Abstract
Substituted tetrahydroquinolines (THQs) have been previously identified as inhibitors of mammalian protein farnesyltransferase (PFT). Previously we showed that blocking PFT in the malaria parasite led to cell death and that THQ-based inhibitors are the most potent among several structural classes of PFT inhibitors (PFTIs). We have prepared 266 THQ-based PFTIs and discovered several compounds that inhibit the malarial enzyme in the sub- to low-nanomolar range and that block the growth of the parasite (P. falciparum) in the low-nanomolar range. This body of structure–activity data can be rationalized in most cases by consideration of the X-ray structure of one of the THQs bound to mammalian PFT together with a homology structural model of the malarial enzyme. The results of this study provide the basis for selection of antimalarial PFTIs for further evaluation in preclinical drug discovery assays.
Introduction
New antimalarial drugs are needed because of widespread resistance to well-established agents such as chloroquine (www.mmv.org). We have been working on inhibitors of malaria protein farnesyltransferase (PFTa) because earlier studies showed that such agents are cytotoxic to Plasmodium falciparum,1–3 the causative agent of falciparum malaria. PFT catalyzes the transfer of the 15-carbon farnesyl group from farnesyl-pyrophosphate to the SH group of the tetrapeptide motif CaaX (C is cysteine, a is usually but not necessarily an aliphatic amino acid, and X is a variety of amino acids) present at the C-terminus of proteins that become farnesylated. Inhibitors of mammalian PFT have been extensively developed as anticancer agents by the pharmaceutical industry,4 and thus, we have the opportunity to extend the medicinal chemistry and preclinical pharmacology of PFT inhibitors toward the development of antimalarial drugs (piggy-back drug development). To this end, we tested many of the known classes of PFT inhibitors and found that tetrahydroquinoline (THQ)-based PFT inhibitors developed at Bristol Myers Squibb5 are the most potent against P. falciparum PFT (Pf-PFT) and against parasite growth in human red blood cells.3 Our previous medicinal chemistry efforts led to the identification of THQ 1 (Figure 1) and a few related compounds that inhibit Pf-PFT in vitro, with IC50 (concentration of inhibitor that 50% inhibits Pf-PFT) values of ~0.6 nM, and inhibit parasite growth in red cells with ED50 (concentration of inhibitor that 50% inhibits the growth of P. falciparum in red blood cells in vitro) values of ~5 nM.3 Studies with mammalian PFT have shown that the 6-cyano group on the THQ ring is important in conferring tight enzyme binding,6 and structural studies show that the imidazole appended to N-1 of the THQ ring directly coordinates the Zn2+ ion at the active site of mammalian PFT.7,8
Figure 1.
Tetrahydroquinoline-based protein farnesyltransferase inhibitors. See main text for discussion.
Continuous dosing of THQ 1 using surgically implanted, osmotic minipumps in mice infected with rodent malaria (P. berghei) led to a dramatic reduction in parasite number.3 Studies of malaria resistance to these Pf-PFT inhibitors provided very strong evidence that blocking Pf-PFT action is the basis for parasite killing.7,8 The reasons that PFT inhibitors are very toxic to malarial cells but not to mammalian cells are not known. One hypothesis is that the malaria parasite appears to lack protein geranylgeranyltransferase type I, the enzyme responsible for the monogeranylgeranylation of proteins in mammalian cells. Thus, some critical housekeeping proteins may be farnesylated in malaria but geranylgeranylated in mammalian cells.
The purpose of the present study is to report our continued efforts to develop THQ-based antimalarials. A number of THQs were prepared with various groups attached to the sulfonyl group of 1 (R1 of THQ 2 in Figure 1) or to the sulfonamide nitrogen (R2 in THQ 2 in Figure 1). Less extensive structure-activity studies were carried out by variation of other groups attached to the THQ core. We were able to identify several new THQ-based Pf-PFT inhibitors with sub- to low-nanomolar potency on Pf-PFT and with low nanomolar potency on P. falciparum growth in vitro.
Chemistry
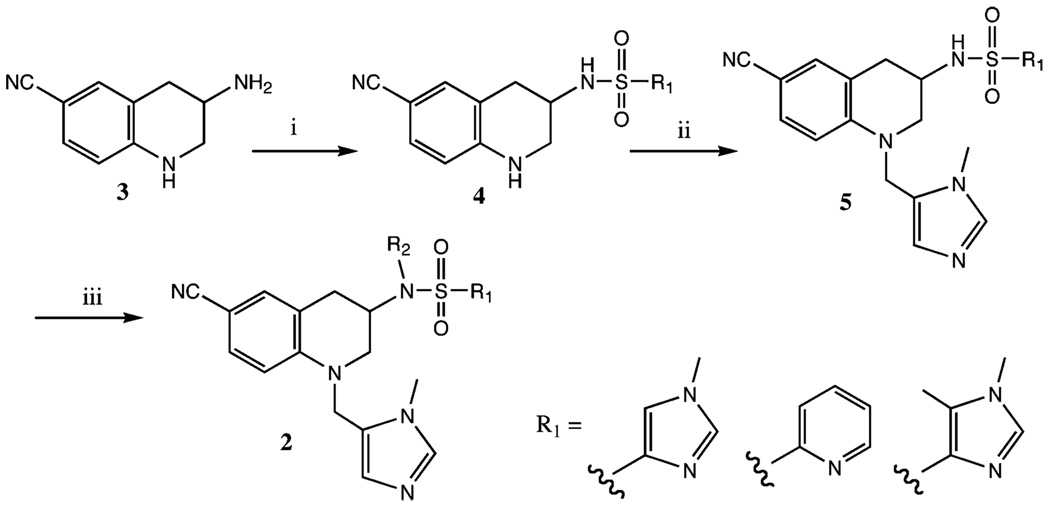
Many of the THQs 2 prepared in this study were prepared by the route shown in Scheme 1 starting from racemic 6-cyano-3-amino-THQ (compound 3, Scheme 1), which was made as described.5 This route is useful for variation of the R2 group, which is added in the last synthetic step. Scheme 2 was used to prepare analogs of 2 in which the R group attached to the piperidine nitrogen is varied. Scheme 3 was used to allow easier variation of both the R1 and R2 groups because, unlike in Scheme 1, the R1 group is introduced later in the synthesis. Scheme 4 was used to prepare THQs, which lack the methyl group on the zinc-binding imidazole. In this case, tritylation of the imidazole was required prior to alkylation of the sulfonamide nitrogen. Scheme 5 shows the synthesis of a THQ analog in which the methylene bridge between the N-1 of the THQ core and the Zn2+-binding imidazole group is replaced with a sulfonyl group or with a CH(CH3) group. THQs containing a 6-phenyl group in place of the 6-cyano group were prepared according to Scheme 6. The key step is the introduction of the phenyl group via Suzuki coupling (conversion of 21 to 22). Scheme 7 shows the synthesis of THQ analogs in which the 6-CN is replaced with carbonyl-bearing functional groups. Scheme 8 was used to prepare the THQ analog 34. The key reaction is nucleophilic displacement between the mesylate derived from the indicated secondary alcohol 32 and secondary amine 33.
Scheme 1a.
a Reagents and conditions: (i) R1SO2Cl, DIPEA, CH3CN; (ii) 1-methyl-1H-imidazole-5-carboxaldehyde, triethylsilane, 1,2-dichloroethane:trifluoroacetic acid (1:1); (iii) R2-Br, Cs2CO3, DMF.
