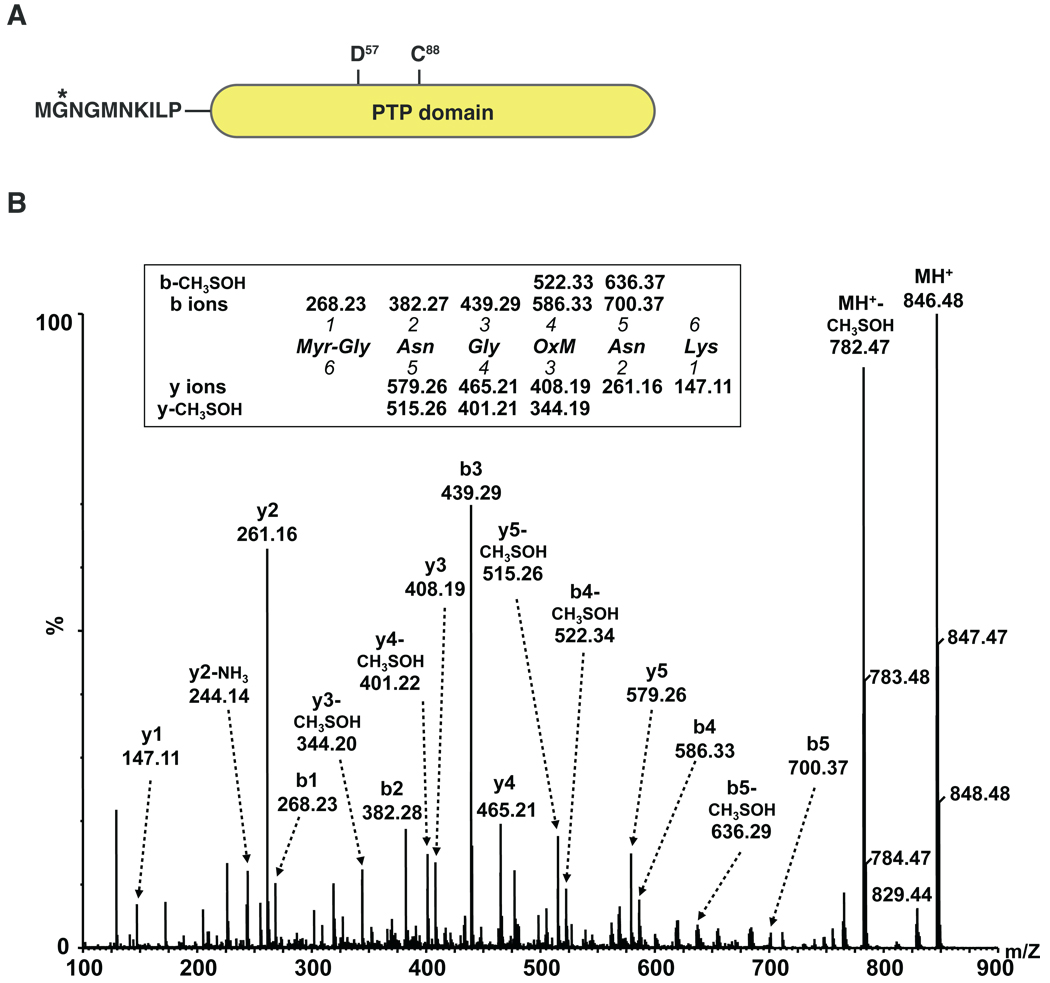
JSP1 contains the putative myristoylation consensus sequence Met-Gly-X3-Asn-Lys- at its N-terminus (Fig. 1A). To test whether JSP1 was myristoylated in cells, the phosphatase was overexpressed and isolated from a human cell line and analyzed by mass spectrometry. We constructed a GFP-tagged version of JSP1, with the tag being added to the C-terminus in order not to interfere with myristoylation. The construct was expressed in 293T cells, GFP-tagged JSP1 was immunoprecipitated from whole cell lysates and analyzed by nanoflow LC/ESI-MS/MS. The tandem mass spectrum of the N-terminal tryptic peptide GNGMNK from wild type JSP1 (JSP1-wt) revealed myristoylation of the first glycine residue (Fig. 1B), which was not detectable in a mutant form of JSP1 in which the myristoylation site (Gly2) was mutated to alanine (JSP1-G2A) (data not shown). The methionine residue in this peptide was oxidized, which occurred either in the cells or during sample preparation. The singly-charged peptide with monoisotopic mass of 846.48, which corresponds to the predicted protonated mass of the myristoylated and oxidized peptide, was selected for sequencing. In addition to all of the predicted b and y fragment ions, abundant peaks due to the neutral loss of CH3SOH (molecular mass of 64 Da) from the oxidized methionine residue were observed which confirmed the interpretation of the MS/MS spectrum [26]. The N-terminal peptide eluted late in the reverse phase HPLC gradient during LC-MS experiments, as would be expected for a hydrophobic (myristoylated) peptide (data not shown).
Fig 1.
JSP1 was myristoylated at its N-terminus. (A) Schematic representation of JSP1. The potential myristoylation site (Glycine 2) is indicated by an asterisk. Amino acids critical for catalysis (D57 and C88) are highlighted. (B) ESI-QTOF tandem mass spectrum of the N-terminal tryptic peptide GNGMNK from wild type JSP1. The singly-charged peptide with monoisotopic mass of 846.48 was selected for sequencing.
