Abstract
The FoxM1 transcription factor plays critical roles in the expression of genes that are essential for cell proliferation. FoxM1 null or depleted cells fail to progress through mitosis, as expression of several mitotic genes depends upon FoxM1. The transcriptional activity of FoxM1 is stimulated by cyclin-cdk-mediated phosphorylation at a site within the transcriptional activation domain. Here, we characterize the role of an N-terminal inhibitory domain in the transcriptional activity of FoxM1. Deletion of the N-terminal 232 amino-acid residues increases the transcriptional and transforming activities of FoxM1. Moreover, while the activity of the full-length FoxM1 is stimulated by growth factors, the activity of the N-terminal deletion mutant is constitutively high in all phases of the cell cycle. The N-terminal deletion also eliminates the requirement for cyclin-cdk to activate FoxM1. We provide evidence that the N-terminal domain interacts with the C-terminal half of the transcription factor to attenuate its transcriptional activity. Moreover, the N-terminal fragment inhibits the transcriptional activity of FoxM1 in G1/S cells, but not in G2/M cells. Our results suggest that cyclin-cdk phosphorylates FoxM1 to counteract the inhibition by the N-terminal domain to fully activate FoxM1 in G2/M phase.
Keywords: FoxM1, cell cycle regulation, FoxM1-inhibitory domain, transforming activity
Introduction
The mammalian forkhead box m1 (FoxM1) transcription factor is expressed in all dividing cells and is required for cell division (Korver et al., 1997, 1998; Ye et al., 1997). Cells depleted of FoxM1 are deficient in mitosis (Laoukili et al., 2005; Wang et al., 2005; Wonsey and Follettie, 2005). Hepatocyte-specific deletion of FoxM1 in FoxM1fl/fl mice (Alb-Cre FoxM1−/−) severely impairs liver regeneration, 80% reduction in DNA replication and a complete inhibition of M phase progression (Wang et al., 2002). Conversely, transgenic expression of FoxM1 accelerates liver regeneration (Ye et al., 1999; Wang et al., 2001a, b, 2002). Moreover, expression of FoxM1 is diminished in senescent cells (Ye et al., 1997), and FoxM1−/− mouse embryonic fibroblasts fail to grow in culture and undergo premature senescence (Wang et al., 2005). These observations are consistent with a role of the FoxM1 transcription factor in the expression of genes that are important for cell cycle progression.
In liver regeneration studies, increased levels of cdk inhibitors p21 and p27 were detected in FoxM1−/− hepatocytes (Wang et al., 2002), which was linked to a diminished expression of Skp2 and Cks1. FoxM1 directly binds to the promoter regions of Skp2 and Cks1 and enhances expression of Skp2 and Cks1 (Wang et al., 2005). FoxM1 is also critical for expression of several mitotic genes, including PLK1, Aurora B, Cdc25B, Survivin, CENPA and CENPB (Laoukili et al., 2005; Wang et al., 2005). Therefore, the FoxM1 target genes identified thus far explain the proliferation defects observed in FoxM1-deficient cells, providing insights into the mechanism by which FoxM1 participates in cell division.
FoxM1 is critical for tumor development (Kalinichenko et al., 2004; Kalin et al., 2006; Kim et al., 2006; Liu et al., 2006). Deletion of FoxM1 inhibits tumor development. Cre-mediated deletion of FoxM1 inhibited growth, suggesting an essential role of FoxM1 in tumor progression (Kalinichenko et al., 2004). More recent studies indicated that deletion of FoxM1 in tumor cells also causes the tumor cells to undergo apoptosis (Wonsey and Follettie, 2005). The increased apoptosis of tumor cells correlated with reduced expression of Survivin in FoxM1-deleted tumor cells (Gusarova et al., 2007).
The C-terminal region of FoxM1 contains a cyclin-cdk phosphorylation site (Major et al., 2004; Weirstra et al., 2006b, c). The cyclin-cdk-mediated phosphorylation stimulates the transcriptional activity of FoxM1. Here, we show that the N-terminal region of FoxM1 contains an inhibitory domain that regulates its transcriptional activity. Deletion of the N-terminal inhibitory domain generates a constitutively active transcription factor that functions independently of cyclin-cdk and exhibits increased transforming activity. The observations are significant with regard to the oncogenic function of FoxM1.