Scheme 2a.

a Reagents and conditions: (i) 10% trifluoroacetic acid, CH2Cl2; (ii) RCOCl or ROCOCl or RNCO or RSO2Cl, CH2Cl2, DIPEA.
Scheme 3a.
a Reagents and conditions: (i) Cbz-Cl, Et3N, CH3CN; (ii) 1-methyl-1H-imidazole-5-carboxaldehyde, triethylsilane, 1,2-dichloroethane:trifluoroacetic acid (1:1); (iii) 10% Pd/C, CH3OH; (iv) R2-Br, Cs2CO3, DMF; (v) R1COOH, EDC, HOBt, Et3N, DMF, or R1COCl, Et3N, CH2Cl2 or R1SO2Cl, Et3N, CH2Cl2.
Scheme 4a.
a Reagents and conditions: (i) 3H-imidazole-4-carboxaldehyde, triethylsilane, 1,2-dichloroethane/trifluoroacetic acid (1:1); (ii) trityl chloride, DIPEA, DMF; (iii) R2-Br, Cs2CO3, DMF; (iv) CH2Cl2, trifluoroacetic acid.
Scheme 5a.
a Reagents and conditions: (i) R2-Br, Cs2CO3, DMF; (ii) 3-methyl-3H-imidazole-4-sulfonyl chloride, DMAP, CH3CN.
Scheme 6a.
a Reagents and conditions: (i) BOC-anhydride, K2CO3, dioxane-water (4:1); (ii) phenylboronic acid, Ba(OH)2, tetrakis triphenylphosphine palladium, DME–water (5:1); (iii) 20% trifluoroacetic acid, CH2Cl2; (iv) R1SO2Cl, DIPEA, CH2Cl2; (v) 1-methyl-1H-imidazole-5-carboxaldehyde, triethylsilane, 1,2-dichloroethane/trifluoroacetic acid (1:1); (vi) R2-Br, Cs2CO3, DMF.
Scheme 7a.
a Reagents and conditions: (i) concd HCl, 80 °C; (ii) H2SO4, R4-OH; (iii) R4-NH2, EDC, DMAP, DMF; (iv) alkyl bromide, Cs2CO3, DMF.
Scheme 8a.
a Reagents and conditions: (i) trityl chloride, Et3N, DMF; (ii) MeMgBr, THF, 0 °C; (iii) MsCl, CH3CN, 60 °C; (iv) trifluoroacetic acid, CH2Cl2.
Inhibition of Pf-PFT and P. falciparum Growth by THQ-Based Pf-PFT Inhibitors
We first give a general description of the potencies of THQ-based Pf-PFT inhibitors on the enzyme and on P. falciparum growth in vitro, and then we attempt to provide a structural rational for some of the activity data. Our initial structure–activity data on THQ-based inhibitors of Pf-PFT led to the discovery of compounds with R1 = 2-pyridyl or N1-methyl-4-imidazolyl as being potent inhibitors of Pf-PFT.3 In Table 1, we report a study of THQs with R1 = 2-pyridyl and with a variety of substituents as R2 groups. A number of THQs were found that inhibited Pf-PFT in vitro by 50% (IC50) in the low nanomolar range and that also inhibited the growth of P. falciparum in erythrocyte cultures in vitro by 50% (ED50) in the low nanomolar range (i.e., 48, 55, 56, 57, 61, and 62). The most potent compound in the series is 55 with an ED50 = 17 nM for the 3D7 strain and 10 nM for the K1 strain. Well-established antimalarial drugs such as chloroquine display ED50 values in the low nanomolar range. Thus, the potency achieved for some of our THQ-based PFT inhibitors is probably sufficient for an antimalarial drug discovery effort. In general, we did not find any compound that inhibited P. falciparum growth in the low nanomolar range that was a relatively poor inhibitor of Pf-PFT.
Table 1.
6-CN-THQs with R1 = 2-Pyridyla
 | ||||||
|---|---|---|---|---|---|---|
| Compound | R2 |
Pf-PFT % Inhibition at |
ED50 (nM) |
|||
| 50 nM | 5 nM | 0.5 nM | 3D7 | K1 | ||
| 35 | 93 | 64 | 18 | 230 | 1005 | |
| 36 | 96 | 65 | 17 | 189 | 932 | |
| 37 |  |
95 | 66 | 18 | 440 | 240 |
| 38 |  |
82 | 37 | 1600 | 2200 | |
| 39 | 81 | 32 | 4 | 300 | 250 | |
| 40 |  |
91 | 48 | 9 | 230 | 240 |
| 41 | 86 | 80 | 35 | 130 | 170 | |
| 42 |  |
97 | 70 | 9 | 133 | 75 |
| 43 | 97 | 80 | 24 | 250 | 375 | |
| 44 | 98 | 78 | 27 | 320 | 270 | |
| 45 |  |
92 | 52 | 6 | 1250 | 2600 |
| 6 |  |
100 | 89 | 19 | 155 | 180 |
| 46 |  |
90 | 47 | 14 | 510 | 400 |
| 47 |  |
96 | 77 | 28 | 100 | 100 |
| 48 | 95 | 68 | 6 | 18 | 45 | |
| 49 | 99 | 84 | 25 | 230 | ||
| 50 |  |
90 | 59 | 15 | 230 | 400 |
| 51 |  |
89 | 69 | 26 | 250 | 600 |
| 52 | 65 | 11 | 6 | 2700 | >5000 | |
| 53 | 32 | 7 | 0 | >5000 | >5000 | |
| 54 | 18 | 6 | 0 | |||
| 55 | 99 | 96 | 74 | 17 | 10 | |
| 56 | 99 | 96 | 68 | 13 | 20 | |
| 57 |  |
95 | 91 | 67 | 12 | 16 |
| 58 | 84 | 39 | 750 | 690 | ||
| 59 | 94 | 62 | 450 | 2800 | ||
| 60 |  |
96 | 93 | 17 | 144 | 965 |
| 61 |  |
98 | 82 | 19 | 51 | 210 |
| 62 |  |
100 | 93 | 42 | 45 | 56 |
| 63 |  |
97 | 66 | 140 | 600 | |
| 64 |  |
98 | 79 | 10 | 370 | 400 |
| 65 | 100 | 93 | 4 | 51 | 100 | |
| 66 | 95 | 89 | 105 | 490 | ||
| 67 |  |
96 | 94 | 66 | 600 | 900 |
| 68 | 66 | 15 | 520 | 750 | ||
| 69 |  |
96 | 84 | 49 | 210 | 405 |
| 70 | 98 | 87 | 32 | 700 | 700 | |
| 71 | 91 | 50 | 11 | 430 | 2500 | |
| 72 |  |
100 | 82 | 19 | 230 | 1250 |
| 73 |  |
91 | 38 | 0 | 650 | 3000 |
| 74 | 96 | 71 | 9 | 200 | 650 | |
| 75 |  |
98 | 81 | 22 | 120 | 600 |
| 76 |  |
95 | 53 | 1 | 300 | 1500 |
| 77 |  |
97 | 69 | 143 | 617 | |
| 78 | 90 | 52 | 850 | 850 | ||
| 79 | 53 | 11 | 450 | 590 | ||
| 80 |  |
85 | 31 | 12 | 175 | 98 |
| 81 |  |
53 | 4 | 3000 | 3000 | |
| 82 |  |
88 | 41 | 10 | 810 | 1450 |
| 83 |  |
88 | 46 | 17 | 1738 | 1315 |
| 84 |  |
55 | 13 | 2750 | ||
| 85 |  |
96 | 77 | 46 | 1875 | |
| 86 |  |
92 | 63 | 820 | 3600 | |
| 87 |  |
84 | 32 | 0 | 2150 | 1425 |
| 88 |  |
90 | 51 | 0 | 350 | 200 |
| 89 |  |
97 | 81 | 24 | 1000 | 800 |
| 90 | 81 | 33 | 6 | 2100 | 2600 | |
All prepared according to Scheme 1.