JSP1 phosphatase activity was not dependent on myristoylation
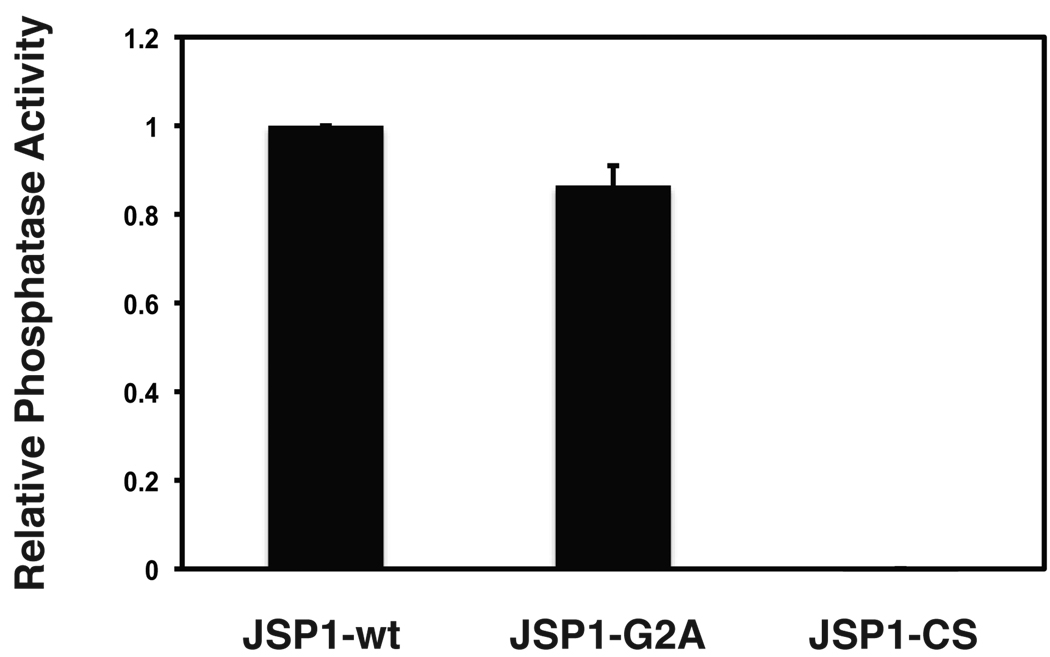
Having confirmed that JSP1 was myristoylated in cells, we tested whether myristoylation was essential for the intrinsic phosphatase activity of JSP1. We examined the activity of JSP1-wt, JSP1-G2A or an inactive mutant, in which the active site cysteine (Cys88) was changed to serine (JSP1-CS). These were expressed with a C-terminal 6xHis-tag in E. coli, and the recombinant protein was purified using Ni-NTA Sepharose. Since JSP1 preferentially dephosphorylates Tyr residues [20], we used 32P-labeled reduced carboxamidomethylated and maleylated lysozyme (RCML) as a substrate to determine phosphatase activity in vitro. Whereas JSP1-CS did not show any phosphatase activity, JSP1-wt and the -G2A mutant displayed comparable specific activities (Fig. 2), indicating that myristoylation was not necessary for the intrinsic phosphatase activity of the enzyme.
Fig 2.
JSP1 phosphatase activity was not dependent on myristoylation. Recombinant wild type JSP1 (JSP1-wt), the myristoylation mutant (JSP1-G2A), or a catalytically inactive mutant (JSP1-CS) were incubated with 32P-labeled reduced carboxamidomethylated and maleylated lysozyme (RCML) as substrate, and phosphatase activity was measured. Results (shown as activity relative to wild type phosphatase) are the mean of two experiments (±SEM).
Myristoylation regulated the subcellular localization of JSP1
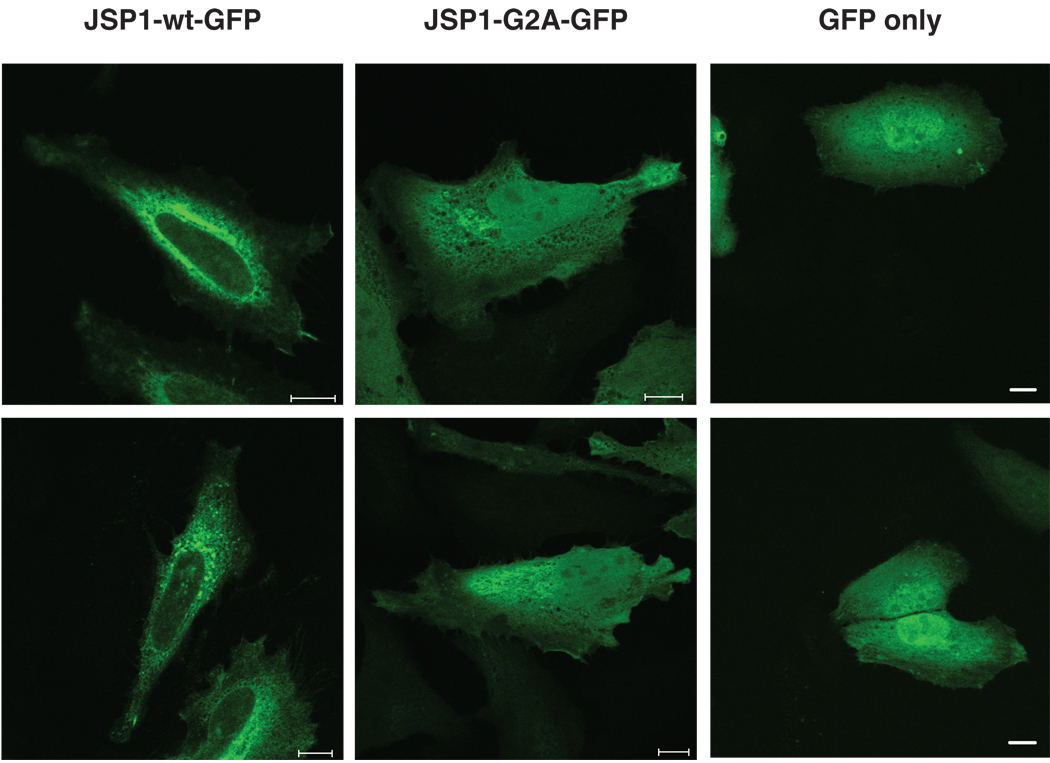
Myristoylation is a post-translational modification that can target proteins to the plasma membrane. In order to determine the subcellular localization of myristoylated versus non-myristoylated JSP1, we expressed GFP-tagged JSP1-wt or -G2A in MCF-7 cells, and analyzed JSP1 localization by confocal laser scanning microscopy (Fig. 3). Whereas JSP1-wt localized to distinct sites in the cytoplasm, and was excluded from the nucleus, JSP1-G2A was uniformly distributed throughout the cell. Cells transfected with a control GFP expression plasmid displayed a uniform distribution of GFP in the cytoplasm and nucleus, excluding the possibility that the distinct localization of JSP1-wt could be due to the attached tag (Fig 3). This suggested that myristoylation determined the subcellular localization of JSP1 in cells. While the JSP1-G2A mutant displayed diffuse cytoplasmic and nuclear localization, the perinuclear pattern of JSP1-wt expressing structures was indicative of localization with intracellular membrane structures. We tested colocalization of JSP1-wt with endosomal and Golgi structures using specific markers (EEA1, TfnR for endosomes and GM130, TGN46 for Golgi) and found that JSP1-wt partially colocalized with the Golgi apparatus and showed minimal colocalization with endosomes (data not shown).
Fig 3.
