Abstract
Summary: We present an open source, platform independent tool, called CisGenome Browser, which can work together with any other data analysis program to serve as a flexible component for genomic data visualization. It can also work by itself as a standalone genome browser. By working as a light-weight web server, CisGenome Browser is a convenient tool for data sharing between labs. It has features that are specifically designed for ultra high-throughput sequencing data visualization.
Availability: http://biogibbs.stanford.edu/∼jiangh/browser/
Contact: jiangh@stanford.edu
1 INTRODUCTION
For the past decade, the advent of high-throughput technologies such as microarrays and the more recent ultra high-throughput sequencing have given rise to fruitful research on large-scale genomic data analysis. As a result, numerous software tools have been developed and widely used. Since genomic data is usually very large and complex, visualization tools are always essential for data examination and interpretation.
There are two major approaches for genomic data visualization. One approach is to provide built-in data visualization components together with data analysis tools. A typical example is DNA-chip analyzer (dChip; Li and Wong, 2003) whose convenient visualization functions contribute greatly to its popularity. This type of approach usually gives the best user experience. Since the data visualization components are specifically designed for the results that are generated by the data analysis tools, they can work together seamlessly. However, only a small fraction of data analysis tools have built-in visualization components because the programming of a good visualization component requires expertise and is also often very time-consuming. For these reasons, most of the data analysis tools use the second approach, where the programs generate output files in specific formats that can subsequently be imported and visualized by external data visualization tools, such as Affymetrix Integrated Genome Browser (Nicol et al., 2009), analyzed by graphical data analysis environments such as Excel, MATLAB or R, or uploaded onto web-based browsers, e.g. UCSC Genome Browser (Karolchik et al., 2008). All these approaches require additional operations or even programming to load and examine the data. If uploading the data onto a web-based browser, the transfer of a large amount of data via the Internet is often a bottleneck, limiting the ability of researchers to examine their data thoroughly.
To better facilitate genomic data visualization, we present a flexible genomic data visualization tool, called CisGenome Browser, which has a specifically designed feature: besides being manually operated by a user, it can also be easily controlled by a third-party program regardless of the programming language in which the third-party program is written. This feature makes CisGenome Browser a flexible and universal data visualization tool. Any data analysis tool can simply output its results in formats that CisGenome Browser can interpret and then calls CisGenome Browser to visually present the results to the users. The whole procedure can be made automatic and transparent, which gives the illusion to the users that the data analysis program and CisGenome Browser work together as a single piece of software.
2 METHODS
2.1 Workflow
The workflow for a third-party program to control CisGenome Browser can be outlined in six steps, as illustrated in Figure 1.
The third-party program prepares data files in formats that CisGenome Browser can interpret.
The third-party program prepares a parameter file in a simple text format (the Windows INI format), which specifies the paths of the data files, the genomic region of interest and other parameters for data visualization.
The third-party program opens a web browser and then calls CisGenome Browser via a HTTP request, with one parameter specifying the name of the parameter file.
Working as a local web server, CisGenome Browser listens on a specific port. When it receives a request, CisGenome Browser parses the HTTP header and retrieves the name of the parameter file. It then opens the parameter file and reads in all the parameters, including the paths of the data files.
CisGenome Browser loads the data files entirely or partially depending on the parameters specified in the parameter file.
CisGenome Browser generates the web page according to the parameters and sends it back to the web browser, which then presents the web page to the user.
Fig. 1.
CisGenome Browser works as a visualization component for a third-party program. The six steps are numbered according to their temporal order in runtime.
2.2 Other features
CisGenome Browser is an open source software. It has a modular design and allows new features to be added conveniently. It was originally written as a data visualization module for CisGenome, a ChIP-chip and ChIP-seq data analysis tool (Ji et al., 2008). CisGenome Browser works with CisGenome core programs to visualize signal associated with genomic loci, raw images for Affemetrix microarrays and motif logos for transcriptional factor binding sites. CisGenome Browser was largely rewritten afterwards in order to serve as a standalone platform-independent visualization tool. As another example for its flexibility, CisGenome Browser is also used by the software JETTA (http://gluegrant1.stanford.edu/∼junhee/JETTA/, Junction and Exon array Toolkits for Transcriptome Analysis) to visualize microarray probe intensities.