Results
Deletion of N-terminal 232 residues enhances transcriptional and transforming activity of FoxM1
FoxM1 contains a fork-head box family DNA-binding domain between residues 232 and 332 (Ye et al., 1997) and a C-terminal region of FoxM1 encompasses the transactivation domain (Figure 1a) (Major et al., 2004). To investigate the role of the N-terminal 232 residues in the transcriptional activity of FoxM1, we generated an N-terminal deletion mutant (N-del). The mutant was compared with the wild-type FoxM1 in transcription assays (Major et al., 2004). The N-del exhibited greater than 20-fold increase in the transcriptional activity compared to the wild-type FoxM1 (Figure 1b), which is consistent with a recent study (Wierstra and Alves, 2006a). To further investigate the properties of N-del, which is similar to the wild-type FoxM1 localizes in the nucleus (Supplementary Figure S1), we analysed the effects on the expression of FoxM1 target genes. U2OS and NIH3T3 cells were transfected with FoxM1 or N-del. Total proteins and RNAs from transfected cells were subjected to immunoblotting and quantitative reverse transcription (RT)–PCR assays to measure the level of FoxM1 target gene expression. Assays for Aurora B, a target of FoxM1 (Wang et al., 2005), by immunoblotting consistently indicated higher expression in the N-del-transfected cells (Figure 1c, upper panel). Under our experimental condition, transient expression of the wild-type FoxM1 caused only a marginal increase in the mRNA level of Skp2 (Figure 1d). N-del, in contrast, induced a significantly higher stimulation of Skp2 expression. These results are consistent with the notion that the N-terminal region of FoxM1 contains a transcriptional inhibitory domain.
Figure 1.
Deletion of the N-terminal 232 residues increases the transcriptional activity of forkhead box m1 (FoxM1). (a) Schematic diagram of FoxM1. (b) U2OS, Saos2 or NIH3T3 cells were transiently transfected with the 6X-FoxM1-TATA luciferase reporter and a cytomegalovirus (CMV)-Renilla luciferase construct in conjunction with the control (CMV), the wild-type FoxM1 (WT) or the N-terminal deletion mutant (N-del)vector. Extracts of the transfected cells were analysed for luciferase activity. Luciferase activity is plotted as a percentage of WT FoxM1B transcriptional activity following normalization to Renilla luciferase activity. (c) U2OS cells were electroporated with the wild-type FoxM1 or the N-del-expressing plasmids. The relative expression of Aurora B was determined by western blot analysis. The numbers above the lanes indicate fold increase. (d) RNA was isolated from NIH3T3 cells electroporated with the wild-type FoxM1 or N-del and was used for QRT–PCR with Skp2 primers. Data are shown as mean ± s.e.m. (**P<0.01; Student’s t-test).
Previous studies indicated an essential role of FoxM1 in tumor cell growth (Kalinichenko et al., 2004). Therefore, we hypothesized that N-del would exhibit increased transforming activity. NIH3T3 cells expressing wild-type FoxM1 or the N-del mutant were subjected to foci formation assay. Clearly, transfection of N-del induced a significantly greater number of foci compared to that induced by the wild-type FoxM1 (Figure 2a). Moreover, compared to the wild-type FoxM1-expressing cells, N-del-expressing cells showed statistically significant increase in the ability to promote anchorage-independent growth (Figure 2b). Also, we examined the invasion ability of the cells transfected with N-del using matrigel-coated transwell assay system. As shown in Figure 2c, the N-del-transfected cells exhibited a significant increase in the migration ability. Further, the N-del-expressing cells grew efficiently in low-serum media (Dulbecco’s modified Eagle’s medium containing 2% fetal bovine serum), whereas empty vector or wild-type FoxM1-expressing cells did not (Figure 2d). Therefore, it appears that the N-terminal region of FoxM1 functions to attenuate the transforming activity of FoxM1 by inhibiting its transcription function.
Figure 2.