Myristoylation regulated the subcellular localization of JSP1. HeLa cells transfected with plasmids encoding GFP-tagged JSP1-wt, JSP1-G2A, or GFP only were analyzed by confocal laser scanning microscopy. Each image represents a single confocal section acquired through the plane of the nucleus. Two representative sections are shown (Scale bar, 10 10µm).
Myristoylation was necessary for JSP1-induced activation of JNK
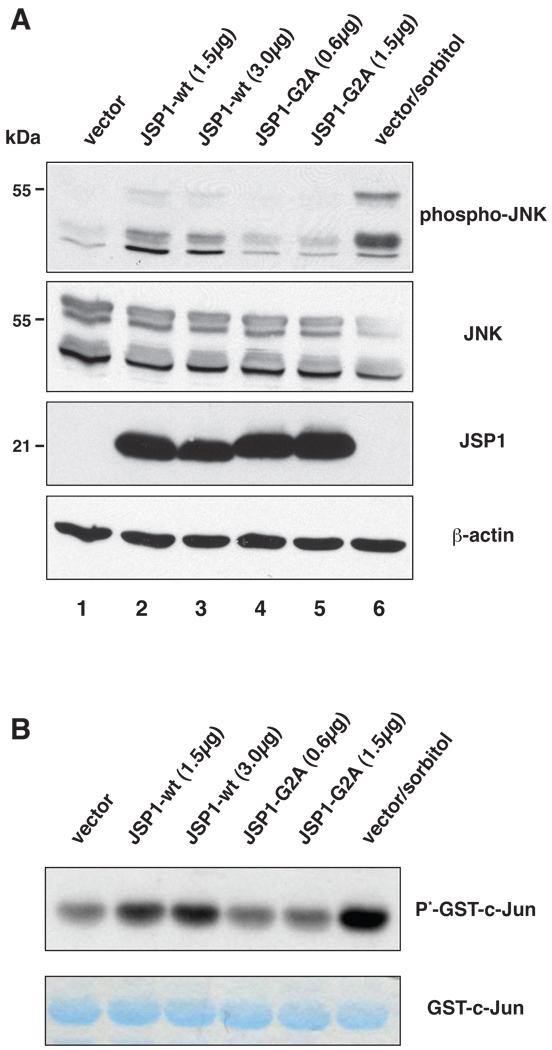
In contrast to most phosphatases, which negatively regulate MAPK signaling, JSP1 was shown to be a positive regulator of the JNK signaling pathway [20, 21]. Since myristoylation determined the subcellular localization of JSP1, we asked whether abrogation of myristoylation would also affect JSP1-induced JNK activation. We transfected Cos-1 cells with expression constructs encoding JSP1-wt or the -G2A mutant, and determined phosphorylation of JNK at Thr183 and Tyr185 using phospho-specific antibodies. In contrast to JSP1-wt, the -G2A mutant failed to stimulate JNK phosphorylation (Fig. 4A, compare lanes 2–3 and 4–5). Notably, JSP1 seemed to stimulate preferentially the phosphorylation of the p46-isoform of JNK, whereas sorbitol treatment induced phosphorylation of both the p46- and p55-isoforms. To confirm functional activation of the JNK signaling pathway, we performed a solid-phase kinase assay using the downstream transcription factor c-JUN as substrate. In accordance with JNK phosphorylation, expression of JSP1-wt, but not the myristoylation mutant, enhanced phosphorylation o f c-JUN (Fig. 4B). Thus, myristoylation was necessary for JSP1-induced activation of the JNK signaling pathway.
Fig 4.
Myristoylation was essential for JSP1-induced activation of JNK. (A) Cos-1 cells were transfected with vector only or different amounts of JSP1-wt or -G2A mutant expression plasmid, and total cell lysates were analyzed by immunoblotting with a phospho-specific antibody to JNK. Sorbitol treatment [500 mM] was included as positive control. Total levels of JNK and JSP1 were analyzed by immunoblotting with a JNK- and JSP1-specific antibody, respectively. The level of β-actin was determined as additional loading control. (B) Cos-1 cells were transfected with vector only or different amounts of JSP1-wt or -G2A mutant expression plasmid, and endogenous JNK was precipitated from total cell lysates using recombinant GST-tagged c-Jun. Precipitates were incubated with [γ-32P]-ATP, and c-Jun phosphorylation was determined by autoradiography. Total GSTc-Jun levels were visualized with GelCode® Blue Stain Reagent.
Wild type JSP1, but not the myristoylation mutant, induced cell death
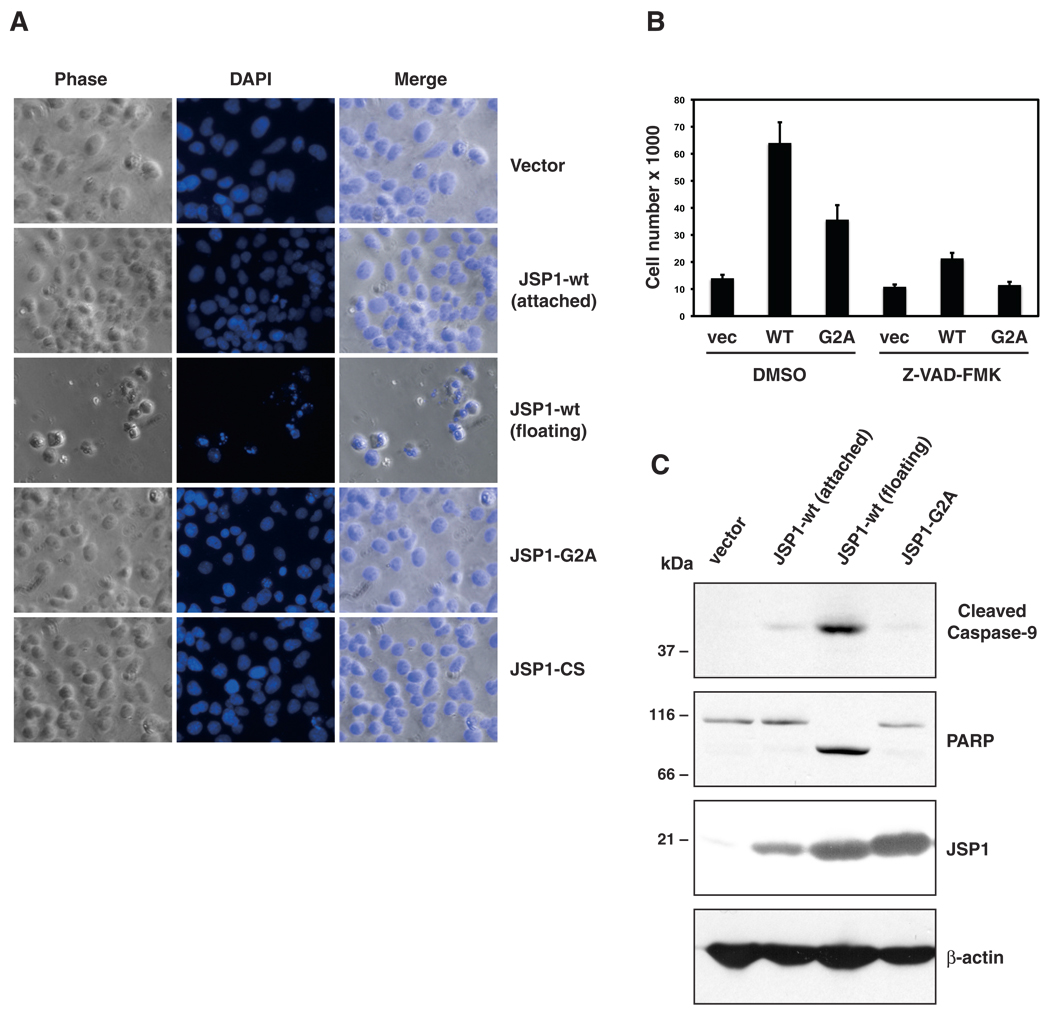
During the course of our experiments, we observed that ~ 30% of the cells transfected with the JSP1-wt expression construct started floating off the dish 24 hrs post-transfection. In contrast, cells expressing JSP1-G2A, or the inactive mutant JSP1-CS, remained attached to the culture dish. Anchorage-dependent cells normally undergo apoptosis after losing contact with neighbouring cells or the extracellular matrix in a process termed anoikis. To test whether the detached cells displayed features of apoptosis, we analyzed the phenotype of transfected cells after staining with DAPI (Fig. 5A). We observed that the floating cells showed condensation of chromatin, a morphological characteristic of apoptosis [27]. This was in contrast to cells expressing JSP1-G2A or -CS, or to those transfected cells expressing JSP1-wt that remained attached to the culture dish. Since detachment of cells and condensation of chromatin also occurs during mitosis [28], we tested the impact of the pan-caspase inhibitor Z-VAD-FMK on cell floating (Fig. 5B). Compared to the DMSO control, Z-VAD-FMK significantly reduced the number of floating JSP1-wt-transfected cells. To confirm further that JSP1-wt-transfected cells undergo apoptosis, we analyzed lysates from transfected cells by immunoblotting with antibodies specific for cleaved Caspase-9 and PARP (Fig. 5C). We detected both cleaved caspase-9 and PARP in JSP1-wt expressing, floating cells. In contrast, cells expressing the -G2A mutant, and those transfected with the JSP1-wt expression construct that remained attached to the dish, did not show cleavage of Caspase-9 or PARP. Interestingly, the floating cells expressed considerably higher levels of JSP1-wt than those cells that remained attached to the culture dish. Furthermore, cells expressing the JSP1-G2A mutant remained attached to the dish despite the fact that the mutant protein was expressed at similar levels to those of the wild type protein that was encountered in the floating cells (Fig. 5C). Taken together, these data indicated that JSP1-wt, but not the -G2A mutant, induced detachment of cells and induction of apoptosis, further demonstrating that the myristoylation mutant was functionally impaired.