Table 2 summarizes results for THQ-based PFT inhibitors with R1 = N1-methyl-4-imidazolyl and with variation of the R2 group. Many compounds were found with IC50s and ED50s in the low nanomolar range. The most potent in the series are 106, 107, 102, 104, 115, and 131 with values of ED50 < 10 nM. Compound 106 shows exceptional potency with an ED50 = 1.8 nM. This is the most potent compound in terms of ED50 that we found in the current study and among the most potent antimalarials ever reported. In general, the compounds with R1 = N1-methyl-4-imidazolyl are more potent than the analogous compounds with R1 = 2-pyridyl.
Table 2.
6-CN-THQs with R1 = 4-(l-Me-1H-Imidazolyl)a
 | |||||||
|---|---|---|---|---|---|---|---|
| Compound | R2 |
Pf-PFT % Inhibition at |
IC50 | ED50 (nM) |
|||
| 50 nM | 5 nM | 0.5 nM | (nM) | 3D7 | K1 | ||
| 91 | 99 | 92 | 25 | 75 | 50 | ||
| 92 | 98 | 87 | 31 | 150 | 73 | ||
| 93 |  |
0.9 | 250 | 130 | |||
| 94 | 72 | 15 | 0 | >5000 | >5000 | ||
| 95 | 60 | 5 | 5 | >5000 | 4600 | ||
| 96 | 64 | 15 | 0 | 2700 | 3100 | ||
| 97 | 34 | 9 | 0 | 2600 | >5000 | ||
| 98 | 99 | 90 | 28 | 460 | 75 | ||
| 99 |  |
100 | 96 | 61 | 19 | 17 | |
| 100 | 97 | 81 | 19 | 125 | 400 | ||
| 101 | 65 | 6 | 0 | >5000 | >5000 | ||
| 102 |  |
97 | 95 | 54 | 9 | 4.5 | |
| 103 |  |
98 | 90 | 24 | 10 | 5.5 | |
| 104 |  |
98 | 96 | 49 | 14 | 7 | |
| 105 |  |
100 | 98 | 46 | 25 | <20 | |
| 106 | 99 | 98 | 54 | 1.8 | 1.8 | ||
| 107 | 99 | 0.4 | 12 | 2 | |||
| 108 | 97 | 96 | 54 | 78 | 75 | ||
| 109 | 99 | 93 | 50 | 40 | 100 | ||
| 110 |  |
100 | 99 | 65 | 18 | 18 | |
| 111 |  |
99 | 93 | 36 | 71 | 75 | |
| 112 | 100 | 95 | 42 | 50 | 65 | ||
| 113 |  |
100 | 98 | 43 | 16 | 16 | |
| 114 |  |
99 | 82 | 8 | 65 | 70 | |
| 115 | 97 | 96 | 82 | 9 | 10 | ||
| 116 | 93 | 62 | 9 | 200 | 300 | ||
| 117 | 99 | 65 | 0 | 87 | 60 | ||
| 118 | 100 | 86 | 26 | 21 | 8.5 | ||
| 119 | 100 | 100 | 86 | 6.2 | 4.2 | ||
| 120 |  |
97 | 91 | 55 | 130 | 130 | |
| 121 |  |
99 | 98 | 69 | 57 | <20 | |
| 122 |  |
11 | 5 | 1 | 2600 | 3600 | |
| 123 | 100 | 95 | 34 | 55 | 20 | ||
| 124 | 96 | 90 | 54 | 70 | 55 | ||
| 125 | 62 | 10 | 1 | ||||
| 126 | 66 | 19 | 0 | >5000 | >5000 | ||
| 127 | 98 | 96 | 25 | 10 | 36 | ||
| 128 | 99 | 89 | 17 | 70 | 210 | ||
| 129 |  |
69 | 9 | 0 | 3000 | 4500 | |
| 130 |  |
100 | 98 | 46 | 620 | 163 | |
| 131 | 100 | 99 | 82 | 6 | 10 | ||
| 132 |  |
100 | 100 | 77 | 40 | ||
| 133 | 93 | 54 | 5 | 330 | 340 | ||
| 134 | 98 | 98 | 56 | 0.5 | 42 | 38 | |
| 135 |  |
99 | 90 | 9 | 68 | 65 | |
| 136 | 99 | 95 | 30 | 56 | 65 | ||
| 137 |  |
90 | 58 | 0 | 3000 | 3750 | |
| 138 |  |
99 | 94 | 18 | 60 | 230 | |
| 139 | 99 | 91 | 37 | 210 | 600 | ||
| 140 |  |
92 | 20 | 0 | 490 | 890 | |
| 141 |  |
98 | 97 | 14 | 18 | ||
| 142 |  |
97 | 96 | 90 | 40 | 130 | |
| 143 |  |
99 | 96 | 57 | 140 | 100 | |
| 144 |  |
99 | 91 | 12 | 355 | 350 | |
| 145 |  |
99 | 90 | 30 | 390 | ||
| 146 |  |
99 | 99 | 66 | 20 | <20 | |
We also carried out side-by-side pharmacokinetic studies of potent THQ-based Pf-PFT inhibitors to discover a compound that could be tested in malaria-infected rodents.9 We measured the rate of flux of THQ-based PFT inhibitors across a tight-junction monolayer of Caco-2 cells and showed that this in vitro assay is a good predictor of oral bioavailability of these compounds in rodents.9 In general, we found that R1 = 2-pyridyl promotes better Caco-2 permeability and oral bioavailability than compounds with R1 = N1-methyl-4-imidazolyl.9 Because it appears that THQ-based PFT inhibitors are cleared in rodents by hepatic metabolism, we measured the half-time for metabolism of THQs by rat and mouse liver microsomes in vitro.9 Based on an overall balance of desirable properties, 6 emerged as a promising antimalarial lead compound.9 Thus, we investigated in detail the structure–activity relationships among 6 analogs. Table 3 shows the results with 6 analogs in which the R group attached to the piperidine N of the R2 group is varied. In general, we found a good correlation between IC50 values for Pf-PFT inhibition and ED50 values for blocking the growth of parasites in vitro.
Table 3.