Cells expressing the N-terminal deletion mutant (N-del) of forkhead box m1 (FoxM1) exhibit increased transforming abilities. NIH3T3 cell lines expressing green fluorescent protein (GFP) (MOCK), GFP-FoxM1 wild type (WT) or GFP-N-del (N-del) were established. (a) For focus formation assay, cells were allowed to grow for 10–14 days and then stained with crystal violet. The number of foci in the plot represents the mean number from three plates. (b) Cells were suspended in 0.35% agarose and plated on 0.7% bottom agar layer. The appearance of colonies was analysed after 21 days. (c) Cell invasion was assayed using Matrigel-coated invasion chamber. After 24 h incubation, cells on the lower surface of the membrane were stained and counted. Data are shown as mean ± s.e.m. (P<0.01; Student’s t-test). (d) Cells were plated on 100 mm dishes in complete medium and incubated overnight. The next day, media were substituted with Dulbecco’s modified of Eagle’s medium containing 2% fetal bovine serum. Then cells were trypsinized and counted at the indicated time point. Data shown are the average of three plates.
The transcriptional function of N-del is independent of activation by cyclin-cdk
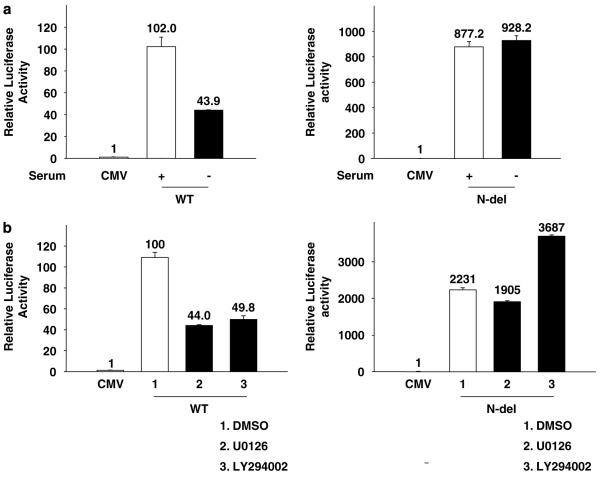
The transcriptional activity of FoxM1 is stimulated by growth factors and cyclin-cdk (Major et al., 2004). We sought to investigate whether the N-terminal inhibitory region of FoxM1 plays any role in that process. U2OS cells were transfected with wild-type FoxM1 or N-del along with the luciferase-reporter plasmid. Following transfection, one set of cells was maintained in serum-free medium, whereas the other set was maintained in serum-containing medium for 72 h. In the absence of serum, the wild-type FoxM1 exhibited decreased transcriptional activity, although it was expressed at a comparable level (data not shown); however, N-del exhibited high constitutive activities (Figure 3a). To further investigate the growth factor response of FoxM1-activated transcription, we analysed the effects of the inhibitors of phosphatidylinositol 3-kinases (LY294002) and mitogen-activated protein kinase (U0126). Both phosphatidylinositol 3-kinase and mitogen-activated protein kinase inhibitors caused a significant inhibition of the transcriptional activity of the wild-type FoxM1. The N-del, in contrast, did not exhibit any significant inhibition (Figure 3b).
Figure 3.
The N-terminal deletion (N-del) of forkhead box m1 (FoxM1) is active independently of growth factor pathways. U2OS cells were transfected with the 6X-FoxM1-TATA luciferase reporter plasmid in conjunction with the empty cytomegalovirus (CMV), the wild-type FoxM1 (WT) or the N-del-expressing plasmid. Sixteen hours after transfection, cells were either serum starved for 72 h (a) or treated with 50 μM phosphatidylinositol 3–kinase (PI3) (LY294002) or 50 μM mitogen-activated protein (MAP) kinase (U0126) inhibitors for 16 h (b). The relative luciferase activity is shown.