Fig 5.
Wild type JSP1, but not the myristoylation mutant, induced cell death. (A) Cos-1 cells were transfected with expression plasmids for JSP1-wt, -G2A or -CS, stained with DAPI, and analyzed by fluorescence microscopy. (B) Cos-1 cells were transfected with vector only (vec), JSP1-wt (WT) or -G2A (G2A) and simultaneously treated with the caspase inhibitor Z-VAD-FMK or DMSO as control. Cells detached from the culture dish were collected, stained with Trypan Blue and counted. Results are the mean of three experiments (±SEM). (C) Cos-1 cells were transfected with vector only, JSP1-wt or -G2A. Cells detached from the culture dish and attached cells were collected separately, and total cell lysates were analyzed by immunoblotting with antibodies specific for Cleaved Caspase-9, PARP and JSP1, respectively.
Discussion
The dual-specificity phosphatase JSP1, and its relative DUSP15, are unique among members of the PTP family in that they contain a potential myristoylation consensus sequence at the N-terminus (MGNGMXK). In their study of VHY/DUSP15, Mustelin’s group demonstrated that the VHY and VHX proteins incorporated 14C in cells metabolically labeled with [14C]-myristic acid [29]. The goal of the present study was to demonstrate directly that JSP1 was myristoylated, to apply a mass spectrometric approach to identify the residue in JSP1 that was modified, and to analyze whether myristoylation had an effect on JSP1 function.
Modification by covalently linked fatty acids, i.e. myristoylation or palmitoylation, has been shown to occur on a wide variety of signaling proteins. These hydrophobic modifications can confer reversible association with membranes and other signaling proteins, which modulates the specificity and efficiency of signal transduction [30]. N-myristoylation is the covalent attachment of myristate, a 14-carbon saturated fatty acid, to the N-terminal glycine of eukaryotic and viral proteins. The process is catalyzed by N-myristoyl transferase (NMT), and generally occurs co-translationally following removal of the initiator methionine residue by methionylaminopeptidases. The consensus sequence for NMT protein substrates is Met-Gly-X3-Ser/Thr-Lys/Arg-, but only the requirement for Gly at the N-terminus is absolute. For example, the tyrosine kinase c-Abl, a myristoylated protein, contains Gly and Lys at positions 2 and 7, respectively, but no Ser/Thr at position 6 [31, 32].
Between 0.5% and 3% of all eukaryotic proteins are N-myristoylated. These proteins have a broad range of functions and include protein kinases and phosphatases, Gα proteins, nitric oxide synthase (NOS), ADP-ribosylation factors (ARFs), and membrane-or cytoskeleton-associated structural proteins (e.g. MARCKS). The myristoyl moiety serves several functions: it can promote reversible binding and localization to membranes, stabilize the conformation of proteins, and regulate protein interactions. For example, myristoylation of Src is required for its localization to the plasma membrane, which is critically important for its proper function. A non-myristoylated mutant of Src, although catalytically active, has no transforming activity [33, 34]. Stabilization of a protein by myristoylation is exemplified by the example of cAMP-dependent protein kinase, where the myristoyl group binds to a hydrophobic cleft in the protein, thus stabilizing its tertiary strucutre [35]. An unusual example for the regulation of protein interaction is NADH-cytochrome b5 reductase (b5R), where myristoylation interferes with binding of the signal recognition particle, resulting in a part of b5R escaping the ER insertion pathway and relocating to the outer mitochondrial membrane [36]. Myristoylation has also been implicated in the regulation of apoptosis. Although normally a co-translational process, several proteins, including the proapoptotic protein Bid, actin and the Ser/Thr kinase Pak2, become myristoylated at newly generated N-terminal glycines after caspase cleavage [32]. In the case of Bid, the myristoylated fragment relocates to the mitochondrial membrane, where it induces oligomerization of Bak and subsequent cytochrome c release [37].