Analogs of 6 with a Variation of the R Group Attached to the Piperidine N
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Syn. | Pf-PFT | |||||||
| Scheme | %inhibition at | ED50 | ||||||
| 50 nM | 5 nM | IC50 | (nM) | |||||
| Compound | R | 0.5 nM | (nM) | 3D7 | K1 | |||
| 147 | 2 | 94 | 65 | 29 | 2.4 | 37 | 95 | |
| 148 | 2 | 99 | 82 | 20 | 0.8 | 16 | 25 | |
| 149 | 2 |  |
98 | 91 | 24 | 0.6 | 18 | 13 |
| 150 | 2 |  |
99 | 76 | 17 | 1.2 | 48 | 75 |
| 151 | 2 | 96 | 88 | 36 | 0.6 | 20 | 76 | |
| 152 | 2 | 95 | 63 | 12 | 3 | 80 | 690 | |
| 153 | 2 |  |
100 | 94 | 39 | 0.75 | 11.5 | 10 |
| 154 | 2 |  |
100 | 94 | 39 | 58 | 50 | |
| 155 | 2 |  |
98 | 77 | 30 | 2 | 43 | 55 |
| 156 | 2 |  |
100 | 92 | 31 | 50 | 60 | |
| 157 | 2 |  |
88 | 37 | 15 | 750 | 690 | |
| 158 | 2 |  |
94 | 63 | 20 | 7 | 150 | 330 |
| 159 | 2 |  |
26 | 7 | 0 | 900 | 460 | 1700 |
| 160 |  |
92 | 92 | 51 | 80 | 93 | ||
| 161 | 2 |  |
99 | 87 | 35 | 70 | 81 | |
| 162 | 2 | 100 | 96 | 61 | 0.58 | 16 | 15 | |
| 163 Isomer (R) |
1 | 100 | 99 | 58 | 0.29 | 11 | 13 | |
| 164 Isomer (S) |
1 | 98 | 70 | 02 | 2.7 | 95 | 75 | |
| 165 | 2 | 100 | 95 | 46 | 0.5 | 55 | 40 | |
| 166 | 2 | 97 | 90 | 36 | 50 | 50 | ||
| 167 | 2 |  |
100 | 88 | 12 | 1.4 | 62 | 62 |
| 168 | 2 | 91 | 91 | 47 | 65 | 75 | ||
| 169 | 2 |  |
98 | 66 | 13 | 550 | 650 | |
| 170 | 2 |  |
98 | 95 | 60 | 0.65 | 67 | 65 |
| 171 | 2 | 100 | 94 | 35 | 13 | 11.5 | ||
| 172 | 2 |  |
100 | 93 | 38 | 65 | 45 | |
| 173 | 2 |  |
98 | 84 | 28 | 320 | 150 | |
| 174 | 2 |  |
99 | 91 | 31 | 35 | 65 | |
| 175 | 2 |  |
94 | 62 | 14 | 140 | 200 | |
| 176 | 2 |  |
38 | 0 | 0 | 3000 | 2800 | |
| 177 | 2 | 99 | 93 | 45 | 0.9 | 62 | 50 | |
| 178 | 2 |  |
98 | 94 | 56 | 75 | 55 | |
| 179 | 2 |  |
96 | 89 | 17 | 75 | 55 | |
| 180 | 2 |  |
96 | 67 | 7 | 275 | 280 | |
| 181 | 2 |  |
92 | 60 | 20 | 700 | 650 | |
When those compounds in Table 3 with values of ED50 < ~100 nM were tested for Caco-2 permeability and microsomal stability,9 162 emerged as a compound with a good balance of potency and desirable pharmacokinetic properties. In Table 4, we summarize results with compounds in which R2 is held as the R2 of 162 and R1 is varied. We also studied compounds with R2 = 2-fluorophenylCH2, 4-MeSO2-phenylCH2, and t-BuNHCOCH2 because these groups led to potent compounds in the early scans. Most compounds in this series were significantly less potent on Pf-PFT and on parasites compared to 162. The exceptions were those with a small heterocyclic R1 group, that is, 191.
Table 4.
6-CN-THQs with a Variation of R1a
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compound | R1 | R2 |
Pf-PFT % Inhibition at |
IC50 | ED50 (nM) |
|||
| 50 nM | 5 nM | 0.5 nM | (nM) | 3D7 | K1 | |||
| 182 | 98 | 93 | 43 | 50 | 70 | |||
| 183 |  |
99 | 93 | 48 | 100 | |||
| 184 |  |
98 | 79 | 14 | 170 | 190 | ||
| 185 |  |
65 | 6 | 0 | 1800 | 2000 | ||
| 186 |  |
97 | 80 | 24 | 55 | 70 | ||
| 187 |  |
93 | 86 | 39 | 90 | 150 | ||
| 188 | 98 | 84 | 20 | 140 | 160 | |||
| 189 |  |
88 | 39 | 2 | 550 | 1700 | ||
| 190 |  |
95 | 80 | 24 | 260 | 400 | ||
| 191 |  |
99 | 99 | 72 | 3 | 1.6 | ||
| 192 |  |
96 | 67 | 15 | 38 | |||
| 193 |  |
100 | 96 | 39 | 60 | 75 | ||
| 194 |  |
95 | 66 | 18 | 55 | 75 | ||
| 195 |  |
99 | 86 | 25 | 62 | 140 | ||
| 196 |  |
 |
92 | 61 | 10 | 2300 | 1900 | |
| 197 | 89 | 49 | 5 | >5000 | 675 | |||
| 198 |  |
100 | 98 | 39 | 42 | 45 | ||
| 199 |  |
66 | 5 | 0 | 2700 | >5000 | ||
| 200 |  |
44 | 00 | 0 | 4100 | >5000 | ||
| 201 | 55 | 22 | 1 | 1100 | 1800 | |||
| 202 | 37 | 3 | 0 | >5000 | 3950 | |||
| 203 |  |
58 | 19 | 0 | 1600 | 2400 | ||
| 204 |  |
99 | 81 | 24 | 138 | 105 | ||
| 205 |  |
99 | 89 | 31 | 135 | 100 | ||
| 206 |  |
68 | 21 | 2 | >5000 | >5000 | ||
| 207 |  |
15 | 0 | 0 | >5000 | >5000 | ||
| 208 | 51 | 7 | 0 | 600 | >5000 | |||
| 209 | 61 | 11 | 0 | 820 | 2100 | |||
| 210 | 41 | 16 | 11 | 1200 | 3050 | |||
| 211 | 62 | 10 | 8 | 360 | 3600 | |||
| 212 | 96 | 76 | 12 | 580 | 500 | |||
| 213 | 94 | 62 | 23 | 175 | 175 | |||
| 214 | 86 | 44 | 10 | 730 | 750 | |||
| 215 | 66 | 22 | 13 | 900 | 1000 | |||
| 216 | 25 | 4 | 4 | 150 | 150 | |||
| 217 |  |
74 | 28 | 4 | 700 | 1000 | ||
| 218 | 99 | 91 | 38 | 165 | 330 | |||
| 219 | 97 | 83 | 17 | 185 | 340 | |||
| 220 |  |
99 | 87 | 16 | 400 | 800 | ||
| 221 | 100 | 100 | 84 | <20 | <20 | |||
| 222 | 97 | 88 | 36 | 75 | 60 | |||
| 223 | 97 | 78 | 10 | 2100 | 3500 | |||
| 224 | 97 | 55 | 13 | 830 | ||||
| 225 | 96 | 67 | 8 | 600 | ||||
| 226 | 80 | 27 | 2 | 8 | 750 | 3100 | ||
| 227 | 15 | 14 | 10 | 800 | >5000 | >5000 | ||
| 228 | 3 | 0 | 0 | >5000 | >5000 | |||
| 229 | 0 | 3 | 2 | >5000 | >5000 | |||
| 230 |  |
11 | 2 | 5 | >5000 | >5000 | ||
| 231 | 11 | 10 | 5 | >5000 | >5000 | |||
| 232 |  |
60 | 21 | 14 | >5000 | >5000 | ||
| 233 | 71 | 33 | 30 | 42 | >5000 | 5000 | ||
All prepared according to Scheme 1.