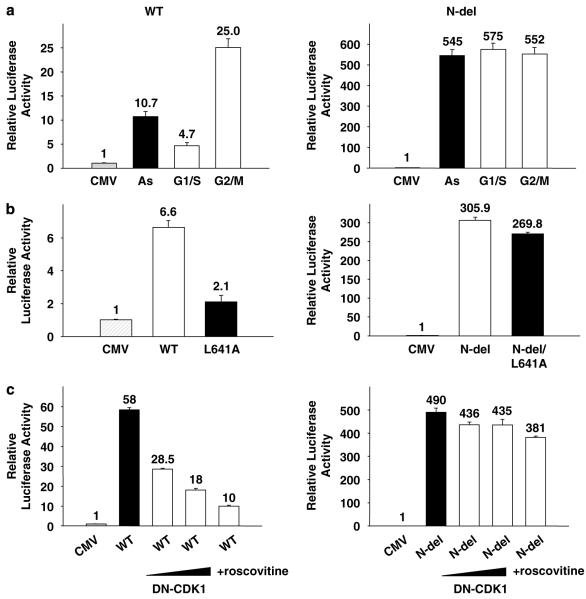
We then investigated the cell cycle regulation of the transcriptional activity of FoxM1. U2OS cells, transfected with FoxM1 or N-del expression plasmid along with a luciferase reporter, were treated with aphidicolin or nocodazole. The transcriptional activity in the arrested cells was compared with that in the asynchronous cells. The wild-type FoxM1 transcriptional activity was significantly inhibited in aphidicolin-treated cells (Figure 4a), but no inhibition was observed in the nocodazole-treated cells. Treatments with aphidicolin (5 μg ml−1)or nocodazole (50 ng ml−1) did not alter the level of FoxM1 (data not shown). The N-del mutant, in contrast, did not exhibit any inhibition in the aphidicolin-treated cells, and it exhibited high constitutive transcriptional activity in arrested and in asynchronous cells (Figure 4a). The differential response of FoxM1 and N-del suggests that the N-terminal region of FoxM1 is critical for the cell cycle regulation of the FoxM1 transcriptional activity.
Figure 4.
The N-terminal deletion (N-del) of forkhead box m1 (FoxM1) is constitutively active during cell cycle. Transactivation of the 6X-FoxM1-TATA luciferase reporter was analysed in U2OS cells transfected with either the empty cytomegalovirus (CMV), the wild-type FoxM1 (WT)or the N-del-expressing plasmids (a). Twelve hours after transfection, cells were treated with 5 μg ml−1 aphidicolin (G1/S) for 24 h or 50 ng ml−1 nocodazole (G2/M) for 16 h. (b) U2OS cells were transfected with plasmids expressing the WT FoxM1, FoxM1/L641A, the N-del or N-del/L641A along with the reporter 6X-FoxM1-TATA luciferase. The relative luciferase activities are shown. (c) U2OS cells transfected with WT (left) or N-del (right) with increasing levels of dominant-negative Cdk1 or 10 μM Cdk inhibitor (roscovitine). The relative luciferase activities are shown.
To further investigate the basis of the constitutive activity of N-del, we mutated the cyclin-binding site by changing the leucine at 641 to alanine (L641A)in a N-del background. The substitution mutant Ndel/L641A was compared with N-del for its ability to stimulate transcription. Interestingly, the mutation of the cyclin-cdk-binding site had very little effect on the transcriptional activity of N-del (Figure 4b), suggesting that N-del mutant is independent of activation by cyclin-cdk. Consistent with this, we observed that, unlike the wild type, N-del is resistant to inhibition by dominant-negative cdk1 or an inhibitor of Cdk (roscovitine) (Figure 4c). Therefore, we suggest that the cyclin-cdk interaction with the C-terminal activation domain functions to overcome the inhibitory effects of the N-terminal regulatory domain.
The N-terminal domain of FoxM1 interacts with the C-terminal activation domain and inhibits function in a cell cycle-regulated manner
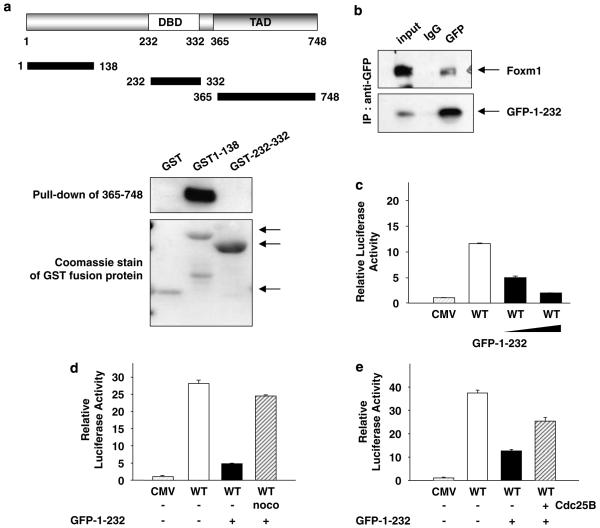
To determine the mechanism by which the N-terminal domain inhibits the transcriptional activation function of FoxM1, we investigated whether the N-terminal domain associates with the C-terminal transactivation domain. We performed pull-down assays using glutathione-S-transferase-fusion proteins containing N-terminal region (1–138) and DNA-binding domain (232–332). The glutathione-S-transferase-fusion proteins on glutathione–sepharose-beads were incubated with cell extracts expressing T7-epitope-tagged transcriptional activation domain of FoxM1. As shown in Figure 5a, the glutathione-S-transferase-fusion protein containing the N-terminal 138 residues of FoxM1 was able to interact with the transactivation domain of FoxM1. The interaction was confirmed by co-immunoprecipitation assays. U2OS cells were transfected with a plasmid expressing a green fluorescent protein (GFP) fusion of the N-terminal inhibitory domain of FoxM1 and T7-FoxM1. Immunoprecipitation was performed with extracts from transfected cells with an antibody against GFP or isotope matched immunoglobulin G. Clearly, the GFP fusion protein containing the N-terminal domain of FoxM1 was able to co-immunoprecipitate FoxM1 (Figure 5b).
