Abstract
Motivation: The growing field of systems biology has driven demand for flexible tools to model and simulate biological systems. Two established problems in the modeling of biological processes are model selection and the estimation of associated parameters. A number of statistical approaches, both frequentist and Bayesian, have been proposed to answer these questions.
Results: Here we present a Python package, ABC-SysBio, that implements parameter inference and model selection for dynamical systems in an approximate Bayesian computation (ABC) framework. ABC-SysBio combines three algorithms: ABC rejection sampler, ABC SMC for parameter inference and ABC SMC for model selection. It is designed to work with models written in Systems Biology Markup Language (SBML). Deterministic and stochastic models can be analyzed in ABC-SysBio.
Availability: http://abc-sysbio.sourceforge.net
Contact: christopher.barnes@imperial.ac.uk; ttoni@imperial.ac.uk
1 INTRODUCTION
In the last decade, modeling of biochemical systems using ordinary and stochastic differential equations (ODE and SDE) has become increasingly popular as quantitative ideas have begun to pervade the biomolecular sciences. Inferring model parameters and ranking alternative models is necessary in order to gather reliable future predictions about the dynamical behavior of such systems. The problem of parameter estimation in deterministic systems has been addressed by using local and global non-linear optimization methods (Mendes and Kell, 1998; Moles et al., 2003) as well as maximum-likelihood estimation (Baker et al., 2005; Bortz and Nelson, 2006; Muller et al., 2004; Timmer and Muller, 2004) and within a Bayesian framework (Banks et al., 2005, Huang et al., 2006, Putter et al., 2002). An approximate Bayesian computation (ABC) scheme based on sequential Monte Carlo (SMC) has been developed for likelihood-free parameter inference in deterministic and stochastic systems (Toni et al., 2009). Furthermore, this approach also allows for model selection, i.e. evidence-based ranking of alternative models.
Because in ABC methods the evaluation of the likelihood is replaced by simulations, the implementation of these methods hides some numerical and technical problems. This includes finding numerically the solution of stiff ODE systems or stochastic systems described by SDEs or a master equation. This requires a flexible and adaptive implementation of ABC methods to address each specific model with its biochemical as well as dynamical problems.
Here we present an extensible Python package, ABC-SysBio, which implements approximate Bayesian computation for parameter inference and model selection in deterministic and stochastic models. The package supports the standard models exchange format, SBML, as well as user-defined models written in Python. In addition, graphical processing unit support is provided via pycuda (Klöckner et al., 2009). User-defined algorithmic parameters allow for the adaptation and tuning of the inference procedures included in the package to suit each specific dynamical model.
Note that in contrast to other packages for parameter inference in a Bayesian framework, e.g. DIYABC (Cornuet et al., 2008) and BioBayes (Vyshemirsky and Girolami, 2008), ABC-SysBio allows for parameter inference and model selection of both deterministic and stochastic models. Its implementation is flexible and user friendly: it supports important model exchange standards and is easily integrated into existing computational pipelines and systems biology frameworks through the flexibility of Python.
2 METHODS
The package ABC-SysBio is implemented as a Python module, abcsysbio. Together with the two Python scripts abc-sysbio-sbml-sum and run-abc-sysbio, it creates a user friendly tool that can be applied to models in SBML format without any further software development. It is advisable to use the package together with the Python Enthought Distribution, though this is not essential. It works on both MacOS and Linux operating systems.
The module, abcsysbio, can be imported into an interactive Python session, and by defining the arguments to the functions in the interactive namespace, they can be used through the Python shell.
When run-abc-sysbio is called, model(s) written in SBML format are parsed to generate a corresponding Python module representing the model. The format of the Python module written depends on the integration type, which also informs the program which solver to use to simulate the model.
We provide algorithms to simulate ODE, SDE and Gillespie models (Press et al., 1992). All algorithms are adapted to the specific requirements of models in the Biomodels database. Beside the possibility of only numerically solving the provided model, one of the three following algorithms can be called.
2.1 ABC rejection sampler for parameter inference
Given a parameter θ, its prior distribution π(θ) and a dataset x, we want to approximate the posterior distribution π(θ|x). The ABC rejection sampler proceeds as follows:
Sample θ* from π.
Simulate a dataset x* from the model with parameter θ*.
If d(x0, x*) ≤ ϵ, accept θ, otherwise reject.
Return to 1.
where d(x0, x*) is a distance function and ϵ is a tolerance. The implemented distance function is the Euclidian distance; however the user can easily define custom distance metrics. To obtain reliable parameter estimates, ϵ should be very small. The ABC rejection sampler should be used only for simple systems that allow a fast simulation, because the rejection rate of this algorithm is usually very high.
The ABC rejection sampler describes the first sampled population of the ABC SMC algorithm and is therefore implemented as part of the ABC SMC algorithms described below.
2.2 ABC SMC for parameter inference
Using ABC in a SMC framework leads to the ABC SMC algorithm (Toni et al., 2009). A number of particles are sampled from the prior distribution π(θ) and propagated through a sequence of intermediate distributions until the population represents an approximated posterior distribution. The intermediate distributions are defined by a sequence of tolerances ϵt in decreasing order, ϵ1 > ϵ2 > … > ϵT ≥ 0. Therefore ABC SMC for parameter inference is automatically invoked when only one model, but more than one ϵ are provided. Additionally, the user needs to provide perturbation kernels, which define how a particle is perturbed after resampling from the previous intermediate distribution.
This algorithm can be considered a special case of the model selection algorithm implemented in ABC-SysBio. The ABC SMC for parameter inference algorithm is nested within ABC SMC for model selection. Therefore, the same ABC-SysBio functions, with the same computational features, are called whichever of the algorithms are run.