Myristoylation can also influence the movement and final destination of a signaling protein within the cell. We observed that myristoylation of JSP1 determined its localization to distinct sites in the cytoplasm. Signaling from internal membranes is now considered to be an important aspect of the spatial and temporal regulation of signaling pathways, e.g. the Ras/MAPK pathway [38, 39]. In order to specify the JSP1-containing structures, we tested colocalization of JSP1 with various marker proteins. Although we found that JSP1 colocalized with Golgi markers, further study is required to ascertain more precisely the distribution of JSP1 within the cell and to define its phosphorylated substrates and, thereby, its mechanism of action.
We have previously reported that JSP1 specifically activated the JNK pathway, hence the name JNK-stimulatory phosphatase 1 [20]. This result was supported by a second study that showed that the murine DSP JNK-pathway associated phosphatase (JKAP), a splice isoform of JSP1, specifically activated JNK when overexpressed in human embryonic kidney 293T cells [21]. Overexpression of a catalytically inactive mutant (JKAP-C88S) blocked tumor necrosis factor-α-induced JNK activation. Moreover, in murine JKAP−/− embryonic stem cells, JNK activation was abolished in response to tumor necrosis factor-α and transforming growth factor-β, but not in response to ultraviolet-C irradiation. These data illustrate that JSP1 is required for cytokine-induced activation of the JNK pathway. In contrast, Ayoama et al. suggested that when overexpressed in Cos-7 cells, JSP1/LMW-DSP2 dephosphorylated and inactivated p38, and, to a lesser extent, JNK after stimulation of the kinases with the appropriate agonists [22]. In addition, Alonso et al. reported a negative effect of JSP1/VHX on T cell receptor-induced activation of ERK2 in transfected Jurkat T cells [23]. The reason for these discrepancies is unclear, but could be due to differences of JSP1 function in the different cell systems used. In the present study, we have confirmed activation of JNK and its downstream transcription factor c-JUN by JSP1, which was dependent on a functional myristoylation site. Since myristoylation was not necessary for the intrinsic phosphatase activity of JSP1, but determined the subcellular localization of the phosphatase, this result suggests that correct localization of JSP1 to specific subcellular compartments is critically important for its functional activity in the JNK signaling pathway.
Overexpression of wild type JSP1, but neither the myristoylation-deficient mutant nor a catalytically inactive mutant, induced ~ 30% of the transfected cells to float off the dish and undergo apoptosis. Interestingly, the cells could tolerate high levels of the myristoylation-deficient mutant and remain attached, whereas similar levels of the wild type protein induced apoptosis. This study illustrates that the toxicity of wild type JSP1 presents a technical challenge that prohibits functional analysis using overexpression systems. Consistent with this, we have not been able to create stable cell lines expressing JSP1-wt constitutively, and almost all of the existing cell lines we have examined do not express detectable levels of JSP1 protein. In fact, in order to generate sufficient quantities of wild type protein for mass spectrometric analysis of the myristoylation site, we used 293T cells as expression system, since the presence of the SV40 large T antigen in these cells enhances their resistance to apoptosis.
Apoptosis is a tightly regulated mechanism for disposal of damaged cells and to remove cells during normal growth and development [27, 40]. Cells that undergo apoptosis initially become rounded, which is accompanied or followed by membrane blebbing, resulting in small vesicles termed apoptotic bodies. Inside the cell, apoptosis is characterized by condensation and fragmentation of the nucleus [27], as well as hydrolysis of nuclear DNA into distinct fragments by endonucleases [41]. Two main pathways lead to caspase-dependent apoptosis. In the extrinsic pathway, binding of death ligands to their respective receptors recruit adaptor proteins, such as Fas-associated death domain protein (FADD), which in turn bind and aggregate caspase-8 molecules, resulting in their autocleavage and activation. Active caspase-8 proteolytically processes and activates downstream caspases, eventually leading to substrate proteolysis, such as the nuclear poly (ADP-ribose) polymerase PARP [40]. In the intrinsic pathway, cell stress or damage activates members of the pro-apoptotic BH3-only protein family, which induce permeabilization of the outer mitochondrial membrane. Release of mitochondrial cytochrome c triggers assembly of a caspase-9-activating complex and subsequent activation of the downstream caspase cascade. These pathways are not mutually exclusive and are connected by caspase-8, which can trigger proteolysis of the BH3-only protein BID. When we analyzed the phenotype of JSP1-transfected, floating cells, we observed typical signs of apoptotic cell death, including condensed chromatin in the nucleus. Further analysis revealed that floating could be inhibited by treating the cells with a pan-caspase inhibitor, Z-VAD-FMK, simultaneously with JSP1 transfection. Induction of apoptosis was further implicated by the presence of cleaved Caspase-9 and PARP in floating cells (with high expression of wild type JSP1), but not in attached cells (low JSP1 expression), or cells with equally high expression of the myristoylation mutant. These results suggest that JSP1 induces apoptosis when overexpressed in cells, and further demonstrate the importance of myristoylation for this functional activity of JSP1.
It is reasonable to suggest that elevated JNK activity may precede the detachment and induction of apoptosis in the sub-population of cells expressing high levels of JSP1. We attempted to test this by treating cells with the JNK-inhibitor SP600125 concomitant with transfection, to determine whether inhibition of JNK abrogated the effect despite JSP1 expression. However, these efforts were frustrated by the lack of specificity of SP600125, which has also been reported by others [42, 43]. Resolution of the importance of JNK activation will require further experimentation.
In summary, JSP1, and VHY/DUSP15, are unique among the members of the PTP family in having a putative N-terminal myristoylation sequence and unusual in light of their potential to promote signaling. In this study, we demonstrate that JSP1 is myristoylated. Although this modification is not required for the intrinsic phosphatase activity of JSP1, we demonstrate that myristoylation is necessary for the ability of JSP1 to activate JNK signaling and to trigger apoptosis upon overexpression in our cell models. Further studies will focus on identification of physiological substrates of JSP1, to reveal the mechanism underlying its effects on JNK signaling and whether this is linked to the observed triggering of apoptosis.