Figure 5.
The N-terminal domain of forkhead box m1 (FoxM1) interacts with its transactivation domain and inhibits its transcriptional activity. (a) Glutathione-S-transferase (GST)-fusion proteins containing various fragments of FoxM1 (GST1–138 or GST 232–335)were used in pull-down assay along with lysates from NIH3T3 cells transfected with a plasmid expressing T7-epitope-tagged transactivation domain of FoxM1 (residues between 365 and 748). The binding of T7-365–748 proteins was determined by western blot with T7 antibody. The lower panel shows a Coomassie staining of the GST-fusion proteins (arrows). (b) U2OS cells were transfected with plasmids expressing green fluorescent protein (GFP1–232) and T7-epitope-tagged FoxM1. The extracts were subjected to immunoprecipitation using GFP or a control antibody. The immunoprecipitates were analysed for T7-FoxM1 by western blot assays. (c and d) U2OS cells were transfected with 6XFoxM1 reporter plasmid and an increasing amount (c) or a constant amount (d) of the plasmid expressing the N-terminal fragment of FoxM1 (GFP1–232) in conjunction with a constant amount of T7-FoxM1. (d) Sixteen hours after transfection, cells were treated with nocodazole (noco) for another 16 h. (e) U2OS cells transfected with 6 × FoxM1-TATA luciferase reporter and plasmids expressing T7-FoxM1B, N-terminal fragment of FoxM1 (GFP1-232) and Cdc25B. Relative luciferase activity is shown.
We then sought to investigate whether the N-terminal domain could inhibit the transcriptional activity of FoxM1 in trans. U2OS cells were transfected with a plasmid expressing FoxM1 along with the reporter plasmid in the presence or absence of a plasmid expressing the N-terminal inhibitory domain. Expression of the inhibitory domain did not alter the expression of the full-length FoxM1 (data not shown). However, the transcriptional activation by the full-length FoxM1 was inhibited in a dose-dependent manner (Figure 5c). Interestingly, the inhibition by the N-terminal domain is cell cycle-dependent. It caused a significant inhibition of FoxM1 in asynchronous cells, but only marginally in cells arrested in early M phase (Figure 5d). Moreover, the inhibitory effect of N-terminal domain was less potent when we transfected Cdc25B in conjunction with the N-terminal fragment (Figure 5e). We speculate that in early M phase, cyclin-cdk-mediated phosphorylation of FoxM1 confers resistance to inhibition by the N-terminal domain.
The N-terminal domain of FoxM1 inhibits expression of the FoxM1 target genes
Because the N-terminal fragment of FoxM1 inhibited the transcriptional activity of FoxM1 in trans (Figure 5c), we further examined whether the N-terminal fragment could inhibit the endogenous target gene expression. We transiently transfected NIH3T3 cells with a plasmid expressing the N-terminal fragment (GFP1–232)of FoxM1 or with an empty vector. The relative expression of Skp2, Cdc25B and Aurora B, which are known FoxM1 target genes, was determined by quantitative real-time RT–PCR. As expected, transfection of the N-terminal fragment caused a significant reduction in the expression of the FoxM1 target genes (Figure 6). The inhibition was specific for the FoxM1 target genes, as there was no inhibition in the expression of the translation initiation factor 4EBP1, which is not a FoxM1 target gene. These observations further confirm a regulatory role of the N-terminal domain of the FoxM1 transcription factor.
Figure 6.
