Abstract
Diet and nutritional status are among the most important, modifiable determinants of human health. The nutritional value of food is influenced in part by a person’s gut microbial community (microbiota) and its component genes (microbiome). Unraveling the interrelationships between diet, the structure and operations of the gut microbiota, and nutrient and energy harvest is confounded by variations in human environmental exposures, microbial ecology and genotype. To help overcome these problems, we created a well-defined, representative animal model of the human gut ecosystem by transplanting fresh or frozen adult human fecal microbial communities into germ-free C57BL/6J mice. Culture-independent, metagenomic analysis of the temporal, spatial and intergenerational patterns of bacterial colonization showed that these humanized mice were stably and heritably colonized, and reproduced much of the bacterial diversity of the donor’s microbiota. Switching from a low-fat, plant polysaccharide-rich diet to a high-fat/high-sugar “Western” diet shifted the structure of the microbiota within a single day, changed the representation of metabolic pathways in the microbiome, and altered microbiome gene expression. Reciprocal transplants involving various combinations of donor and recipient diets revealed that colonization history influences the initial structure of the microbial community, but that these effects can be rapidly altered by diet. Humanized mice fed the Western diet have increased adiposity; this trait is transmissible via microbiota transplantation. Humanized gnotobiotic mice will be useful for conducting proof-of-principle “clinical trials” that test the effects of environmental and genetic factors on the gut microbiota and host physiology.
INTRODUCTION
Advances in ‘next-generation’ DNA sequencing have dramatically reduced costs, and markedly increased capacity, allowing culture-independent, metagenomic methods to be readily deployed to characterize microbial communities (microbiota) associated with human body habitats, at various stages of the human lifecycle, and in various populations (1–5). These metagenomic surveys not only capture the microbial organismal and genetic diversity associated with humans, but also begin to investigate the functional contributions that our microbes make to our physiologic phenotypes, our health, and our disease predispositions. The distal gut is home to our largest number of microbes. A recent study of the fecal microbiota of adult mono- and dizygotic pairs of twins and their mothers revealed that no single, identifiable abundant (defined as representing >0.5% of the microbiota) bacterial species was shared by all 154 individuals surveyed. Nonetheless, family members had a more similar community structure than unrelated individuals. In addition, the degree of relatedness of the fecal microbiota of adult monozygotic twin pairs was not greater than that of dizygotic twin pairs (5), suggesting that early environmental exposures are important determinants of community structure. Despite interpersonal differences in bacterial species content, large collections of microbial genes are shared among unrelated individuals. This core microbiome encodes metabolic traits that benefit the host, including activities related to processing of otherwise indigestible components of the host diet, such as polysaccharides (5).
A major goal of comparative human metagenomic studies is to move beyond a description of microbial species or genes present in particular habitats, and link the structure and dynamic operations of microbial communities reciprocally to human biology and pathobiology. Addressing these issues directly in humans is challenging because of numerous uncontrolled variables, both genetic and environmental.
Mice raised in germ-free environments, without any exposure to microbes, and then colonized at specific life stages with different microbial communities are referred to as gnotobiotic animals, and provide an excellent system for controlling host genotype, microbial community composition, diet, and housing conditions. Microbial communities can be harvested from donor mice having defined genotypes and physiologic phenotypes and used to determine how these communities impact formerly germ-free recipients, and how the recipient affects the transplanted microbiota/microbiome. In this respect, gnotobiotic mice provide an opportunity to marry comparative metagenomic studies of donor communities with functional assays of community properties. However, there is a caveat about applying results obtained from gnotobiotic mice carrying mouse-derived microbial communities to humans: culture-independent comparisons have revealed that, although the distal gut microbiota of mice and humans harbor the same bacterial phyla (Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, Cyanobacteria, TM7, Fusobacteria, and Spirochaeates), most bacterial genera and species found in mice are not seen in humans (6).
To develop a more applicable mouse model, we transplanted an adult human fecal microbiota into germ-free C57BL/6J mice. A stable human gut community became established in the recipient mice similar to that of the donor. Using diet-induced obesity as a model, we show how these humanized animals can be used to conduct controlled proof-of-principle “clinical” metagenomic studies of host-microbiome interrelationships.
RESULTS
Establishment of a humanized mouse gut microbiota
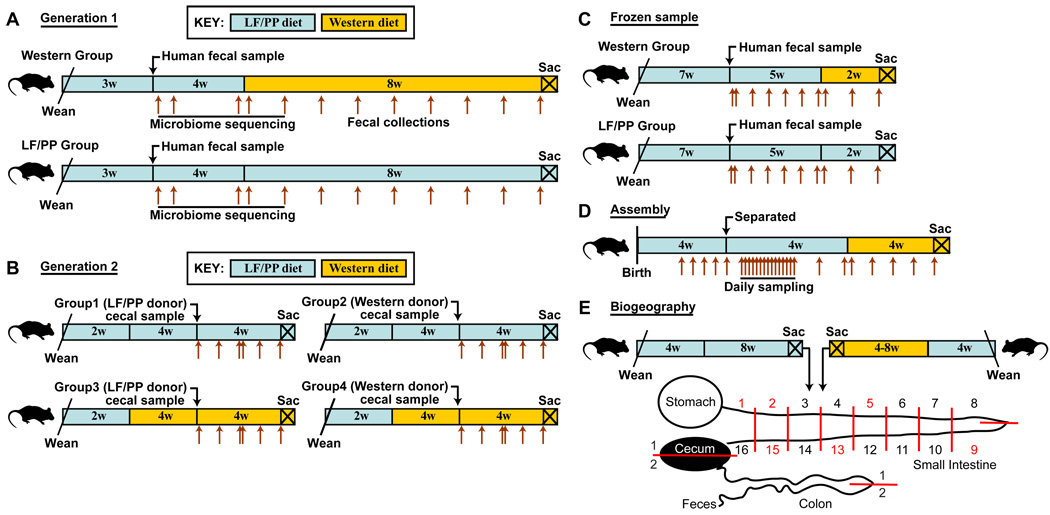
We performed an initial colonization of young adult (5–7 week-old), male C57BL/6J mice using the microbial community present in a freshly voided fecal sample from a healthy adult human (fig. 1A). The sample was immediately placed in an anaerobic environment (Coy chamber). One aliquot of the sample was frozen, while a second was diluted in reduced PBS and an aliquot of this dilution was introduced by gavage into each of 15 recipient animals. The procedure was completed within 1h after the fecal sample was produced. The recipient mice were maintained on a standard low-fat, plant polysaccharide-rich diet (LF/PP), in separate cages within a gnotobiotic isolator. Fecal samples were collected from these animals one day, one week, and one month after colonization, at which point half of the mice were switched to a high-fat/high-sugar Western diet. Mice from the LF/PP group and the Western diet group were kept on their respective diets for two additional months, with weekly fecal sampling, at which point they were sacrificed.
Fig. 1. Design of human microbiota transplant experiments.