Experimental procedures
Mammalian expression constructs
Full-length human JSP1 (UniProt accession number: Q9NRW4) was cloned into the mammalian expression vector pDEST12.2 (Invitrogen, Carlsbad, CA, U.S.A.). JSP1 mutants were generated by site-directed mutagenesis using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, U.S.A.). For GFP-tagged JSP1, JSP1 wild type or mutants were cloned into the eukaryotic expression vector pEGFP-N1 (Clontech, Mountain View, CA, U.S.A.).
Cell culture, transfection and lysate preparation
Cos-1, 293T and HeLa cells were maintained at 37°C and 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone, Logan, Utah, U.S.A.), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen). Cells were transfected with pDEST12.2-JSP1-wt, -G2A or -CS, and pEGFP-N1-JSP1-wt or -G2A, respectively, using TransIT-LT1 Transfection Reagent (Mirus, Madison, WI, U.S.A.), FuGENE 6 Transfection Reagent (Roche Applied Science, Indianapolis, IN, U.S.A.) or Effectene Transfection Reagent (Qiagen, Valencia, CA, U.S.A.) according to the manufacturer's protocol. After 24 hrs, floating cells were collected by centrifugation and resuspended in lysis buffer (PBS/1% Triton X-100/25 µg/ml Aprotinin/25 µg/ml Leupeptin/1 mM Na3VO4/5 mM NaF). Attached cells were collected directly in lysis buffer, and lysates were cleared by centrifugation.
Immunoblot analysis
Protein concentration of whole cell lysates was determined by Bradford assay, and equal amounts of total protein were subjected to SDS-PAGE, followed by transfer to nitrocellulose membrane (Whatman, Florham Park, NJ, U.S.A.). The membrane was blocked in 5% non-fat dried milk in Tris-buffered saline containing 0.05% Tween 20 (TBST) and incubated with primary antibody (Phospho-SAPK/JNK (Cell Signaling Technology, Danvers, MA, U.S.A.), JNK1/JNK2 (BD Biosciences, San Jose, CA, U.S.A.), JSP1 (generated in this lab), Cleaved Caspase-9 (Cell Signaling Technology), PARP (kind gift of Dr. Yuri Lazebnik, CSHL) or β-actin (Sigma, Saint Louis, MO, U.S.A.)). Bands were visualized with horseradish peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA, U.S.A.) and ECL Western Blotting Detection Reagent (GE Healthcare, Piscataway, NJ, U.S.A.).
Protein expression and purification
Full-length human JSP1 constructs were cloned into the bacterial expression vector pET-21b (Novagen, Gibbstown, NJ, U.S.A.). JSP1-His6 constructs were expressed in E. coli-BL21, cells were lysed by sonication in lysis buffer (50 mM Tris (pH 7.0)/100 mM NaCl/Complete protease inhibitors (Roche)), and recombinant protein was purified from the cleared lysate using Ni-NTA Superflow (Qiagen). The eluted protein was dialyzed against storage buffer (25 mM Tris (pH 7.0)/50 mM NaCl/5 mM DTT/0.02% NaN3/50% glycerol) and stored at −80°C.
Assay of protein phosphatase activity
32P-labeled reduced carboxamidomethylated and maleylated lysozyme (RCML) substrate was prepared as described previously [44]. Protein phosphatase assays were performed according to standard protocols [20] using 5 µM labeled substrate and 0.5 or 1 µg of recombinant JSP1.
Counting of floating cells
Cos-1 cells were treated with 50 µM pan-caspase inhibitor Z-VAD-FMK (BIOMOL, Plymouth Meeting, PA, U.S.A.), or DMSO as control, for 30 min prior to transfection, and then transfected with JSP1-wt or JSP1-G2A expression constructs in the presence of inhibitor. 24 hrs post-transfection, cells that had detached from the culture dish were collected by centrifugation at 2,000 rpm, for 10 min, at 4°C. Pellets were resuspended in 100 µl of PBS, stained with Trypan Blue (Invitrogen) and counted using a haemocytometer.
Microscopic analysis
For JSP1 localization studies, HeLa cells were seeded on glass coverslips 24 hrs prior to transfection. Transfections with pEGFP-N1-JSP1-wt or JSP1-G2A were carried out using Effectene transfection reagent (Qiagen), according to the manufacturer’s protocol. 24 hrs post-transfection, cells were fixed with 3% (w/v) paraformaldehyde, mounted with Mowiol (Calbiochem, Gibbstown, NJ, U.S.A.) and analyzed by confocal laser scanning microscopy. For the analysis of JSP1-induced cell death, Cos-1 cells were seeded on glass coverslips 24 hrs prior to transfection. Transfections with pDEST12.2-JSP1-wt, -G2A or -CS were carried out using FuGENE 6 Transfection Reagent (Roche Applied Science), according to the manufacturer’s protocol. 24 hrs post-transfection, cells were fixed with 5% (w/v) paraformaldehyde, permeabilized in 0.5% Triton X-100/PBS, and stained with 1 µg/ml DAPI (Sigma). Floating cells were collected separately, treated as described above, and transferred onto slides. Cells were mounted with ProLong® Antifade reagent (Invitrogen), and analyzed by fluorescence microscopy.
Mass spectrometry
293T cells were transfected with pEGFP-N1-JSP1-wt or -G2A using TransIT-LT1 Transfection Reagent (Mirus), according to the manufacturer’s protocol. 48 hrs post-transfection, whole cell lysates were prepared in immunoprecipitation (IP)-lysis buffer (50 mM Tris (pH 7.4)/150 mM NaCl/1% NP-40/0.5% sodium deoxycholate/25 µg/ml Aprotinin/25 µg/ml Leupeptin), and 2.5 mg of total protein was precleared with Protein G Sepharose 4B Fast Flow (GE Healthcare) for 1 h at 4°C. The precleared supernatant was incubated with polyclonal anti-GFP antibody (Invitrogen) coupled to Protein G Sepharose beads for 1 h at 4°C. After washes with IPlysis buffer, beads were resuspended in 2x Laemmli sample buffer, and proteins were resolved by SDS-PAGE. The gel was fixed in 40% Methanol/10% acetic acid, and stained with SYPRO Ruby Protein Gel Stain (Invitrogen) according to the manufacturer's protocol. Bands containing JSP1-wt or -G2A were excised and digested using mass spectrometry grade trypsin (Promega, Madison, WI, U.S.A.) at 12.5 ng/µl in 25 mM NH4HCO3 buffer according to a modified version of the protocol of Shevchenko et al. [45]. The resulting peptides were extracted, dried under vacuum, and resuspended in 10 µl of 0.1% formic acid/20% acetonitrile. 4 µl of peptide mixtures were analyzed using nanoflow LC/ESI-MS/MS, with a NanoAquity UPLC coupled directly to a QTOF Premier mass spectrometer (Waters, Milford, MA, U.S.A.). Peptides were separated by a 100 µm i.d.×10 cm column (Waters) packed with 1.7 ⎧m BEH C18 beads using a linear gradient from 5 to 85% acetonitrile in 0.1% formic acid over 100 min at 300 nl/min. Data acquisition involved MS survey scans followed by 3 automatic data-dependent MS/MS acquisitions per survey scan.