The N-terminal fragment inhibits forkhead box m1 (FoxM1) target genes. Exponentially growing NIH3T3 cells were electroporated with either 5 μg cytomegalovirus (CMV) empty vector or vector expressing the N-terminal fragment of FoxM1 green fluorescent protein (GFP-1-232). RNA was isolated and used for QRT–PCR with primers specific to Skp2, Cdc25B, Aurora B or 4EBP1. Data are shown as mean ± s.e.m. (**P<0.01; Student’s t-test).
Discussion
FoxM1 is a critical cell cycle transcription factor involved in tumor progression (Kalinichenko et al., 2004). Therefore, an understanding of the mechanisms that control the activity of FoxM1 is important. The observations that an N-terminal inhibitory domain controls the transcriptional and the transforming activities of FoxM1 are significant in that regard.
Regulation of FoxM1 in the cell cycle is expected from its involvement in the transcription of the cell cycle-regulated genes. In this study, we show that its transcriptional activity is stimulated by growth factors and reaches a peak level in the G2/M phase of the cell cycle. Nocodazole-arrested cells exhibited higher transcriptional activity compared to the aphidicolin-arrested cells. Both G2/M and G1/S cyclins associate with FoxM1 (Major et al., 2004; Wierstra and Alves, 2006b, c). We provide further insights into the mechanism by which cyclin-cdks activate the transcription function of FoxM1. Deletion of the N-terminal domain confers growth factor independence and renders FoxM1 constitutively active throughout the cell cycle. Moreover, we show that N-del is fully active in the absence of cyclin-cdk interaction. Mutation of residue L641 within the ‘LXL’ cyclin-binding motif that eliminates binding to cyclin-cdk had no effect on the transcriptional activity of N-del. These observations are consistent with the notion that the N-terminal inhibitory domain confers cyclin-cdk dependence to the transcriptional activity of FoxM1.
The N-terminal domain of FoxM1 associates with the C-terminal activation domain to inhibit its transcriptional activity. Interestingly, the inhibition by the N-terminal fragment of FoxM1 could be significantly reversed by nocodazole treatment, suggesting that G2/M-specific factors may counteract N-terminus-mediated inhibition of FoxM1. Moreover, co-transfection of Cdc25B also potently interfered with the inhibitory effect of the N-terminal inhibitory domain. We speculate that the phosphorylation by cyclin B-cdk1 disrupts the interaction between the N-terminal inhibitory domain and the C-terminal activation domain to then allow binding of the co-activator CREB binding protein to the activation domain of FoxM1.
We suppose that the N-terminal inhibitory domain of FoxM1 is significant in maintaining the normal cell phenotype, because we find that N-del possesses a much higher transforming activity compared to the full-length protein. It is likely that the N-terminal inhibitory domain plays a role in regulating the transcriptional activity of FoxM1 in early phases of the cell cycle. Clearly, the activity of FoxM1 is significantly lower in aphidicolin-arrested cells compared to nocodazole-arrested cells. We suppose that the relatively low activity of FoxM1 in G1 phase might be important for regulated entry into and progression through S phase. FoxM1 is not inactive in G1/S phases. It is important for entry into S phase because FoxM1−/− cells exhibit deficiency and delay in entering S phase (Z Wang and RH Costa, unpublished observation). Moreover, in liver regeneration studies, we demonstrated a role for FoxM1 in S-phase entry (Ye et al., 1999). In this regard, it is noteworthy that HPV16 oncoprotein E7, which enhances progression through G1/S phases, binds to FoxM1 and stimulates FoxM1-activated transcription (Luscher-Firzlaff et al., 1999). The non-transforming mutants of E7 or E7 encoded by the low-risk types of HPV do not activate FoxM1. It will be interesting to determine whether the HPV16 E7 oncoprotein activates FoxM1 by deregulating the inhibitory domain in the N-terminal region of FoxM1.
Materials and methods
Cell lines and transformation assay
Cells were transfected with plasmids expressing GFP, GFP-FoxM1 and GFP-N-del and then selected in medium containing hygromycin for 3 weeks. Following selection, the cell lines were assayed for focus forming ability by plating in 100 mm tissue culture dishes. The medium was replaced every 3 days with medium containing 5% serum for 10–14 days. To analyse the foci, cultures were stained with crystal violet and counted. To examine anchorage-independent growth, cells were suspended in medium containing 0.35% agarose and then poured onto 60 mm dishes coated with 0.7% agarose. The culture was maintained for 3 weeks and colonies larger than 0.1 mm were counted. Invasion assay was performed using Matrigel invasion chamber system (BD Biosciences, Bedford, MA, USA). Cells (5 × 104) were trypsinized and seeded onto the upper chamber in serum-free Dulbecco’s modified Eagle’s medium. Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum was placed in the bottom well. After 24 h incubation, non-invasive cells in the upper chamber were removed by a cotton-tipped swap. The migrated cells on the lower surface were fixed and stained for counting.