Molecular modeling studies described below suggested that addition of a Me group to R1 = N1-methyl-4-imidazolyl at the 5-position would better fill the R1-binding pocket on Pf-PFT. Table 5 shows a scan of R2 groups keeping R1 as 4-(N1-methyl-5-Me-imidazolyl). Compound 234 with the same R2 as in 162 emerged as the best compound.
Table 5.
6-CN-THQs with Dimethyl Imidazolea
 | ||||||
|---|---|---|---|---|---|---|
| R2 |
Pf-PFT % Inhibition at |
ED50 (nM) |
||||
| Comp. | 50 nM | 5 nM | 0.5 nM | 3D7 | K1 | |
| 234 |  |
100 | 97 | 49 | 35 | <20 |
| 235 |  |
98 | 87 | 18 | 140 | 75 |
| 236 |  |
92 | 70 | 23 | 320 | 450 |
| 237 |  |
97 | 76 | 10 | 640 | |
| 238 |  |
98 | 87 | 13 | 850 | |
All were prepared according to Scheme 1.
Table 6 gives the activity of compounds in which the sulfonyl is replaced by carbonyl (sulfonamide to amide change). In general, these compounds are much poorer inhibitors of Pf-PFT and of P. falciparum growth than the corresponding sulfonyl-containing THQ compounds. Thus, we prepared only a limited set of these amide-containing THQ compounds.
Table 6.
6-CN-THQs with Amidesa
 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compound | R1 | R2 |
Pf-PFT % Inhibition at |
IC50 | ED50 (nM) |
|||
| 50 nM | 5 nM | 0.5 nM | (nM) | 3D7 | K1 | |||
| 239 | >50 | >5000 | >5000 | |||||
| 240 | >50 | >5000 | >5000 | |||||
| 241 | >50 | >5000 | >5000 | |||||
| 242 | 26 | 9 | 6 | 2900 | ||||
| 243 | 56 | 19 | 11 | 290 | ||||
| 244 | 33 | 0 | 0 | 690 | 750 | |||
| 245 | 43 | 1 | 0 | 650 | 730 | |||
| 246 | 33 | 0 | 0 | 950 | 2000 | |||
| 247 | 52 | 9 | 0 | 270 | 450 | |||
| 248 | 16 | 0 | 0 | 2700 | ||||
| 249 |  |
92 | 51 | 0 | 260 | |||
| 250 |  |
66 | 13 | 0 | 340 | |||
| 251 |  |
88 | 39 | 0 | 75 | |||
All were prepared according to Scheme 3.
Table 7 shows THQ-based compounds with alteration to the N-Me-imidazoleCH2 group attached to the THQ ring N. This substituted imidazole is a direct ligand to the Zn2+ at the active site of Pf-PFT (see below). Removal of the Me group from the imidazole has only a minimal effect on binding to Pf-PFT (as predicted by modeling) but greatly increases ED50 for killing parasites, suggesting that methylation of the Zn2+-binding imidazole promotes penetration of compound across erythrocyte and parasite membranes. Replacement of the CH2 that links the imidazole to the THQ ring N by SO2 obliterates binding to Pf-PFT and antiparasite activity. Addition of a Me group to this CH2 bridge (mixture of four stereoisomers) also greatly reduces activity as does replacement of the imidazole by a 3-pyridylCH2 group.
Table 7.
6-CN-THQs with a Variation of R3
 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Syn. Scheme |
R1 | R2 | R3 |
Pf-PFT % Inhibition at |
ED50 (nM) |
||||
| Comp | 50 nM | 5 nM | 0.5 nM | 3D7 | K1 | ||||
| 252 | 4 | 94 | 120 | ||||||
| 253 | 4 | 93 | 120 | ||||||
| 254 | 4 |  |
45 | 22 | 150 | 140 | |||
| 255 | 5 |  |
0 | 0 | 0 | >5000 | 3200 | ||
| 34 | 8 |  |
96 | 79 | 12 | 650 | 650 | ||
| 256 | 4 | 35 | 12 | 3 | 3400 | 3000 | |||
THQ compounds with a 6-phenyl instead of 6-CN were available to us from a different drug discovery program. Some of these compounds maintain modest potency as inhibitors of Pf-PFT and as antimalarials (266, 267), but in general, we could not find a 6-phenyl THQ with a potency approaching our best compounds in the 6-CN-THQ series. It may be noted that the length of the phenyl is about the same as the length of the CN group, and thus, compounds like 267 may be binding to Pf-PFT in the same way as the 6-CN-THQ compounds, but we have no experimental evidence for this.
The data in Table 9 shows that replacement of the 6-CN group on the THQ core with various amides or a carboxylate is very detrimental to Pf-PFT binding and parasite killing.
Table 9.
6-Acyl-THQsa
 | ||||||
|---|---|---|---|---|---|---|
| Compound | R4 |
Pf-PFT % Inhibition at |
ED50 (nM) |
|||
| 50 nM | 5 nM | 0.5 nM | 3D7 | K1 | ||
| 285 | 25 | 0 | 4 | >5000 | >5000 | |
| 286 | 46 | 7 | 14 | 4000 | 2600 | |
| 287 | 66 | 17 | 5 | 2800 | 2300 | |
| 288 | 35 | 2 | 0 | >5000 | ||
| 289 | 80 | 29 | 0 | 3500 | 1425 | |
| 290 | 69 | 20 | 12 | 1200 | ND | |
| 291 | 59 | 1 | 0 | 4600 | ND | |
| 292 | 73 | 23 | 0 | 2800 | 2600 | |
All were prepared according to Scheme 7.
Structural Rational of the Structure–Activity Results for THQ-Based PFT Inhibitors
To date, we have not been able to obtain Pf-PFT in amounts sufficient for crystallization trials. However, the X-ray structure of rat and human PFTs are available alone and with a number of bound inhibitors, including the structure of the rat-PFT/162 complex.8 As described in General Methods, we made a homology model of Pf-PFT based on this experimental structure. Most of the residues in the 162 binding site are conserved between rat- and Pf-PFT. The only differences are that Pro-150β of rat-PFT, which resides in the vicinity of the terminal methyl of the R2 carbamate of 162, is replaced with Thr-512β in Pf-PFT. Tyr-93β, adjacent to the 6-CN of 162, is Leu-443β in Pf-PFT. Two more conservative changes, somewhat further away from 162, play a role in the binding of other THQ-based inhibitors discussed in this paper: Tyr-166α (Phe-151α in Pf-PFT) and Phe-360β (Tyr-836β in Pf-PFT). Thus, the crystal structure of the complex of 162 with the rat enzyme is of great value despite only 23% sequence identity for rat- versus Pf-PFT α-subunits and 37% identity between the β-subunits.
The elucidation of the structure–activity relationships of 162 and other THQ-based PFT inhibitors is complicated by the use of racemates. Each compound is a mixture of two enantiomers due to the chiral center at carbon-3 of the THQ ring (Figure 1). Moreover, each enantiomer can exist in two conformations, with the 3-substituent either axial or equatorial. Hence, it is crucial to know which of the two enantiomers block the enzyme. To gain insight, we first determined the IC50 of the individual enantiomers of 162. This was carried out by coupling 6-cyano-l,2,3,4-tetrahydro-quinolin-3-ylamine hydrochloride to S-mandelic acid and separating the diastereomers as described.5,10 The two enantiomeric amines were then converted to the two enantiomers of 162, 163, and 164, as shown in Scheme 1. The R-enantiomer of 162, 163, displayed an IC50 against Pf-PFT that is 10 times below that of the S-enantiomer, 164 (Table 3). This difference is recapitulated in the observed ED50S for blocking P. falciparum growth. The 10-fold difference in enzyme binding is modest, corresponding to a difference in binding energy of 1.3 kcal/mol. Superposition of the two enantiomers shows that the first substituent atoms of the two possible equatorial enantiomers at C3 of the THQ ring are only 0.8 Å apart, thereby, allowing for the C3 substituents to adopt similar positions in the binding site regardless of the stereochemistry (Figure 2).