Acknowledgements
This work was supported by NIH Grant CA112534 and a Grant from the Hartman Foundation (to N.K.T.), and NIH-NCRR Grant S10 RR017990 (to T.A.N.). U.S. was the recipient of a postdoctoral fellowship from the Cold Spring Harbor Laboratory (CSHL). We thank Dr. Yuri Lazebnik (CSHL) for providing the PARP antibody.
Abbreviations
- DSP
dual specificity phosphatase
- JNK
c-JUN N-terminal kinase
- JSP1
JNK stimulatory phosphatase 1
- MAPK
mitogen-activated protein kinase
- MAPKK or MEK
MAPK kinase
- MAPKKK or MEKK
MAPK kinase kinase
- PTP
protein tyrosine phosphatase
References
- 1.Chang L, Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. doi: 10.1038/35065000 35065000 [pii]. [DOI] [PubMed] [Google Scholar]
- 2.Kyriakis JM, Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 3.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 4.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. doi: nrc2694 [pii] 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 5.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. doi: S0092-8674(00)00116-1 [pii]. [DOI] [PubMed] [Google Scholar]
- 6.Yan M, Dai T, Deak JC, Kyriakis JM, Zon LI, Woodgett JR, Templeton DJ. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 7.Tournier C, Whitmarsh AJ, Cavanagh J, Barrett T, Davis RJ. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci U S A. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tournier C, Dong C, Turner TK, Jones SN, Flavell RA, Davis RJ. MKK7 is an essential component of the JNK signal transduction pathway activated by proinflammatory cytokines. Genes & development. 2001;15:1419–1426. doi: 10.1101/gad.888501. doi: 10.1101/gad.888501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ip YT, Davis RJ. Signal transduction by the c-Jun N-terminal kinase (JNK)--from inflammation to development. Current opinion in cell biology. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. doi: S0955-0674(98)80143-9 [pii]. [DOI] [PubMed] [Google Scholar]
- 10.Bhalla US, Ram PT, Iyengar R. MAP kinase phosphatase as a locus of flexibility in a mitogen-activated protein kinase signaling network. Science. 2002;297:1018–1023. doi: 10.1126/science.1068873. doi: 10.1126/science.1068873 297/5583/1018 [pii]. [DOI] [PubMed] [Google Scholar]
- 11.Hornberg JJ, Bruggeman FJ, Binder B, Geest CR, de Vaate AJ, Lankelma J, Heinrich R, Westerhoff HV. Principles behind the multifarious control of signal transduction. ERK phosphorylation and kinase/phosphatase control. The FEBS journal. 2005;272:244–258. doi: 10.1111/j.1432-1033.2004.04404.x. doi: EJB4404 [pii] 10.1111/j.1432-1033.2004.04404.x. [DOI] [PubMed] [Google Scholar]
- 12.Julien SG, Dube N, Read M, Penney J, Paquet M, Han Y, Kennedy BP, Muller WJ, Tremblay ML. Protein tyrosine phosphatase 1B deficiency or inhibition delays ErbB2-induced mammary tumorigenesis and protects from lung metastasis. Nature genetics. 2007;39:338–346. doi: 10.1038/ng1963. doi: ng1963 [pii] 10.1038/ng1963. [DOI] [PubMed] [Google Scholar]
- 13.Bentires-Alj M, Neel BG. Protein-tyrosine phosphatase 1B is required for HER2/Neu-induced breast cancer. Cancer research. 2007;67:2420–2424. doi: 10.1158/0008-5472.CAN-06-4610. doi: 0008-5472.CAN-06-4610 [pii] 10.1158/0008-5472.CAN-06-4610. [DOI] [PubMed] [Google Scholar]
- 14.Myers MG, Jr, Mendez R, Shi P, Pierce JH, Rhoads R, White MF. The COOH-terminal tyrosine phosphorylation sites on IRS-1 bind SHP-2 and negatively regulate insulin signaling. J Biol Chem. 1998;273:26908–26914. doi: 10.1074/jbc.273.41.26908. [DOI] [PubMed] [Google Scholar]
- 15.Shi ZQ, Lu W, Feng GS. The Shp-2 tyrosine phosphatase has opposite effects in mediating the activation of extracellular signal-regulated and c-Jun NH2- terminal mitogen-activated protein kinases. J Biol Chem. 1998;273:4904–4908. doi: 10.1074/jbc.273.9.4904. [DOI] [PubMed] [Google Scholar]
- 16.Jeffrey KL, Camps M, Rommel C, Mackay CR. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat Rev Drug Discov. 2007;6:391–403. doi: 10.1038/nrd2289. doi: nrd2289 [pii] 10.1038/nrd2289. [DOI] [PubMed] [Google Scholar]
- 17.Patterson KI, Brummer T, O'Brien PM, Daly RJ. Dual-specificity phosphatases: critical regulators with diverse cellular targets. Biochem J. 2009;418:475–489. doi: 10.1042/bj20082234. [DOI] [PubMed] [Google Scholar]
- 18.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nature reviews. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 19.Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. doi: 1210412 [pii] 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Luche R, Wei B, Gordon ML, Diltz CD, Tonks NK. Activation of the Jnk signaling pathway by a dual-specificity phosphatase, JSP-1. Proc Natl Acad Sci U S A. 2001;98:13613–13618. doi: 10.1073/pnas.231499098. doi: 10.1073/pnas.231499098 98/24/13613 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen AJ, Zhou G, Juan T, Colicos SM, Cannon JP, Cabriera-Hansen M, Meyer CF, Jurecic R, Copeland NG, Gilbert DJ, et al. The dual specificity JKAP specifically activates the c-Jun N-terminal kinase pathway. J Biol Chem. 2002;277:36592–36601. doi: 10.1074/jbc.M200453200. doi: 10.1074/jbc.M200453200 M200453200 [pii]. [DOI] [PubMed] [Google Scholar]
- 22.Aoyama K, Nagata M, Oshima K, Matsuda T, Aoki N. Molecular cloning and characterization of a novel dual specificity phosphatase, LMW-DSP2, that lacks the cdc25 homology domain. J Biol Chem. 2001;276:27575–27583. doi: 10.1074/jbc.M100408200. doi: 10.1074/jbc.M100408200 M100408200 [pii]. [DOI] [PubMed] [Google Scholar]