Cell proliferation in low serum media
Cells (5 × 105) were plated in complete medium onto 100 mm dishes and incubated overnight. The next day, the medium was substituted with Dulbecco’s modified Eagle’s medium containing 2% fetal bovine serum. Cells were trypsinized and counted at different time points.
Real-time PCR
U2OS cells or NIH3T3 cells were harvested at 24 h following transfection for total RNA preparation using RNA-STAT-60 (Tel-TestB Inc., Friendswood, TX, USA). Following DNase I digestion, we used the Bio-Rad cDNA synthesis kit to synthesize 1 μg of total DNA. The following reaction mixture was used for all PCR samples: 1 × IQ SybrGreen supermix (Bio-Rad, Carlsbad, CA, USA), 100 nM of each primer and 1 μl of cDNA of 25 μl total volume. Reactions were amplified and analysed in triplicate using the MyiQ single-color real-time PCR detection system (Bio-Rad).
Supplementary Material
Acknowledgements
While this work was in progress, RHC died in his fight against pancreatic cancer. The authors dedicate this work to his memory. This work was supported by NIH grants to RHC. LFL is supported by NIH Grant CA46565, AT is supported by NIH Grant DK044525 and PR is supported by NIH Grants CA124488 and CA100035.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, et al. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin TV, Wang IC, Ackerson TJ, Major ML, Detrisac CJ, Kalinichenko VV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66:1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IM, Ackerson T, Ramakrishna S, Tretiakova M, Wang IC, Kalin TV, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66:2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- Korver W, Roose J, Clevers H. The winged-helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res. 1997;25:1715–1719. doi: 10.1093/nar/25.9.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korver W, Schilham MW, Moerer P, Hoff MJ, Dam K, Lamers WH, et al. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor Trident. Curr Biol. 1998;8:1327–1330. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- Liu M, Dai B, Kang SH, Ban K, Huang FJ, Lang FF, et al. FoxM1B is overexpressed in human glioblastomas and critically regulates the tumorigenicity of glioma cells. Cancer Res. 2006;66:3593–3602. doi: 10.1158/0008-5472.CAN-05-2912. [DOI] [PubMed] [Google Scholar]
- Luscher-Firzlaff JM, Westendorf JM, Zwicker J, Burkhardt H, Henriksson M, Muller R, et al. Interaction of the fork head domain transcription factor MPP2 with the human papilloma virus 16 E7 protein: enhancement of transformation and transactivation. Oncogene. 1999;18:5620–5630. doi: 10.1038/sj.onc.1202967. [DOI] [PubMed] [Google Scholar]
- Major ML, Lepe R, Costa RH. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol Cell Biol. 2004;24:2649–2661. doi: 10.1128/MCB.24.7.2649-2661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1)ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001a;33:1404–1414. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci USA. 2002;99:16881–16886. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc Natl Acad Sci USA. 2001b;98:11468–11473. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierstra I, Alves J. Despite its strong transactivation domain, transcription factor FOXM1c is kept almost inactive by two different inhibitory domains. Biol Chem. 2006a;387:963–976. doi: 10.1515/BC.2006.120. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. FOXM1c is activated by cyclin E/Cdk2, cyclin A/Cdk2, and cyclin A/Cdk1, but repressed by GSK-3alpha. Biochem Biophys Res Commun. 2006b;348:99–108. doi: 10.1016/j.bbrc.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Wierstra I, Alves J. Transcription factor FOXM1c is repressed by RB and activated by cyclin D1/Cdk4. Biol Chem. 2006c;387:949–962. doi: 10.1515/BC.2006.119. [DOI] [PubMed] [Google Scholar]
- Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- Ye H, Holterman AX, Yoo KW, Franks RR, Costa RH. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S phase. Mol Cell Biol. 1999;19:8570–8580. doi: 10.1128/mcb.19.12.8570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Kelly TF, Samadani U, Lim L, Rubio S, Overdier DG, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17:1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.