CisGenome Browser is written in C++ using platform-independent libraries. As a result, it can run on Windows, Linux and the Mac platforms. The executable has no dependence on other packages or libraries, which makes the installation procedure very straightforward. It provides a light-weight web server (the whole program is <10 MB in size), which runs locally so that there is no need to upload the data via the Internet. The browser prohibits remote connection in the default configuration for safety reasons. It can also be configured so that remote users can browse the data sessions prepared by local users, which makes it a convenient tool for data sharing between labs.
CisGenome Browser is capable of visualizing a wide variety of genomic data, including genome sequences, conservation scores, gene annotations or any signal associated with genomic loci or regions such as those generated by microarrays, sequencing or other types of biological experiments. It can visualize the signal in bar plot, dot plot, line plot or heatmap plot, and allows several datasets to appear in one track in different colors. Almost every aspect of the visualization procedure is configurable in parameter files, such as picture height, width, color, axes, etc. The interface of CisGenome Browser is very similar in its look and feel to other genome browsers, such as the UCSC Genome Browser (Karolchik et al., 2008). One can easily shift or scale the genomic region, search for genes, and add, delete or customize tracks. It also integrates links to external websites such as NCBI (Wheeler et al., 2005), UCSC (Karolchik et al., 2008) and Ensembl (Flicek et al., 2008) on the webpage or in convenient pop-up menus. (See Table 1 for a summary of its features.)
Table 1.
CisGenome Browser feature list
| Feature category | Features |
|---|---|
| Runtime | Light-weight local web server, OS independent |
| Input file format | Tab-delimited text formats, UCSC BED, WIG and refFlat formats, BAR format, BAM format |
| Navigation | Panning, zooming, searching by gene names or genomic regions |
| Track management | Adding, deleting or customizing tracks |
| Plot types | Bar plot, dot plot, line plot, heatmap plot, gene structures, conservation scores, genome sequences, sequencing reads |
| Others | Visualizing Affymetrix microarray image, motif logos, integrated pop-up links to NCBI, UCSC and Ensembl, selected zooming |
The input data file formats for CisGenome Browser include tab-delimited text format, UCSC BED, WIG and refFlat formats, Affymetrix BAR format and BAM format (Li et al., 2009). When a text format file is loaded for the first time, CisGenome Browser automatically converts the file into binary format with a built-in index. Later on only the binary format file will be used for fast data retrieval.
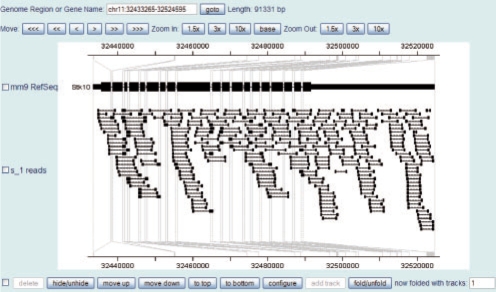
There are features specifically designed in CisGenome Browser for ultra high-throughput sequencing data visualization. For example, CisGenome Browser can visualize a genomic region by rescaling different parts of it so that the part of particular interest (such as exons) will be amplified, while the rest part (such as introns) will be compressed (Figure 2).
Fig. 2.
Paired-end RNA-seq reads mapped to mouse gene Stk10 are visualized in CisGenome Browser. The gene structure is folded so that all the intronic regions are compressed.
ACKNOWLEDGEMENTS
We thank Hongkai Ji and Wenxiu Ma for helpful advice.
Funding: National Institute of Health grant R01-HG004634 (to W.H.W.); National Institute of Health grant 2P01-HG000205 (in part); Agilent Technologies (gift grant).
Conflict of Interest: none declared.
REFERENCES
- Flicek P, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC genome browser database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. DNA-chip analyzer (dChip) In: Parmigiani G, et al., editors. The Analysis of Gene Expression Data: Methods and Software. New York: Springer; 2003. pp. 120–141. [Google Scholar]
- Li H, et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol JW, et al. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]