Abstract
Summary: Computational gene function prediction can serve to focus experimental resources on high-priority experimental tasks. FuncBase is a web resource for viewing quantitative machine learning-based gene function annotations. Quantitative annotations of genes, including fungal and mammalian genes, with Gene Ontology terms are accompanied by a community feedback system. Evidence underlying function annotations is shown. For example, a custom Cytoscape viewer shows functional linkage graphs relevant to the gene or function of interest. FuncBase provides links to external resources, and may be accessed directly or via links from species-specific databases.
Availability: FuncBase as well as all underlying data and annotations are freely available via http://func.med.harvard.edu/
Contact: fritz_roth@hms.harvard.edu
1 INTRODUCTION
Computational prediction—e.g. of gene function, gene phenotype, protein interactions or genetic interactions—offers a statistically sound form of triage for reducing experimental tasks that would be prohibitive otherwise. For example, in genetic disease mapping, a candidate gene approach can reduce the study size required to establish significance. This is critically important, since large association studies are costly and may be infeasible for rare diseases. Functions are commonly represented by Gene Ontology (GO; Ashburner et al., 2000) terms, which encompass molecular functions, cellular locations and biological processes.
Experimentalists differ in their requirements for function prediction. To maximize new discoveries, some will wish to cast a wide net that may include many false positives. Others, for whom follow-up experiments are more resource-intensive, will wish to proceed conservatively. Therefore, FuncBase displays quantitative confidence measures by which predictions may be ranked. Because users typically have additional domain knowledge that they can draw upon to filter out unlikely predictions, FuncBase shows predictions in the context of underlying evidence.
FuncBase currently displays function annotations for several species. For each species, annotations are based on machine learning algorithms applied to an integrated data collection including protein motif annotation, phenotype and disease association, phylogenetic profiles, protein interactions and gene expression. Full descriptions for the underlying machine learning algorithm are provided in Tian et al. (2008), Pena-Castillo et al. (2008) and Taşan et al. (2008).
2 BACKGROUND
For each gene-function pair examined, a gene function prediction algorithm may provide a binary ‘black or white’ classification, a ranking or a quantitative confidence measure.
Interfaces displaying gene function predictions currently take one of three forms. In the first form, binary calls are incorporated into an existing species-specific database, such as the Saccharomyces Genome Database (SGD; Cherry et al., 1998) or the Mouse Genome Informatics resource (MGI; Bult et al., 2008). While ‘black or white’ calls are useful for archiving accepted knowledge about gene function, they are incomplete guides to grey areas of current knowledge.
The second form of interface enables users to apply prediction algorithms to datasets provided by the user. This second form is taken by such websites as GeneMANIA (Mostafavi et al., 2008) and VIRGO (Massjouni et al., 2006).
A third form, represented by FuncBase , STRING (von Mering et al., 2007) and BioPIXIE (Myers et al., 2005), is a browser of precalculated predictions ranked by confidence score, together with their literature verification status. Relaxing the requirement that quantitative predictions be generated ‘on the fly’ allows use of more computationally intensive prediction algorithms.
3 FEATURES
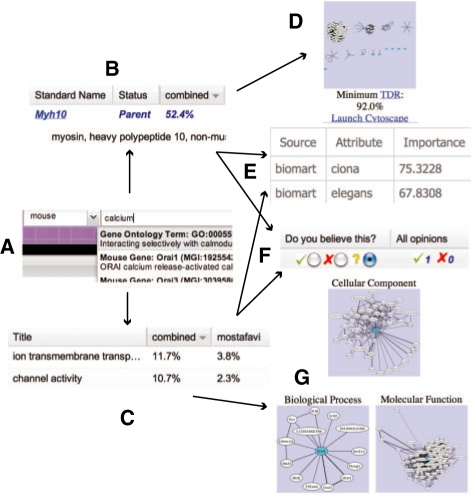
View predictions by gene or function: Predictions in FuncBase can be viewed either by function (GO term) or by gene. Users may search for their gene or function using a rich search syntax (Section 4) permitting entry of gene or protein synonyms from multiple identifier systems, and text-matching within gene or function descriptions (Fig. 1A).
Fig. 1.
Search (A) for an annotation report of a GO term (B) or gene (C). GO term reports show evidence of functional relationships (D) and function-related gene properties (E). The user may provide opinions (F) on any quantitative annotation. Gene reports also present evidence based on functional relationships (G).