(A) The initial (first-generation) humanization procedure, including the diet shift. Brown arrows indicate fecal collection time points. (B) Reciprocal microbiota transplantations. Microbiota from first-generation humanized mice fed LF/PP or Western diets were transferred to LF/PP or Western diet-fed germ-free recipients. (C) Colonization of germ-free mice starting with a frozen human fecal sample. (D) Characterization of the postnatal assembly and daily variation of the humanized mouse gut microbiota. (E) Sampling of the humanized mouse gut microbiota along the length of the gastrointestinal tract.
Sequencing of the 16S rRNA genes in an ecosystem is used to identify microbes present, without the need to culture them, and to classify their evolutionary relationships. Therefore, we performed multiplex pyrosequencing of amplicons, generated by PCR from variable region 2 (V2) of bacterial 16S rRNA genes (5, 7, 8) to characterize the taxa present in the fecal communities of the two groups of humanized gnotobiotic mice (N=153 fecal samples analyzed, collected at the time points shown in fig. 1A; tables S1 and S2). Fecal and cecal samples obtained at the time of sacrifice were also characterized, as were six subsamples obtained from the original human fecal sample.
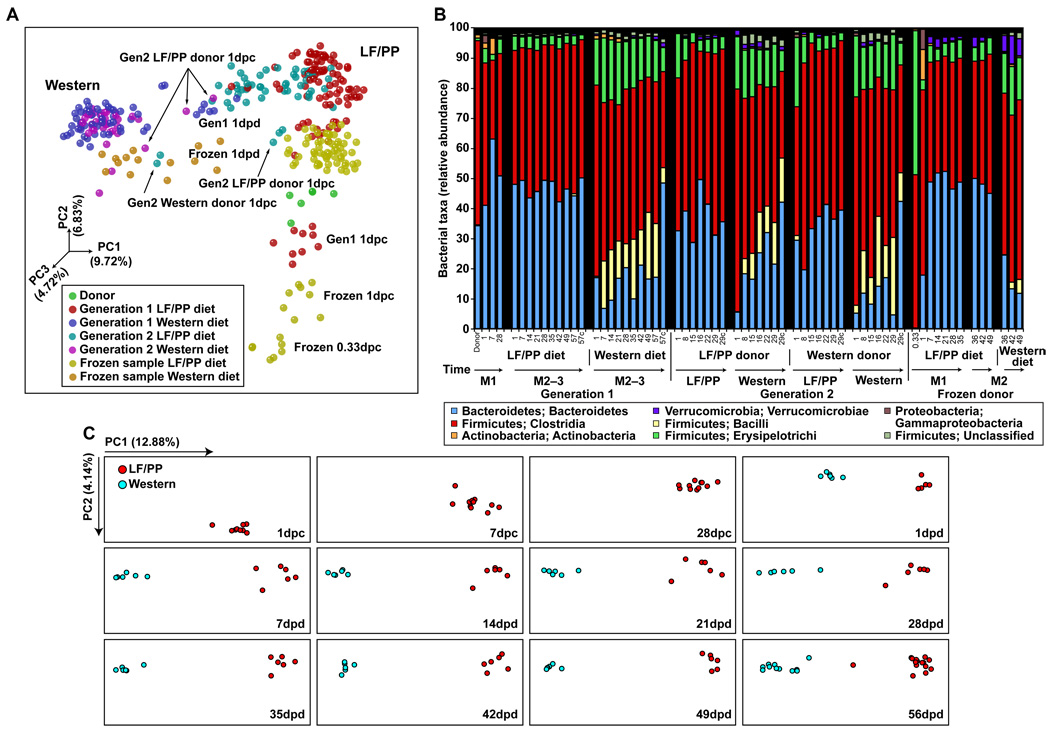
We assessed the degree of similarity of the fecal microbial communities with the UniFrac metric, which is based on the degree to which any two communities share branch length on a ‘master’ phylogenetic tree constructed from all 16S rRNA sequences obtained from all communities surveyed in a study (9). Principal coordinates analysis (PCoA) of UniFrac-based, pairwise comparisons of community structures revealed that in mice consuming a LF/PP diet throughout the 12 week period of surveillance, the microbiota attained a sustained configuration by 28 days (d) after transplantation (fig. 2A and C). Administration of the Western diet to mice 4 weeks after colonization resulted in a shift in gut community structure that was evident after a single day and that stabilized by 7d (fig. 2A–C). Assuming an intestinal transit time of 4–6 hours (h) (10), this means that the initial diet-induced shift was evident after microbes had been exposed to the altered diet for less than 18–20h.
Fig. 2. The effects of switching from the LF/PP diet to the Western diet on the humanized mouse gut microbiota.
(A) 16S rRNA gene surveys [analyzed by unweighted UniFrac-based principal coordinates analysis (PCoA)] from the human donor (green), first-generation humanized mice fed LF/PP (red) or Western (blue) diets, second-generation microbiota transplant recipients consuming the LF/PP (light blue) or Western (purple) diets, and mice humanized with a frozen sample fed LF/PP (yellow) or Western (orange) diets (total of 340 samples with >800 sequences/sample). Weighted UniFrac resulted in a similar overall clustering pattern (data not shown). Principal coordinate 1 (PC1) and PC2 are the x- and y-axis, respectively, and have been scaled on the basis of percent variance. PC3 is depicted by the shading of each point. Percent variance is shown in parentheses. dpc, days post colonization with a human donor sample; dpd, days post diet switch. (B) Taxonomic distribution [RDP level 3 (class level taxa) (27)] of two generations of humanized mice fed a LF/PP or Western diet. Values represent the average relative abundance across all samples within the indicated group. C, cecal samples while all other samples are fecal; M, month. (C) 16S rRNA gene sequences (analyzed by unweighted UniFrac-based PCoA) from the mice in the experiment described in fig. 1A. The x- and y-axes are scaled based on the percent variance accounted for by each component (shown in parentheses). Each box corresponds to a single timepoint. dpc, days post colonization with the human donor sample; dpd, days post diet switch.
The Western diet-associated humanized mouse microbiota had an increased relative representation of the Erysipelotrichi class of bacteria within the Firmicutes phylum when compared to mice fed the LF/PP diet (table S3). Phylogenetic analysis (fig. S1) indicated that organisms with increased representation were most closely related to Clostridium innocuum, Eubacterium dolichum, and Catenibacterium mitsuokai (all Erysipelotrichi previously isolated from the human gut). In mice fed the Western diet, Erysipelotrichi represented on average, 15.88±0.74% of 16S rRNA sequences (fig. 2B), an abundance that is significantly different than that of the community associated with mice maintained on the LF/PP diet (3.25±0.18%; P<0.01; Student’s t-test). Consumption of the Western diet was also associated with a significant increase in the relative abundance of another class of Firmicutes, the Bacilli (mainly Enterococcus) and a significant decrease in the proportional representation of members of the Bacteroidetes (P<0.01; fig. 2B and tables S3 and S4).