2.3 ABC SMC for model selection
An algorithm to select between several deterministic or stochastic dynamical models for a given dataset has been implemented; here the model identifiers/labels are treated as an additional parameter, as described in Toni and Stumpf, 2010. Therefore, a prior distribution over models as well as perturbation kernels for model transitions need to be defined by the user. The implementation of the model selection algorithm using ABC SMC provides the framework for the above mentioned nested algorithms.
2.4 Options
Users can define several algorithmic parameters, for example, prior distributions or perturbation kernels. Several distributions are already implemented, e.g. uniform distribution, Gaussian distribution and log-normal distribution, but further distributions can be added easily. The user should define the ϵ schedule, because it is strongly dependent on the biochemical/dynamical system under investigation, as well as on the noise in the provided data. Note that the dataset does not necessarily need to include data for all species defined in the model, but can be a subset or even a combination of several species.
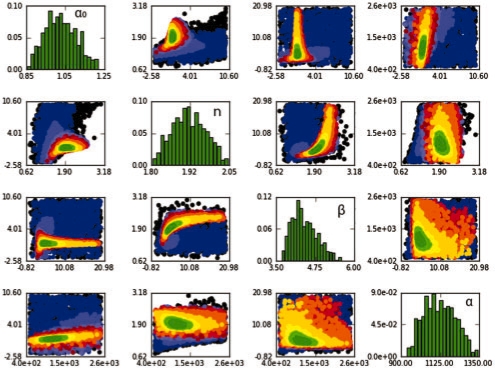
After each sampled population, ABC-SysBio provides the user with information about the algorithm's progress. The rejection rate per population, as well as the sampled particles from intermediate distributions (Fig. 1) are saved in accessible text files. Furthermore, a graphical output including scatterplots of pairwise parameter combinations and histograms summarizing parameter and model distributions are provided. This allows the user to follow the progress of the algorithm. Additionally, all data are copied into a binary file to allow the algorithm to be restarted from a previous population, with changed algorithm parameters.
Fig. 1.
ABC-SysBio output. This example shows the results of the deterministic repressilator model (as described in Toni et al., 2009). Scatterplots of inferred parameters for 11 populations are displayed (from black for the first population to dark green for the 9th population). The approximate posterior over model parameters is shown on the diagonal.
3 SUMMARY
ABC-SysBio is a flexible, extendable and user-friendly Python package that can import models described in the SBML standard combined with experimental data. Our package approximates the posterior parameter distribution and compares different model structures to select the model that represents the data best.
ACKNOWLEDGEMENTS
We thank the members of the Theoretical Systems Biology Group at Imperial College London, Sylvia Richardson, David Balding, Mark Beaumont, Christian Robert and Scott Sisson for discussions on ABC methodology. We are particularly grateful to Justina Norkunaite for becoming an early adopter of ABC-SysBio.
Funding: Wellcome Trust (to J.L., E.C., P.K. and K.E.); Biotechnology and Biological Sciences Research Council (to C.B. and M.P.H.S.); Medical Research Council (to T.T.). M.P.H.S. is a Royal Society Wolfson Research Merit Award holder.
Conflict of Interest: none declared.
REFERENCES
- Baker C, et al. Ludewig computational approaches to parameter estimation and model selection in immunology. J. Comput. Appl. Math. 2005;184:5076. [Google Scholar]
- Banks H, et al. A hierarchical Bayesian approach for parameter estimation in HIV models. Inverse Probl. 2005;21:1803–1822. [Google Scholar]
- Bortz DM, Nelson PW. Model selection and mixed-effects modeling of HIV infection dynamics. Bull. Math. Biol. 2006;68:2005–2025. doi: 10.1007/s11538-006-9084-x. [DOI] [PubMed] [Google Scholar]
- Cornuet J.-M, et al. Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics. 2008;24:2713–2719. doi: 10.1093/bioinformatics/btn514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, et al. Hierarchical Bayesian methods for estimation of parameters in a longitudinal HIV dynamic system. Biometrics. 2006;62:413–423. doi: 10.1111/j.1541-0420.2005.00447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klöckner A, et al. PyCUDA: GPU run-time code generation for high-performance computing. 2009 [Google Scholar]
- Mendes P, Kell D. Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics. 1998;14:869–883. doi: 10.1093/bioinformatics/14.10.869. [DOI] [PubMed] [Google Scholar]
- Moles C, et al. Parameter estimation in biochemical pathways: a comparison of global optimization methods. Genome Res. 2003;13:2467–2474. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller TG, et al. Tests for cycling in a signalling pathway. J. R. Stat. Soc. Ser. C. 2004;53:557. [Google Scholar]
- Press WH, et al. Numerical Recipes in C: The Art of Scientific Computing. 2. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- Putter H, et al. A Bayesian approach to parameter estimation in HIV dynamical models. Stat. Med. 2002;21:2199–2214. doi: 10.1002/sim.1211. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- Timmer J, Muller T. Modeling the nonlinear dynamics of cellular signal transduction. Int. J. Bifurcat. Chaos. 2004;14:2069–2079. [Google Scholar]
- Toni T, et al. Approximate Bayesian computation scheme for parameter inference and model selection in dynamical systems. J. R. Soc. Interface. 2009;6:187–202. doi: 10.1098/rsif.2008.0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni T, Stumpf MPH. Simulation-based model selection for dynamical systems and population biology. Bioinformatics. 2010;26:104–110. doi: 10.1093/bioinformatics/btp619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyshemirsky V, Girolami M. Biobayes: a software package for bayesian inference in systems biology. Bioinformatics. 2008;24:338–1934. doi: 10.1093/bioinformatics/btn338. [DOI] [PubMed] [Google Scholar]