Both function and gene views (examples shown in Figs 1B and C) allow predictions to be sorted by the confidence score from any available prediction method. GO annotations previously assigned by the corresponding species-specific authority are displayed next to each prediction.
View supporting evidence: Users may wish to further filter quantitative annotations based on their domain knowledge. Therefore, FuncBase displays key pieces of evidence underlying annotations.
Some annotation algorithms take a guilt-by-profiling approach—e.g. genes involved in ‘negative regulation of microtubule polymerization or depolymerization’ (GO:0031111) tend to contain a DH protein domain (InterPro pattern IPR000219). Therefore, each function view displays the gene properties that are most predictive of that function. A table (Fig. 1E), available by clicking an annotation row, indicates all properties held by the corresponding gene.
Some annotation algorithms take a guilt-by-association approach, in which GO annotations are ‘transferred’ between genes with evidence of a functional relationship (e.g. physical interaction between the corresponding proteins). Different variants of the functional linkage graphs are appropriate for different GO terms (see Taşan et al., 2008 and Tian et al., 2008), so in function views one graph is displayed (Fig. 1D), and in gene views FuncBase three functional linkage graph versions are shown that correspond to the three branches of the GO (Fig. 1G). Functional linkage graphs can be viewed in FuncBase as static images, or manipulated within Cytoscape (Shannon et al., 2003).
Quantitative annotations from multiple sources: A unique feature of FuncBase is its ability to accommodate prediction sets from multiple bioinformatics teams differing by input data or algorithm. For example, 10 prediction sets are available for Mus musculus. We invite others to submit predictions associated with peer-reviewed publications for sharing via FuncBase.
User feedback: FuncBase is governed by the philosophy that annotation in general and predictive annotation in particular is a work in progress, and that users will often bring domain knowledge that supersedes current or predicted annotation. Therefore, for every gene/function combination displayed, a form invites expert users to provide feedback on whether they agree, disagree or are uncertain about this annotation (Fig. 1F). Free text notes can be attached to any opinion. Current tallies of true and false responses are shared among all users and made available in summary form to the appropriate species authority. Community feedback on predictions gathered and shared in real time is novel to the FuncBase quantitative annotation resource.
4 IMPLEMENTATION
The back-end of FuncBase consists of the Pylons MVC framework, the Lucene search provider and the PostgreSQL database server. The front-end uses ExtJS (Javascript) and a modified version of Cytoscape 2.6. Most web site actions are accomplished through asynchronous browser–server communication. Functional linkage graph layout is via BioLayoutKK within Cytoscape, using linkage certainties as edge weights.
ACKNOWLEDGEMENTS
For their advice, we thank SGD members, including J. Park, J.M. Cherry and E. Hong; MGI members, including J. Blake and D. Hill; Roth lab members, including G. Berriz and R. Deo.
Funding: National Institues of Health (grants NS054052, NS035611, HL081341, HG001715, HG004233 and HG003224); A Canadian Institute for Advanced Research Fellowship (to F.P.R.).
Conflict of Interest: none declared.
REFERENCES
- Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, et al. The mouse genome database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36(Database issue) doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry J, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglott D, et al. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2005;33(Database issue) doi: 10.1093/nar/gki031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massjouni N, et al. VIRGO: computational prediction of gene functions. Nucleic Acids Res. 2006;34(Suppl. 2):W340–W344. doi: 10.1093/nar/gkl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, et al. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl. 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers CL, et al. Discovery of biological networks from diverse functional genomic data. Genome Biol. 2005;6 doi: 10.1186/gb-2005-6-13-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Castillo L, et al. A critical assessment of Mus musculus gene function prediction using integrated genomic evidence. Genome Biol. 2008;9(Suppl. 1):S2. doi: 10.1186/gb-2008-9-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşan M, et al. An en masse phenotype and function prediction system for Mus musculus. Genome Biol. 2008;9(Suppl. 1):S8. doi: 10.1186/gb-2008-9-s1-s8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taşan M, et al. Quantitative functional annotation of H. sapiens genes. Unpublished results. 2010 [Google Scholar]
- Tian W, et al. Combining guilt-by-association and guilt-by-profiling to predict Saccharomyces cerevisiae gene function. Genome Biol. 2008;9(Suppl. 1):S7. doi: 10.1186/gb-2008-9-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, et al. STRING 7–recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 2007;35(Database issue) doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]