Figure 2.
Near coincidence of the sulfonamide nitrogen positions of the C3 enantiomers of THQ inhibitors like 162. The figure shows the superposition of the two possible envelope conformations of the THQ ring, resulting in the positions of the exocyclic sulfonamide nitrogens being only 0.8 Å apart (only the equatorial conformations are shown; they are 2.7 kcal/mol more stable than the axial conformations). The R/S designation is specifically for the sulfonamide substitution. All figures of 3-dimensional molecular scenes were made with PyMOL (http://www.pymol.org).
Figure 3 (top panels) shows the experimental, ternary structure of rat-PFT with bound 162 inhibitor and farnesyl-pyrophosphate substrate.8 The THQ ring of 162 stacks face-on-face and at an angle of about 25° with Tyr 361β (Tyr 837β in the Pf-PFT homology model), projecting the N-methyl-imidazole so that it coordinates the catalytic Zn2+ of the enzyme (Figure 3). The 6-CN sits in a narrow groove made by Leu-96β (Leu 446β in Pf-PFT)), Trp-106β (Trp-456β in Pf-PFT), Asp-359β (Asp-835 β in Pf-PFT), and Tyr-361β (Tyr-837β in Pf-PFT). In this way, the 6-CN-benzo group of 162 adopts a similar but not identical position as the group of 3-benzyl-1-(3H-imidazol-4-ylmethyl)-4-(thiophene-2-sulfonyl)-2,3,4,5-tetrahydro-1H-benzo[e][l,4]-diazepine-7-carbonitrile (BMS-214662), a tetrahydrobenzodiazepine-based PFT inhibitor for which an X-ray structure of its complex with rat PFT has been published;11 the difference in 6-CN-benzo group atoms ranges from 0.5 to 0.9 Å (Figure 3, bottom left panel). The X-ray structure of bound 162 (obtained by soaking the rat PFT crystals with racemic 162) shows that only one enantiomer binds; the data is consistent with our observation that the enantiomer 163 is the better inhibitor (see above). The C3 substituent is equatorial, reflecting the 2.7 kcal/mol calculated preference for this conformation over the axial one in solution. The 2-pyridyl R1 substituent makes a hydrogen bond with Tyr-361β (Tyr-837β in Pf-PFT) and stacks against the second isoprene unit of farnesyl-pyrophosphate. The 2-pyridyl also stacks against the Zn2+-binding N-Me-imidazole of 162. The R2 substituent resides in a large, mainly hydrophobic pocket, which is normally occupied by the side chain of the X residue of the CaaX motif of proteins that are farnesylated (Figure 3, bottom right panel). The R2 group only partially fills this pocket. The carbonyl oxygen of the carbamate portion of the R2 substituent of 162 accepts hydrogen bonds with the side chains of Ser-99β and Trp-102β of rat-PFT (Ser-449β and Trp-452β in Pf-PFT). The terminal Me of the carbamate portion of the R2 group makes hydrophobic contacts with the side chains of His-149β (His-509β in P. fal.) and Ala-151β (Ala-511β in Pf-PFT). The R2 piperidine contacts Trp-102β (Trp-452β in Pf-PFT). Remarkably, the two oxygens of the sulfonamide do not make direct hydrogen bonds to the protein.
Figure 3.
Experimental binding mode of 162 to rat PFT (top left); molecular surface of rat-PFT (left; C, gray; N, blue; O, red; S, orange), with farnesyl-pyrophosphate (red, orange, and cyan) and Zn2+ (green) in CPK representation and 162 in stick model representation; equivalent all-stick model (top right); comparison of the binding modes of 162 (yellow carbons) and 3-benzyl-1-(3H-imidazol-4-ylmethyl)-4-(thiophene-2-sulfonyl)-2,3,4,5-tetrahydro-1H-benzo[e][1,4]diazepine-7-carbonitrile (cyan carbons, see ref 11 for the structure) to rat PFT (bottom left); superposition of 162 (green carbon atoms) and the C-terminal tetrapeptide fragment of the Rap2a peptide farnesyl acceptor substrate (cyan carbon atoms) as bound in PFT (bottom right; catalytic Zn2+ shown).
We used the 162/PFT structural information as a starting point for constructing models of many of the other THQ-based PFT inhibitors prepared in this paper. Rather than discuss each compound, we describe the trends that are supported by the structural information. Figure 4 (top panel) shows the predicted binding mode of 162 in the active site of Pf-PFT. As already noted, THQ-based compounds in Table 1 have a 6-CN group, R1 = 2-pyridyl, and variation of the R2 group. Most of the compounds in Table 1 are predicted to be able to bind with R2 in the a2X region of the Ca1a2X pocket, while maintaining the pose of the THQ scaffold and R1 and R3 groups as seen experimentally. R2 groups that end in a polar group are poor Pf-PFT binders because they cannot interact favorably with the largely hydrophobic environment (38, 39, 52, 53, 54, 90). Moreover, THQs with small R2 groups (38, 52, 53, 90) barely fill the pocket and therefore display poor enzyme inhibition. On the other hand, the R2 group of 87 is too large to fill the pocket. Long, linear R2 groups, some with a small branch, that provide several apolar atoms work better as Pf-PFT inhibitors as they can reach Trp-456β, Trp-452β, Ala-511β and His-509β, but this comes at a cost of the loss in conformational entropy when the flexible side chain rigidifies in the active site of Pf-PFT (35, 36, 40, 41, 42, 43, 44, 45, 48, 46, 47, 49, 50, 38).
Figure 4.
Predicted binding mode of 162 in the active site of Pf-PFT (top left); molecular surface of PFT (left; C, gray; N, blue; O, red; S, orange), with farnesyl-pyrophosphate (red, orange, and cyan) and Zn2+ (green); predicted binding mode of 55 in the active site of Pf-PFT (top right). Note how the R2 pyrrole group is wedged between C4 of the THQ ring and Trp-452β and Trp-456β.
The R2 groups of all other THQs contain one or more rings. The best R2 groups have a short ethylene linker (thus minimizing entropic cost of enzyme binding) followed by a five-membered, aromatic heterocycle (55, 56, 57). They are predicted to bind in the same way as 162 (Figure 4, top right), engaging in extensive hydrophobic interactions with Leu-446β, Trp-456β, Trp-452β, and C4 of their own THQ scaffold; the latter probably stabilizes the enzyme-bound R2 conformation for the inhibitor free in solution. Two of the R2 ring atoms remain solvent exposed, which explains why those two positions can be replaced by nitrogen atoms (56, 57). However, a slightly larger benzene ring cannot be accommodated in the same position (74, 75, 76), but further para-substitution with MeSO2 leads to two hydrogen bonds of this group with Arg-564β and Gln-152α, plus hydrophobic contacts with His-509β (69), explaining the decent potency of this compound. Expansion of the pyrrole ring to an indole (64) leads to reduced inhibition because the R2 conformation cannot be maintained due to steric clash with Leu-446β. Expansion of the pyrrole ring to phthalimide (85) also leads to a different conformation because of steric reasons, though the benzo part is now predicted to interact with His-509β.