16S rRNA surveys of fecal samples obtained from mice in both diet groups revealed that engraftment of the human gut microbiota was largely successful: all bacterial phyla, 11/12 bacterial classes, and 88% (58/66) of genus-level taxa detected in the donor sample were detected among the recipient mice (tables S3 and S4). All eight genus-level taxa that were missing from the humanized mice were present at low abundance in the donor sample (0.008% on average). Diversity in the fecal microbiota of the transplant recipients one month after colonization was not significantly (Student’s t-test) lower than in the donor sample [Shannon index of 4.53±0.15 (donor) versus 4.09±0.05 (LF/PP) and 4.51±0.04 (Western); Chao1 estimate of 1,316±295 (donor) versus 920±24 (LF/PP) and 1,055±29 (Western); a subsample of 951 sequences were randomly selected and analyzed from each dataset to control for differences in sequencing depth; also see fig. S2A].
Propagation of the humanized mouse gut microbiota
To determine whether the humanized mouse microbiota could be transmitted to a second generation of animals, and to test the relative impact of legacy effects (source of the community) versus current host diet, we used 19 week-old humanized mice maintained on the LF/PP diet or the Western diet as donors for a new set of cecal microbiota transplants. The 10 week-old germ-free C57BL/6J recipients had either been maintained on the LF/PP diet since weaning or switched to the Western diet at 6 weeks of age for four weeks, after which time they were colonized by gavage. All possible combinations of donor and recipient diet groups were examined (N=3–5 mice/treatment group; fig. 1B). Fecal microbial community structure was monitored in all recipients with multiplex V2-directed 16S rRNA gene pyrosequencing beginning one day after colonization and continuing weekly for a month (N=89 samples; table S1).
The results indicated that the human gut microbiota could be successfully transferred from generation to generation without a significant drop in diversity, as judged by rarefaction curves (fig. S2A), the Shannon index [4.35±0.06 (LF/PP) and 4.60±0.06 (Western) compared to 4.09±0.05 (LF/PP) and 4.51±0.04 (Western) for the first-generation humanized donors], and Chao1 estimates [977±24 (LF/PP) and 1,084±36 (Western) compared to 920±24 (LF/PP) and 1,055±29 (Western) for the first-generation humanized donors; N=951 sequences per randomly selected subsample]. The overall depth of sequencing of samples obtained from second-generation animals (that is the number of V2 16S rRNA gene pyrosequencer reads collected) was 2-fold lower compared to samples from first-generation samples. Nonetheless, the second-generation microbiota shared at least 83% of the class-level taxa and 73% of the genus-level taxa with the first-generation community (tables S3 and S4).
UniFrac-based PCoA revealed that the microbiota in recipients consuming a Western diet assumed a phylogenetically similar composition regardless of the diet of the donor mouse. Moreover, samples taken from mismatched donor/recipient diet groups (LF/PP donor and Western recipient or vice versa) displayed intermediate clustering and taxonomy after the first day, but clustered according to recipient diet after one week (fig. 2A,B and fig. S3). As with the first-generation recipients, the Western diet-associated microbiota had significantly higher representation of members of the Firmicute classes Erysipelotrichi and Bacilli (Enterococcus), accompanied by a reduction in the proportional representation of the Bacteroidetes (fig. 2B; tables S3 and S4). Together, these results indicate that legacy effects (that is, colonization history) can play a role in determining the initial community structure after transfer, but that these effects are quickly replaced by the diet of the recipient host.
Establishment of a humanized mouse gut microbiota from frozen samples
We next determined whether humanization with a frozen fecal sample would result in similar diversity as colonization with a fresh sample. Therefore, ten 10 week-old germ-free C57BL/6J mice were colonized with the same fecal sample used for the initial humanization except that it had been stored at −80°C for over one year. Five weeks after colonization, half of the mice were switched to the Western diet and sacrificed 2 weeks later (N=5 mice/group) (study design illustrated in fig. 1C). Fecal microbial community structure was monitored in all recipients using multiplex V2-directed 16S rRNA gene pyrosequencing, beginning 8h after colonization (N=92 samples; table S5).
Fecal samples obtained from recipients 8h after inoculation had low diversity (fig. S2B), clustered separately (fig. 2A and fig. S3), and had a higher representation of Erysipelotrichi (47.97±1.99% of 16S rRNA sequences; fig. 2B) compared to recipients of the fresh sample. However, as with humanization using the fresh sample, the microbiota stabilized after 7d, with a level of diversity approaching that of the donor (fig. 2A,B and fig. S2B). All bacterial phyla, 11/12 bacterial classes, and 85% (56/66) of genus-level taxa detected in the frozen donor sample were found in the recipient mouse population (tables S3 and S4). Of the ten genus-level taxa that were missing from the humanized mice, one was found at 0.1% relative abundance in the original donor sample (Firmicutes; Clostridia; Clostridiales; Veillonellaceae; Acidaminococcus) and the rest were all at low abundance in the sample (0.006% on average). Switching mice to a Western diet significantly increased the Erysipelotrichi and significantly decreased the Bacteroidetes; these changes were evident within 1d day of the diet switch (fig. 2A,B; table S3). Our results show that it is possible to humanize germ-free animals by using frozen fecal samples.
Temporal and spatial features of the human gut microbiota
We used our humanized gnotobiotic mice to characterize two aspects of the human gut microbiota that are not currently feasible in humans: (i) postnatal assembly of the microbial community under controlled conditions; and (ii) the impact of a diet perturbation on the spatial organization of the microbial community along the entire length of a gut undisturbed by any of the bowel cleansing protocols commonly used before human endoscopic sampling.
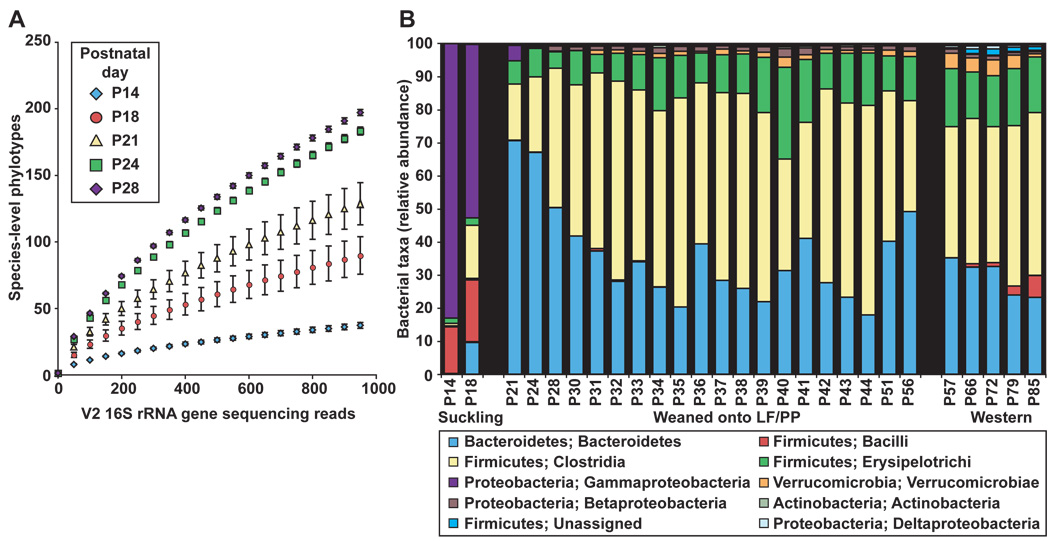
We therefore performed a 16S rRNA gene-based survey of fecal specimens obtained from humanized mice beginning at postnatal day 14 (P14), a time when adequate amounts of (fecal) microbial biomass are first available for characterization of individual samples, through P85 (N=146 samples with ≥500 sequences/sample; table S6). The study design is summarized in fig. 1D. Mice (N=6) delivered by a humanized mother were subsequently co-housed with their mother, weaned onto the LF/PP diet beginning at P21 and, once weaned (P28), individually caged in the same gnotobiotic isolator until P57, at which time they were switched to the Western diet, and samples taken for another 28d.