- 23.Alonso A, Merlo JJ, Na S, Kholod N, Jaroszewski L, Kharitonenkov A, Williams S, Godzik A, Posada JD, Mustelin T. Inhibition of T cell antigen receptor signaling by VHR-related MKPX (VHX), a new dual specificity phosphatase related to VH1 related (VHR) J Biol Chem. 2002;277:5524–5528. doi: 10.1074/jbc.M107653200. doi: 10.1074/jbc.M107653200 M107653200 [pii]. [DOI] [PubMed] [Google Scholar]
- 24.Sekine Y, Tsuji S, Ikeda O, Sato N, Aoki N, Aoyama K, Sugiyama K, Matsuda T. Regulation of STAT3-mediated signaling by LMW-DSP2. Oncogene. 2006;25:5801–5806. doi: 10.1038/sj.onc.1209578. doi: 1209578 [pii] 10.1038/sj.onc.1209578. [DOI] [PubMed] [Google Scholar]
- 25.Sekine Y, Ikeda O, Hayakawa Y, Tsuji S, Imoto S, Aoki N, Sugiyama K, Matsuda T. DUSP22/LMW-DSP2 regulates estrogen receptor-alpha-mediated signaling through dephosphorylation of Ser-118. Oncogene. 2007;26:6038–6049. doi: 10.1038/sj.onc.1210426. doi: 1210426 [pii] 10.1038/sj.onc.1210426. [DOI] [PubMed] [Google Scholar]
- 26.Mo W, Ma Y, Takao T, Neubert TA. Sequencing of oxidized methionine-containing peptides for protein identification. Rapid Commun Mass Spectrom. 2000;14:2080–2081. doi: 10.1002/1097-0231(20001115)14:21<2080::AID-RCM120>3.0.CO;2-P. doi: 10.1002/1097-0231(20001115)14:21<2080::AID-RCM120>3.0.CO;2-P [pii] 10.1002/1097-0231(20001115)14:21<2080::AID-RCM120>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 27.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa J, Kitten GT, Nigg EA. A somatic cell-derived system for studying both early and late mitotic events in vitro. J Cell Sci. 1989;94(Pt 3):449–462. doi: 10.1242/jcs.94.3.449. [DOI] [PubMed] [Google Scholar]
- 29.Alonso A, Narisawa S, Bogetz J, Tautz L, Hadzic R, Huynh H, Williams S, Gjorloff-Wingren A, Bremer MC, Holsinger LJ, et al. VHY, a novel myristoylated testis-restricted dual specificity protein phosphatase related to VHX. J Biol Chem. 2004;279:32586–32591. doi: 10.1074/jbc.M403442200. doi: 10.1074/jbc.M403442200 M403442200 [pii]. [DOI] [PubMed] [Google Scholar]
- 30.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. doi: nchembio834 [pii] 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 31.Resh MD. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochimica et biophysica acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. doi: S0167-4889(99)00075-0 [pii]. [DOI] [PubMed] [Google Scholar]
- 32.Wright MH, Heal WP, Mann DJ, Tate EW. Protein myristoylation in health and disease. J Chem Biol. 2009 doi: 10.1007/s12154-009-0032-8. doi: 10.1007/s12154-009-0032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buss JE, Kamps MP, Gould K, Sefton BM. The absence of myristic acid decreases membrane binding of p60src but does not affect tyrosine protein kinase activity. Journal of virology. 1986;58:468–474. doi: 10.1128/jvi.58.2.468-474.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamps MP, Buss JE, Sefton BM. Rous sarcoma virus transforming protein lacking myristic acid phosphorylates known polypeptide substrates without inducing transformation. Cell. 1986;45:105–112. doi: 10.1016/0092-8674(86)90542-8. doi: 0092-8674(86)90542-8 [pii]. [DOI] [PubMed] [Google Scholar]
- 35.Zheng J, Knighton DR, Xuong NH, Taylor SS, Sowadski JM, Ten Eyck LF. Crystal structures of the myristylated catalytic subunit of cAMP-dependent protein kinase reveal open and closed conformations. Protein Sci. 1993;2:1559–1573. doi: 10.1002/pro.5560021003. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colombo S, Longhi R, Alcaro S, Ortuso F, Sprocati T, Flora A, Borgese N. N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. The Journal of cell biology. 2005;168:735–745. doi: 10.1083/jcb.200407082. doi: jcb.200407082 [pii] 10.1083/jcb.200407082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zha J, Weiler S, Oh KJ, Wei MC, Korsmeyer SJ. Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science. 2000;290:1761–1765. doi: 10.1126/science.290.5497.1761. doi: 9030 [pii]. [DOI] [PubMed] [Google Scholar]
- 38.Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ. Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf-1 by EGF but not insulin in liver parenchyma. The EMBO journal. 1994;13:4269–4277. doi: 10.1002/j.1460-2075.1994.tb06747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mor A, Philips MR. Compartmentalized Ras/MAPK signaling. Annu Rev Immunol. 2006;24:771–800. doi: 10.1146/annurev.immunol.24.021605.090723. doi: 10.1146/annurev.immunol.24.021605.090723. [DOI] [PubMed] [Google Scholar]
- 40.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nature reviews. 2008;9:231–241. doi: 10.1038/nrm2312. doi: nrm2312 [pii] 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 41.Williams JR, Little JB, Shipley WU. Association of mammalian cell death with a specific endonucleolytic degradation of DNA. Nature. 1974;252:754–755. doi: 10.1038/252754a0. [DOI] [PubMed] [Google Scholar]
- 42.Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. doi: 10.1042/BJ20021535 BJ20021535 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. doi: BJ20070797 [pii] 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng TC, Hsu SF, Tonks NK. Development of a modified in-gel assay to identify protein tyrosine phosphatases that are oxidized and inactivated in vivo. Methods. 2005;35:28–36. doi: 10.1016/j.ymeth.2004.07.005. doi: S1046-2023(04)00168-9 [pii] 10.1016/j.ymeth.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]