Most R2 groups with a single methylene linker to an aromatic ring are unable to make an efficient contact with the hydrophobic residues in the pocket of Pf-PFT as the ethylene-linked ones, even when they in turn carry a hydrophobic substituent (58, 59, 60, 61, 62, 63, 65, 66, 68, 70, 71, 69, 73, 77, 78, 79, 80, 81, 83, 6, 84). Longer linkers to a ring also do not allow for efficient contact of hydrophobic residues in the pocket (82, 86, 88).
Replacement of the sulfonamide group of THQs with amides universally leads to a loss of binding to Pf-PFT (Table 6). In the crystal structure of rat PFT with 162, and by homology with Pf-PFT, the sulfonamide moiety adopts a known low-energy conformation. Sulfonamides, which have a pyramidalized nitrogen atom, adopt two low-energy conformations because the N–S bond has partial double character. One is eclipsed, thereby allowing for maximal overlap of the nitrogen lone pair electrons and the S=O bonds (Figure 5). The other one is obtained by nitrogen inversion, thereby reducing the steric interactions.12 Ab initio calculations at the MP2/6–31+G* level for the model compound N,N-dimethylmethanesulfonamide, the closest analog to the sulfonamide substructure in our molecules, shows that this compound prefers the inverted conformation with a C–N–S=O torsion angle of 30°.13 This conformation is similar to what is observed in the X-ray structure of 162 bound to rat PFT (Figure 5). Simple stereochemical considerations indicate why the amide analogs of our sulfonamide THQ PFTIs are poor inhibitors. The amide function has two trigonal centers, and thus, a planar arrangement of all substituents. This leads to an orientation of the R1 group in the amides that is 90° off from the orientation of the R1 of 162 (Figure 5), and this altered orientation is sterically disallowed by the active site of the PFT.
Figure 5.
Comparison of the experimentally observed conformation of THQ PFTIs with a sulfonamide linker (left) and the conformation accessible to the analogous amides (right). The latter projects the R1 group into a sterically poorly accessible area of PFT (not shown).
Modeling showed that there is space in the active site of Pf-PFT to accommodate groups other than N-methyl on the Zn2+-binding imidazole. A benzyl (254, Table 7) was not very effective probably because of the extra entropy loss due to two extra rotatable bonds. Replacement of the methylene linker in the Zn2+-binding arm (attached to N1 of the THQ ring) by an SO2 (255, Table 7) leads to an inactive compound. This is most likely because of the increased electron-withdrawing character of the SO2 linker, thereby decreasing the basicity of the imidazole N and, thus, the coordination affinity to the active site Zn2+. Using the SPARC v3.1 program,14,15 we calculated that the pKa of the protonated imidazole drops from 6.8 to 5.1 when the CH2 linker is replaced with SO2. If the drop in proton affinity is the same for Zn2+, this drop in pKa of 1.7 would give a drop in IC50 by 50-fold.
Replacement of the 6-CN group with phenyl is tolerated (Table 8). Replacement with more polar substituents (Table 9) is not tolerated, which is consistent with the lack of apporpriate hydrogen bond donors and acceptors in the pocket on Pf-PFT that binds the 6-CN group.
Table 8.
6-Phenyl-THQsa
 | |||||||
|---|---|---|---|---|---|---|---|
| Compound | R1 | R2 |
Pf-PFT % Inhibition at |
ED50 (nM) |
|||
| 50 nM | 5 nM | 0.5 nM | 3D7 | K1 | |||
| 257 | 80 | 31 | 14 | 3500 | >5000 | ||
| 258 | 88 | 38 | 10 | >5000 | >5000 | ||
| 259 | 86 | 29 | 10 | 5000 | >5000 | ||
| 260 | 98 | 84 | 27 | 3000 | 3700 | ||
| 261 | 95 | 74 | 14 | >5000 | 3700 | ||
| 262 | 98 | 84 | 34 | 1500 | |||
| 263 | 99 | 89 | 22 | 640 | 475 | ||
| 264 | 97 | 89 | 21 | 380 | 360 | ||
| 265 | 97 | 91 | 28 | 750 | 950 | ||
| 266 | 99 | 87 | 37 | 375 | 350 | ||
| 267 |  |
97 | 73 | 23 | 381 | 395 | |
| 268 | 99 | 91 | 29 | 400 | 700 | ||
| 269 | 98 | 81 | 18 | 2400 | 750 | ||
| 270 |  |
32 | 0 | 0 | 450 | 350 | |
| 271 |  |
33 | 10 | 0 | 860 | ||
| 272 | 90 | 38 | 0 | 460 | 150 | ||
| 273 | 88 | 46 | 0 | 4600 | 3500 | ||
| 274 | 79 | 26 | 0 | 3000 | 3700 | ||
| 275 | 99 | 90 | 9 | 450 | 380 | ||
| 276 | 46 | 0 | 0 | 2900 | 1800 | ||
| 277 | 98 | 81 | 12 | 300 | 500 | ||
| 278 | 98 | 84 | 32 | 270 | 450 | ||
| 279 | 19 | 14 | 11 | >5000 | >5000 | ||
| 280 |  |
11 | 6 | 6 | >5000 | 3000 | |
| 281 |  |
22 | 15 | 0 | 2700 | ||
| 282 | 2 | 0 | 0 | >5000 | >5000 | ||
| 283 | 10 | 1 | 4 | 3500 | >5000 | ||
| 284 | 8 | 0 | 0 | 2800 | 2800 | ||
All were prepared according to Scheme 6.
Inhibition of Mammalian PFT
A subset of THQ-based PFTIs were tested for inhibition of rat PFT, and results are summarized in Table 10 and compared to those obtained with Pf-PFT. Most compounds displayed comparable potency on both enzymes. The exceptions are 84 and 129, which are about 10-fold more potent on Pf-PFT than on rat PFT, and 126, which shows the reverse preference. As noted in our earlier publications, clinical trials have shown that PFTIs are well tolerated in man after several weeks of continuous dosing. Thus, specificity toward Pf-PFT versus mammalian PFT is probably not required for a drug that would be used to treat malaria over the course of a few days.
Table 10.
Potency on Mammalian versus Pf-PFT
| cmpd | IC50 on mammalian PFT (nM) |
Approximate IC50 on Pf-PFTa (nM) |
|---|---|---|
| 81 | 10 | 50 |
| 65 | 3.8 | 2 |
| 84 | 440 | 50 |
| 55 | 1.7 | 0.2 |
| 62 | 3 | 0.5 |
| 6 | 7 | 2 |
| 226 | 5 | 20 |
| 151 | 7.8 | 2 |
| 152 | 6.5 | 4 |
| 162 | 3.2 | 0.4 |
| 158 | 16 | 4 |
| 167 | 25 | 3 |
| 165 | 5.5 | 0.5 |
| 134 | 2.6 | 0.5 |
| 129 | 1000 | 40 |
| 91 | 7.5 | 2 |
| 101 | 105 | 40 |
| 126 | 4.8 | 40 |
| 123 | 3.4 | 1 |
Conclusions
In this study we show that THQ-based PFTIs are potent inhibitors of Pf-PFT activity and of erythrocytic stage P. falciparum growth. Several compounds were found with growth inhibition potency down in the low nanomolar range, with several compounds blocking parasite growth at concentrations <5 nM. A good deal of the structure–activity data for the inhibition of Pf-PFT can be accounted for based on the structural consideration of one of the compounds, 162, bound to mammalian PFT. In a companion study, we have carried out detailed preclinical pharmacokinetic studies of our most potent Pf-PFT inhibitors.9 Together, these two studies provide the basis for further development of Pf-PFT inhibitors as novel antimalarial drugs.