From P14 to P24, the gut microbiota exhibited an increase in bacterial diversity (fig. 3A) and a marked shift in the relative abundance of multiple bacterial classes. Notably, there was a decrease in γ-Proteobacteria and Bacilli, which together comprise 81.44±8.48% of the fecal microbiota between P14-P18 and an increase in the Bacteroidetes (fig. 3B and fig. S4A,B). The microbiota assumed an adult-like configuration by P21-P28 (fig. 3B). This configuration stably persisted as judged by pairwise comparisons of UniFrac distances between fecal samples collected daily from P30-P44 (fig. 1D and fig. S4C).
Fig. 3. Postnatal assembly of the humanized gut microbiota.
(A) Rarefaction curves measuring bacterial diversity in the fecal communities (species-level phylotypes defined by ≥97% identity). The curves are based on V2 16S rRNA gene sequences obtained from mice prior to weaning (P14) and after weaning (P28). Values are mean±95% CI. (B) Taxonomic distribution [RDP level 3 (27)] of the gut microbiota sampled from mice from postnatal days P14 to P85. Values represent the average relative abundance across all samples within a given group.
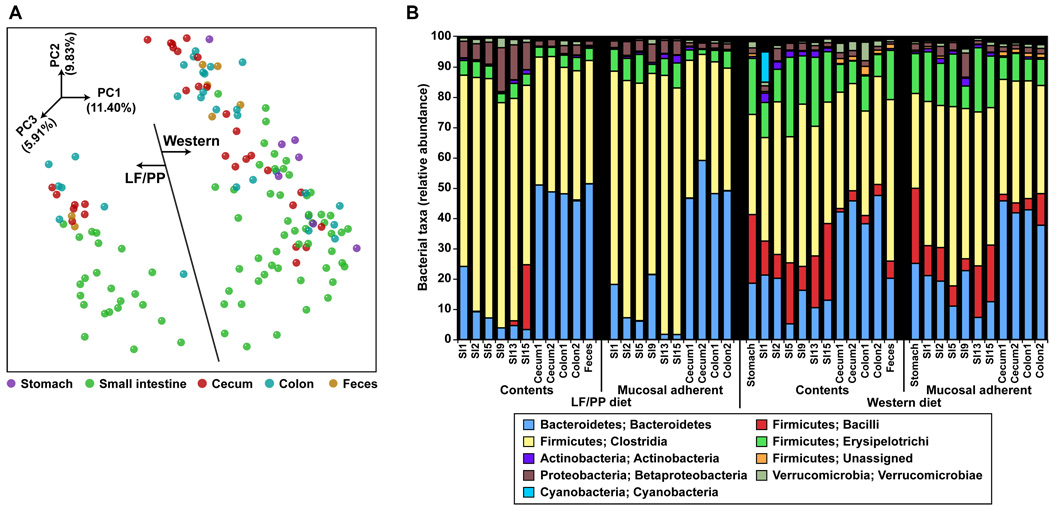
We next defined spatial variations in the microbiota along the length of the gastrointestinal tract of humanized mice consuming a LF/PP or a Western diet, as outlined in fig. 1E. Adult mice were sacrificed, and their small intestines were divided into 16 equal length segments and their ceca and colons were separated into two halves. We obtained fecal samples, and samples from the luminal contents of the stomach, small intestinal segments 1 (most proximal), 2, 5, 9, 13, and 15, and the proximal and distal halves of the cecum and colon. Luminal contents were sampled by perfusing the segments with buffer. The luminal surface of each perfused segment was then swabbed with a culture loop to sample the mucosa-adherent community (see SM and table S7; N=148 samples subjected to multiplex pyrosequencing of V2-derived 16S rRNA amplicons; >500 reads/sample; total of 8 animals surveyed).
Communities from comparably positioned segments of the gut in mice consuming the same diet were similar (P<10−4 based on a G-test of shared phylotypes, defined as ≥97% sequence identity) but we did not detect a significant difference between samples taken from perfused luminal contents and from the swabbed mucosa (P>0.05, G-test of shared phylotypes). Microbial community structure was impacted by location along the cephalocaudal axis (fig. 4a). Notably, there was a dramatic increase in the percent representation of Bacteroidetes as one moved from the small intestine to the cecum, colon, and feces (13.71±1.40% for the stomach/small intestine samples versus an average of 42.33±1.67% for the cecal/colonic/fecal samples; P<10−10, Student’s t-test). Additionally, the overall level of diversity was region-dependent, ranging from lowest levels in the proximal intestine, to the highest levels in the colon and feces (fig. S2C). The Western diet was associated with significantly increased levels of Bacilli and Erysipelotrichi along the entire length of the gut compared to the LF/PP diet (fig. 4B; P<10−6, Student’s t-test). Although the relative abundance of the Bacteroidetes in the cecum and feces was comparable on the LF/PP diet [40.15±2.17% (feces) versus 45.43±1.83% (cecum)], it was significantly lower in fecal samples taken from mice fed the Western diet (fig. 2B and 4B) (16.56±2.26% in the feces versus 44.50±2.05% in the cecum across three experiments; P<10−10, Student’s t-test).
Fig. 4. Clustering and taxonomic analysis of the gut microbiota of humanized mice consuming a LF/PP or Western diet.
(A) 16S rRNA gene surveys (analyzed by unweighted UniFrac PCoA) of the humanized microbiota along the length of the gut (N=148 samples with >500 sequences/sample). Weighted UniFrac results in a similar overall clustering pattern. PC1 and PC2 are the x- and y-axis respectively. Percent variance is shown in parentheses. (B) Taxonomic distribution [RDP level 3 (27)] in communities distributed along the length of the gut. Values represent the average relative abundance across all samples within a given group.
The humanized mouse gut microbiome
The studies described above, including the reciprocal microbiota transplants, allowed us to rationally select time points to sample the gut microbiome in two dietary contexts. In addition, by demonstrating the consistency in microbiota composition between humanized adult mice on a given diet, they provided justification for pooling fecal samples obtained at given time points before and after diet switches so that we could obtain sufficient microbial biomass for direct shotgun sequencing of the microbiome. Therefore, to characterize microbiome gene composition samples were analyzed in 10 pools, each pool representing 3–5 mice sampled at 1d, 7d, or 28d after the human fecal transplant, plus 1d and 1 week after the switch to the Western diet (the same sampling/pooling protocol was applied to members of the control group who remained on the LF/PP diet). DNA isolated from the freshly voided human sample used for the microbiota transplant was also analyzed. Combined, 479,495 reads were generated (average of 43,590±2,246 reads per sample; average length 224 nt/read; table S8). After removing low quality and replicate sequences, microbiomes were annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [version 44 (11)] and a custom database of 122 reference human gut genomes (table S9 and SM).
Clustering of microbial communities based on their gene content revealed a pattern similar to that observed with the 16S rRNA datasets (fig. 2A and fig. 5A). Samples taken one day after humanization had the most divergent microbiome gene content, while samples taken one week after colonization clustered with the human donor sample. Remarkably, a clear shift in gene content was evident 1d after the switch to the Western diet (fig 5A). The Western diet-associated fecal microbiome was enriched for a number of KEGG pathways involved in nutrient processing, including those for ABC transporters and Phosphotransferase Systems (PTS) (table 1). ABC transporters included those predicted to be involved in sugar, amino acid, and cofactor import, while PTS included predicted transporters for fructose, N-acetylgalactosamine, cellobiose, and mannose (table S10).
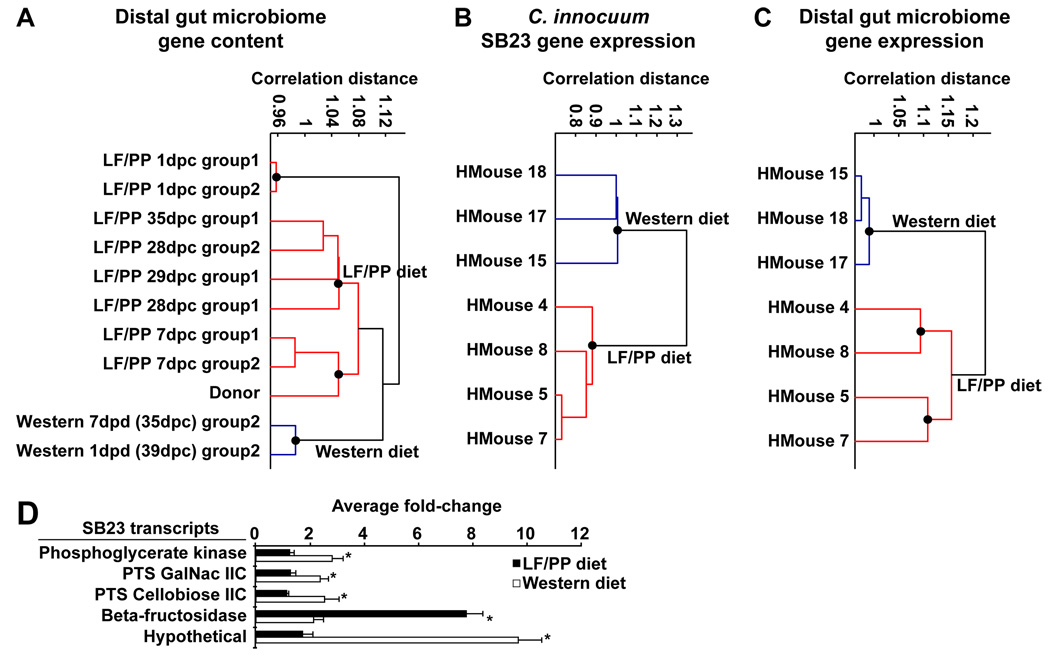
Fig. 5. Clustering of the distal gut microbiome, the C. innocuum SB23 transcriptome, and the community meta-transcriptome in the ceca of humanized mice.
Microbiome/transcriptome profiles were normalized by z-score, used to construct a correlation distance matrix, clustered with UPGMA, and visualized (see SM; Matlab v7.7.0). (A) Clustering of fecal microbial gene content in the human donor’s microbiome and two groups of humanized mice starting at day one post colonization (N=3–5 mice/group; fecal DNA was pooled prior to sequencing). All mice were maintained on the LF/PP diet (red) for 28 days, at which point group2 was transferred to the Western diet (blue). (B) Clustering of C. innocuum SB23 gene expression in humanized mice fed the LF/PP (red) or Western (blue) diet. (C) Clustering of the gut microbiome’s meta-transcriptome in humanized mice fed a LF/PP (red) or a Western (blue) diet. Black circles represent validated clusters (inconsistency threshold=0.75, ‘cluster’ function in Matlab v7.7.0). dpc, days post colonization with a human donor sample; dpd, days post diet switch. (D) qRT-PCR validation of C. innocuum SB23 gene expression in humanized mice (N=3–5 samples/group; see SM). Mean values±SEM are plotted (*P<0.05, Student’s t-test).
Table 1.
Metabolic pathways altered by diet in the humanized mouse gut microbiome*.
| KEGG Pathways Enriched on LF/PP diet | Cyanoamino acid metabolism |
| Glycosaminoglycan degradation | |
| Glycosphingolipid biosynthesis - ganglioseries | |
| N-Glycan degradation | |
| Nucleotide sugars metabolism | |
| Other ion-coupled transporters | |
| Pentose and glucuronate interconversions | |
| Phenylpropanoid biosynthesis | |
| Sphingolipid metabolism | |
| Starch and sucrose metabolism | |
|
KEGG Pathways Enriched on Western diet |
ABC transporters |
| Phosphotransferase system (PTS) | |
| Transporters |
Based on a bootstrap analysis of gut microbiome datasets from humanized mice fed the LF/PP diet (7–35 dpc) or the Western diet (29–35dpc; CI=0.95, 10,000 samples; inclusion criterion, pathways must be found at ≥0.6% relative abundance in at least two samples) dpc, days post colonization with a human fecal sample
The LF/PP diet-associated microbiome was enriched for pathways that are also enriched in the Bacteroidetes (5), including N-glycan degradation, sphingolipid metabolism, and glycosaminoglycan degradation (table 1). Thus, this profile is consistent with the increased representation of Bacteroidetes in the LF/PP compared to Western diet fecal microbiome (fig. 2B).
Microbial transmission of adiposity
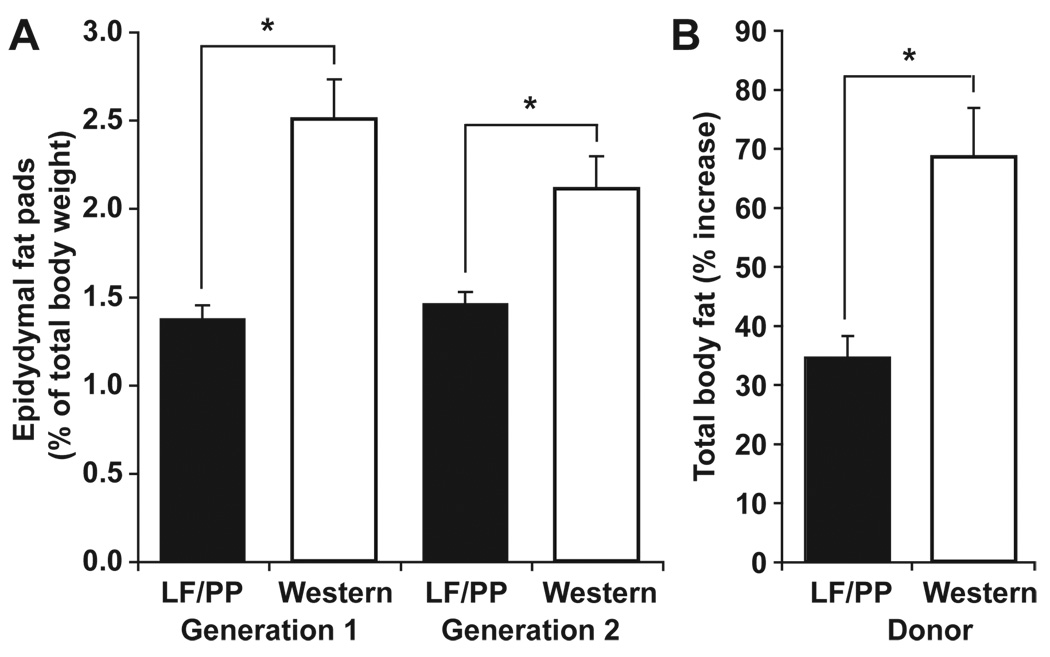
Administration of a Western diet to humanized mice produced a significant increase in adiposity compared to age- and gender-matched humanized mice consuming the LF/PP diet (epididymal fat pad weight, 2.29±0.15% versus 1.42±0.05% of body weight; N=5–8 mice/treatment group; 2 independent experiments; P<0.05, Student’s t-test; figure 6a).
Fig. 6. Transmissibility of adiposity from humanized mice to germ-free recipients.
(A) The effects of Western and LF/PP diets on epididymal fat pad weight (expressed as a percentage of total body weight) in humanized gnotobiotic mice (N=5–8 mice/group; N=2 independent groups). (B) Percent increase in total body fat (measured by DEXA) after colonization of germ-free mice with a microbiota harvested from humanized donors fed the Western or the LF/PP diet (N=4–5 mice/group). Recipients were fed a LF/PP diet. Mean values±SEM are plotted (*P<0.05, Student’s t-test).
Our previous microbiota transplantation studies demonstrated that distal gut communities harvested from genetically obese ob/ob mice (12), or conventionally-raised wild-type C57BL6/J mice who were obese because of consumption of the Western diet (13), produced a greater increase in adiposity in adult germ-free recipient mice, within two weeks, compared to communities that had been donated by lean LF/PP-fed controls.
We performed a comparable experiment to functionally characterize the gut microbiota of humanized mice with diet-induced obesity. Germ-free C57BL/6J recipients were colonized for two weeks with cecal microbial communities harvested from humanized donors fed the LF/PP or Western diets (N=1 donor and N=4–5 recipient mice/group). The two groups of recipients were matched at the start of the experiment for age, weight, body fat (21.9±0.2 vs. 22.1±0.3 g body weight, and 9.6±0.3 vs. 9.5±0.6% total body fat) and diet (LF/PP). There was no significant difference in chow consumption between the two groups of recipient mice (15.3±0.3 vs. 15.2±0.4 kcal/d over the entire experiment; there was also no significant difference during the first or second week when considered separately). Mice colonized with a microbiota from obese Western diet-fed humanized donors gained significantly more adiposity during the two weeks following transplantation than did mice colonized with the humanized microbiota from LF/PP-fed donors (68.5±8.2% versus 34.5±3.7% increase in total body fat; P<0.05, Student’s t-test) (fig. 6B).
Recovery of a Western diet-associated Erysipelotrichi
Our humanized gnotobiotic mouse model also allows the identification of previously uncharacterized lineages of the human gut microbiota. We sought to (i) recover a microbe belonging to the Erysipelotrichi taxa that bloomed in Western diet-fed mice, (ii) define its genome sequence, (iii) compare the isolate’s sequence to that of members of this population in the humanized mouse gut microbiome in order to identify polymorphic regions and, finally, (iv) compare the isolate’s transcriptome and the meta-transcriptome expressed by the gut microbial community of humanized mice consuming a Western or LF/PP diet.
After humanized mice were on the Western diet for over two months, cecal contents were isolated under anaerobic conditions, diluted, and plated on rich medium under strictly anaerobic conditions (see SM). RFLP (Restriction Fragment Length Polymorphism) screening, followed by 16S rRNA gene sequencing allowed us to identify four isolates belonging to Erysipelotrichi out of a total of 17 colonies screened. We sequenced the genome of one of the isolates, which shared 98% identity with the 16S rRNA gene of Clostridium innocuum ATCC14501 (table S11). Annotation of this strain’s genome (designated C. innocuum strain SB23) with the RAST annotation server (14) revealed 245 predicted PTS and 115 ABC transporter genes. Annotation of carbohydrate active enzymes (CAZymes) using procedures implemented for the CAZy database (15) yielded 78 predicted CAZymes, including 21 genes in the glycoside hydrolase (GH) family 1 (candidate 6P-β-glycosidases), 3 genes in glycoside hydrolase family 4 (candidate 6P-glycosidases), 10 genes in glycosyltransferase family 2 (candidate β-glycosyltransferases), and 2 genes in glycoside hydrolase family 32 (candidate β-fructosidases potentially capable of degrading sucrose, a major component of the Western diet). The enrichment for GH1 in C. innocuum SB23 is particularly striking when compared to other annotated human gut-associated bacterial genomes (no genes in 13 sequenced human gut Bacteroidetes; ≤5 genes from each of 14 human gut Firmicutes (P<0.01, z-score=21; table S12). In contrast, the profile of glycosyltransferases in this strain is more typical of other gut Firmicutes.
To investigate genome-wide sequence variation in the in vivo SB23 population, we compared our gut microbiome shotgun sequences to the cultured SB23 isolate’s genome. 96% of the reads that aligned to the reference genome aligned at ≥90% identity. Moreover, 97% of the aligned reads were aligned across ≥90% of their length (see SM and fig. S5). Together, these findings indicate that there is only a low level of sequence variation for the SB23 population in the humanized mouse microbiome.
Using a method for rRNA depletion of total cecal RNA together with an Illumina GAII sequencer, we compared mRNAs expressed by the genes represented in the SB23 genome in the LF/PP versus Western diet-associated cecal microbiota (N=3–4 mice/group; see SM and table S13). The results revealed distinct clustering of the SB23 transcriptomes expressed in humanized mice consuming the Western versus the LF/PP diet (fig 5B). Cyber-T analysis (16) disclosed 69 genes that were significantly upregulated on the Western diet (table S14). Two upregulated PTS genes were included in this dataset (SB23peg3102, 4.8-fold; SB23peg2651, 8-fold). Sequence homology suggests that SB23peg2651 may preferentially import simple sugars (glucose and fructose that together comprise sucrose, a common component of the Western diet) and/or sugars associated with host mucosa (N-acetyl-galactosamine). Genes involved in basic carbohydrate metabolism were also upregulated, including pyruvate formate-lyase (SB23peg2932; 9.5-fold), phosphoglycerate kinase (SB23peg1427; 8.8-fold), and 6-phosphofructokinase (SB23peg2154; 3.5-fold; see table S14). qRT-PCR assays were used to confirm several of these results, including the significant upregulation of phosphoglycerate kinase and two PTS genes on the Western diet, plus a significant upregulation of β-fructosidase on the LF/PP diet (fig. 5D and table S15).
To obtain a broader view of the transcriptional responses of the microbiome to diet shifts, we compared cDNA sequences generated from rRNA-depleted RNAs isolated from the ceca of humanized mice sacrificed 8 weeks after shifting to a Western diet to those from control animals maintained on the LF/PP diet, using a custom database of 122 sequenced human gut genomes (all protein-encoding genes from the 122 genomes were grouped into clusters; table S13 and SM). Clustering of meta-transcriptomes revealed a clear difference between gene expression in the microbiomes from animals on the LF/PP versus Western diet (fig. 5C). A Cyber-T analysis (see SM) revealed 520 gene clusters that were differentially expressed between the two diets (fold-change ≥4; table S16). 148 clusters containing C. innocuum SB23 genes were upregulated on the Western diet, including clusters encoding the previously noted Western diet-associated transcripts [pyruvate formate-lyase (SB23peg2932), a PTS predicted to be involved in fructose/mannose/N-acetygalactosamine import (SB23peg2651-2652), plus phosphoglycerate kinase (SB23peg1427)]. Additionally, 349 clusters without predicted C. innocuum SB23 gene products were upregulated on the Western diet, including 10 Firmicute gene clusters encoding ABC-type sugar transport systems. Twenty-three clusters (21 from Bacteroides genomes) were upregulated on the LF/PP diet, including a predicted β-glucosidase, β-xylosidase, and malate/lactate dehydrogenase (table S16).
Together, these results demonstrate that humanized gnotobiotic mice can be used to conduct controlled, comparative metagenomic studies of how the human gut microbiota and its microbiome adapts in composition and function to defined changes in diet.
DISCUSSION
The idea of transplanting human gut communities into germ-free animals is longstanding (17–22). However, we have now used newly available metagenomic methods to demonstrate that (i) a human distal gut microbiota can be transferred to germ-free mice with remarkable preservation of structure and diversity, even if the starting material is frozen feces, (ii) the community can subsequently be reliably transmitted from mothers to their offspring, (iii) the microbiota in these humanized mice has characteristic and reproducible variations in its composition along the length of the gut, and (iv) the configuration of the microbiota, its microbiome and meta-transcriptome change in a rapid, dramatic and reproducible fashion after switching from a plant polysaccharide-rich/low fat diet to a high-fat/high-sugar Western diet. We also show that a host phenotype, Western diet-induced increased adiposity, can be transmitted for a period of time to recipient gnotobiotic mice via transplantation of their gut microbiota. Together, these findings establish the justification for a translational medicine pipeline for conducting proof-of-principle and proof-of-mechanism clinical metagenomic studies where the impact of representative human microbiomes on host biology can be initially characterized in mice consuming diets resembling those of the humans being studied, and where the effects of various environmental exposures (e.g., foods, xenobiotics, etc.), and host genotypes on the dynamic operations of their gut microbial communities can be modeled under controlled conditions.
The phylogenetic tree of bacterial life in the gut consists primarily of shallow twigs that represent species- or genus-level diversity: these twigs stem back to a few deep splits near the base of the tree that represent the dominant phyla present in this body habitat (Firmicutes, Bacteroidetes, and Actinobacteria). This phylogenetic structure contrasts with other ecosystems such as the soil, where there are many deep branches of the tree represented (23), and suggests that the gut selects for diversification in the few deep evolutionary lineages that flourish there. Microbiota transplants into germ-free mice living in gnotobiotic isolators where inadvertent contact with microorganisms living in the world outside of the isolator is avoided allow us to consider the selective forces that operate to assemble a gut community. We had previously attempted to transplant a foreign gut microbiota into adult germ-free mouse recipients, where the donors were conventionally raised zebrafish. Members of the donor gut community that most closely resembled the phylogenetic lineages normally present in the mouse gut microbiota survived in the mouse intestine, as judged by 16S rRNA surveys of the input (donor) community and selected recipient communities (24). The factors that operated to select these phylotypes were not defined. The results from reciprocal microbiota transplants performed in the current study, in which various combinations of donor and host diets were tested, demonstrate that although the initial environmental exposure (legacy effect) affects early community composition, diet can supersede legacy effects and shape the composition of the microbiota and microbiome as well as its expressed gene repertoire. The ability of a fecal human community to establish and sustain itself in the intestines of adult germ-free mice with a unanticipated level of preservation of microbial diversity likely reflects the confluence of a number of factors, ranging from the initial openness of the germ-free mouse’s gut ecosystem, diet (including endogenous nutrient substrates available in the gut habitat such as mucus glycans which are readily foraged by members of the Bacteroidetes and shared with other components of the community) (25, 26), the relative immaturity of the innate and adaptive immune systems at the time of initial colonization, the fact that mice are coprophagic which allows repeated microbial inoculation, and the implicit notion that there must be a large number of shared features of the biochemical milieus of the mouse and human gut that have yet to be defined.
Our metagenomic analyses of humanized gnotobiotic mice also disclosed that in adult mice, community composition is dramatically altered over a time scale of hours when animals are switched from mouse chow to a Western diet. This bloom in members of the Erysipelotrichi and Bacilli classes of the Firmicutes occurred along the length of the gut. These findings have several implications. First, they indicate that it is possible to identify diets having large effects on the gut microbiota and microbiome over short time intervals using humanized mice, and that subsequent testing of the effects of these diets on humans could involve study designs where diet exposures do not have to be prolonged. Second, the ability to apply a diet selection to identify organisms that bloom in the microbiota of humanized mice, and then to recover these organisms and characterize their attributes in silico, in vitro and in vivo (in gnotobiotic mice) represents an attractive pipeline for the discovery of new classes of probiotics that affect nutrient harvest in a given diet context.
A previous study demonstrated that the ability to produce equol from a soy-isoflavone containing diet could be transmitted to germ-free rats upon colonization with a fecal sample from a high equol-producing human subject, but not with a sample from a low equol-producing individual (22). Our experiments show the feasibility of applying metagenomic methods, including microbial community mRNA profiling (meta-transcriptomics) to microbiota transplantation experiments in order to begin to understand the microbial organismal and microbial genetic basis of how a human metabolic phenotype (metabotype) can be transferred via the gut microbiota. A key finding from our study is that similar microbial communities can be formed in gnotobiotic recipients using fresh and frozen aliquots of a human donor’s fecal sample. The fact that diversity can be captured after collected samples are quickly frozen and then stored at −80°C for almost a year not only reflects the capacity of strict anaerobes to survive freeze-thaw cycles but also speaks to the ability of the gut to select and support the rapid expansion of adapted organisms that may be present in only small numbers in a sample subjected to prolonged storage at this temperature. The ability to use frozen fecal samples has broad implications for human microbiome projects focusing on the gut ecosystem, since it means that humans populations with various physiologic or pathophysiologic states can be sampled, their biospecimens archived, and the functional attributes of their microbial communities compared and contrasted both in silico based on metagenomic datasets, and experimentally in humanized gnotobiotic mice. In cases where the focus is on the interrelationships between diet, gut microbial ecology and nutrient/energy harvest in obese or underfed populations living in various cultural contexts, recreating both the microbiotas and diets of the studied populations in humanized gnotobiotic mice should yield more relevant and personalized animal models. Microbial-based biomarkers can be discovered and validated, and therapeutic tests performed in these animals before they are translated to human studies, or in more sublime way, these mice can become part of a clinical study. Finally, the successful transfer of a human gut microbiota across generations of mice without a significant drop in diversity creates a model for addressing a very intriguing question in genetics: does intergenerational transfer of a microbiome, like methylation, RNAi, and other short-term heritable influences, explain effects ‘induced by the environment’ that last a small number of generations?
MATERIALS AND METHODS
All experiments involving mice were performed using protocols approved by the Washington University Animal Studies Committee. Germ-free adult male C57BL/6J mice were maintained in plastic gnotobiotic isolators under a strict 12 hr light cycle and fed an autoclaved low-fat, polysaccharide-rich chow diet (LF/PP; B&K autoclavable diet 7378000) ad libitum. Humanization was performed by diluting a freshly voided human fecal sample (1g) in 10mL reduced PBS, under anaerobic conditions. The fecal material was then suspended by vortexing, and 0.2mL of the suspension introduced by gavage, into each germ-free recipient. A similar procedure was used for a frozen fecal subsample, which was homogenized with a mortar and pestle while frozen prior to dilution. Mice were subsequently maintained in separate cages in a gnotobiotic isolator, and fed a LF/PP or a high-fat/high-sugar Western diet (Harlan-Teklad TD96132) ad libitum.
To define total body fat content, animals were anesthetized with an intraperitoneal injection of ketamine (10 mg/kg body weight) and xylazine (10 mg/kg), and subjected to dual-energy X-ray absorptiometry [DEXA; Lunar PIXImus Mouse, GE Medical Systems; (12)]. Epidydymal fat pad weights were also used as a biomarker of adiposity.
Procedures used for (i) gut microbial community DNA preparation, (ii) sequencing of 16S rRNA gene amplicons, (iii) isolation of C. innocuum strain SB23, (iv) pyrosequencing of total community DNA and the C. innocuum SB23 genome, (v) C. innocuum strain SB23 genome annotation and metabolic reconstruction, (vi) database searches and in silico microbiome metabolic reconstructions, (vii) meta-transcriptomics (RNA-Seq), (viii) qRT-PCR, and (ix) statistical analyses, are described in SM.
Supplementary Material
Acknowledgements
We thank D. O’Donnell and M. Karlsson for husbandry of gnotobiotic mice, S. Wagoner, J. Manchester, and J. Hoisington-López for outstanding technical support, M. Hamady for software support for the 16S rRNA analysis, A. Goodman, B. Muegge, and A. Reyes for many helpful suggestions during the course of these studies, and B. Henrissat for his generous help with annotation of human gut genomes through the Carbohydrate Active enZymes (CAZy) database.
Funding: Supported in part by grants from the NIH (DK70977) and the Crohn’s and Colitis Foundation of America.
Footnotes
Author contributions: PJT and JIG designed the experiments; PJT, VKR, JJF and RER performed the experiments; PJT, VKR, JJF, RER, RK and JIG interpreted the results; PJT and JIG wrote the manuscript.
Competing interests: The authors declare no competing financial interests.
Accession numbers: Datasets from shotgun sequencing projects have been deposited at DDBJ/EMBL/GenBank under accession number 39857 (C.innocuum strain SB23) and 39859 (humanized mouse gut microbiome). 454 and Illumina sequencing reads have been deposited in the NCBI Short Read Archive. Nearly full-length 16S rRNA gene sequences are deposited in GenBank under the accession numbers GQ491120-GQ493997.
REFERENCES
- 1.Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci U S A. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods. 2008;5:235–237. doi: 10.1038/nmeth.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abrams GD, Bishop JE. Effect of the normal microbial flora on gastrointestinal motility. Proc Soc Exp Biol Med. 1967;126:301–304. doi: 10.3181/00379727-126-32430. [DOI] [PubMed] [Google Scholar]
- 11.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 17.Hazenberg MP, Bakker M, Verschoor-Burggraaf A. Effects of the human intestinal flora on germ-free mice. J Appl Bacteriol. 1981;50:95–106. doi: 10.1111/j.1365-2672.1981.tb00874.x. [DOI] [PubMed] [Google Scholar]
- 18.Pang X, et al. Inter-species transplantation of gut microbiota from human to pigs. Isme J. 2007;1:156–162. doi: 10.1038/ismej.2007.23. [DOI] [PubMed] [Google Scholar]
- 19.Mallett AK, et al. The use of rats associated with a human faecal flora as a model for studying the effects of diet on the human gut microflora. J Appl Bacteriol. 1987;63:39–45. doi: 10.1111/j.1365-2672.1987.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 20.Hirayama K. Ex-germfree mice harboring intestinal microbiota derived from other animal species as an experimental model for ecology and metabolism of intestinal bacteria. Exp Anim. 1999;48:219–227. doi: 10.1538/expanim.48.219. [DOI] [PubMed] [Google Scholar]
- 21.Kibe R, et al. Movement and fixation of intestinal microbiota after administration of human feces to germfree mice. Appl Environ Microbiol. 2005;71:3171–3178. doi: 10.1128/AEM.71.6.3171-3178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowey E, Adlercreutz H, Rowland I. Metabolism of isoflavones and lignans by the gut microflora: a study in germ-free and human flora associated rats. Food Chem Toxicol. 2003;41:631–636. doi: 10.1016/s0278-6915(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 23.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martens EC, Roth R, Heuser JE, Gordon JI. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J Biol Chem. 2009;284:18445–18457. doi: 10.1074/jbc.M109.008094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahowald MA, et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A. 2009;106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole JR, et al. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–D296. doi: 10.1093/nar/gki038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.