Experimental Section
Synthesis of Compounds. General Methods
Unless otherwise indicated, all anhydrous solvents were commercially obtained and stored under nitrogen. Reactions were performed under an atmosphere of dry nitrogen in oven-dried glassware and were monitored for completeness by thin layer chromatography (TLC) using silica gel 60 F-254 (0.25 mm) plates with detection with UV light. 1H NMR spectra were recorded on dilute solutions in CDC13, CD3-OD, or DMSO-d6 at 300 or 500 MHz. Chemical shifts are reported in parts per million (δ) downfield from tetramethylsilane (TMS). Coupling constants (J) are reported in Hz. Electrospray ionization mass spectra were acquired on an Bruker Esquire LC00066. Flash chromatography was carried out with silica gel (40–63 µm). Preparative reverse phase HPLC was performed on an automated Varian Prep star system using a gradient of 20% MeOH to 100% MeOH (with 0.1% trifluoroacetic acid) over 30 min using a YMC S5 ODS column (20 × 100 mm, Waters, Inc.). All final compounds (those tested on Pf-PFT and on parasites cultures) were purified by HPLC as above to single eluting peaks.
General Procedure for Synthesis of Compounds According to Scheme 1. Sulfonation
A solution of 6-cyano-1,2,3,4-tetrahy-dro-quinolin-3-ylamine hydrochloride 3 (5 mmol), sulfonyl chloride (10 mmol; for example, ref 16, and N,N-diisopropylethyl amine (15 mmol) in 25 mL of anhydrous CH3CN was stirred at room temperature overnight. A light-colored precipitate of 4 was isolated by vacuum filtration. More often, product 4 was obtained by flash chromatography on a silica gel column, eluting with 50% ethyl acetate/hexane: yields 85–95%.
1-Methyl-1 H-imidazole-4-sulfonic Acid (6-Cyano-1,2,3,4-tetrahydro-quinolin-3-yl)-amide (4)
1H NMR (500 MHz, DMSO-d6) δ 7.80 (s, 1H), 7.78 (s,1H), 7.70 (br s, 1H), 7.25 (d, J = 8.7 Hz, 1H), 7.20 (s, 1H), 6.81 (s, 1H), 6.51 (d, J = 8.6 Hz, 1H), 3.78 (s, 3H), 3.52–3.43 (m, 1H), 3.25–3.20 (m, 1H), 3.15–2.9 (m, 1H), 2.81–2.73 (m, 1H), 2.65–2.53 (m, 1H). MS m/z 318.5 (M + H+).
Pyridine-2-sulfonic Acid (6-Cyano-1,2,3,4-tetrahydro-quinolin-3-yl)-amide (4)
1H NMR (300 MHz, methanol-d4) δ 8.51 (d, J = 7.2 Hz, 1H), 8.02 (td, J = 2.1, 7.8 Hz, 1H), 7.92 (dt, J = 1.8, 7.5 Hz, 1H), 7.47 (ddd, J = 1.2, 4.8, 7.5 Hz, 1H), 7.12 (dd, J = 2.1, 8.4 Hz, 1H), 7.0 (d, J = 1.8 Hz, 1H), 6.3 (d, J = 8.4 Hz, 1H), 5.87 (d, J = 7.8 Hz, 1H), 3.94–4.01 (m, 1H), 3.36 (dd, J = 2.7, 12 Hz, 1H), 3.25 (ddd, J = 2.1, 4.5, 12.3 Hz, 1H), 2.87 (dd J = 4.2, 16.5 Hz, 1H), 2.71 (ddd, J = 2.1, 4.8, 16.5 Hz, 1H). MS m/z 315 (M + H+).
Reductive Amination
A mixture of sulfonamide 4 (5 mmol), 1-methyl-1H-imidazole-5-carboxaldehyde (10 mmol),16 and 20 mL of 50% trifluoroacetic acid in dichloroethane was warmed at 50 °C under argon. After 2 h, triethylsilane (20 mmol) was added. After 48 h, the solvent was removed under reduced pressure, and the crude product was partitioned between methylene chloride and 1 N NaOH (45 mL). The organic layer was separated, and the aqueous layer was extracted with CH2Cl2 (3 × 25 mL). The combined organic layers were dried over MgSO4, filtered, and concentrated. The crude residue was either recrystallized with dichloromethane or purified on a flash silica gel chromatography to afforded 5 in 45–50% yield.
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide Trifluoro Acetate Salt (5)
Recrystallized from CH2Cl2 afforded the product as a white solid (70%). 1H NMR (300 MHz, methanol-d4) δ 8.89 (s, 1H), 7.82 (s, 1H), 7.72 (s, 1H), 7.41 (s, 1H), 7.36 (d, J = 8.2 Hz, 1H,), 7.22 (s, 1H), 6.80 (d, J = 8.6 Hz, 1H), 4.80 (d, J = 16.80 Hz, 1H), 4.65 (d, J = 16.80 Hz, 1H), 3.87 (s, 3H), 3.85–3.79 (m, 1H), 3.78 (s, 3H), 3.55 (dd, J = 11.9 Hz, 1H), 3.42 (dd, J = 12.4 Hz, 1H), 3.05 (dd, J = 15.78 Hz, 1H), 2.71 (dd, J = 15.78 Hz, 1H). MS m/z 412.5 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (5)
1H NMR (300 MHz, DMSO-d6) δ 8.70 (d, J = 4.8 Hz, 1H), 8.14 (d, J = 6.6 Hz, 1H), 8.06 (dt, J = 1.8, 7.5 Hz, 1H), 7.99 (s, 1H), 7.67 (ddd, J = 1.2, 4.8, 7.8 Hz, 1H), 7.37 (dd, J = 2.1, 8.7 Hz, 1H), 7.27 (d, J = 1.8 Hz, 1H), 6.88 (s, 1H), 6.8 (d, J = 8.7 Hz, 1H), 4.62 (d, J = 16.5 Hz, 1H), 4.59 (d, J = 16.5 Hz, 1H), 3.73–3.85 (m, 1H), 3.60 (s, 3H), 3.38 (dd, J = 2.4, 11.7 Hz, 1H), 3.17 (dd, J = 8.7, 12.9 Hz, 1H), 2.88 (dd, J = 4.2, 15.9 Hz, 1H), 2.70 (dd, J = 8.7, 15.9 Hz, 1H). MS m/z 409.3 (M + H+).
N-[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-4-fluoro-benzenesulfonamide (5)
1H NMR (500 MHz, methanol-d4) δ 8.35 (s, 1H), 7.95–7.93 (m, 2H), 7.46 (s, 1H), 7.39 (d, J = 8.5 Hz, 1H), 7.35 (t, J = 8.0 Hz, 2H), 7.24 (s, 1H), 6.82 (d, J = 8.5 Hz, 1H), 4.82 (d, J = 17.0 Hz, 1H), 4.65 (d, J = 17.0 Hz, 1H), 3.95–3.85 (m, 4H), 3.69–3.65 (m, 1H), 3.55–3.45 (m, 1H), 2.97–2.96 (m, 1H), 2.71–2.61 (m, 1H). MS m/z 426.12 (M + H+).
N-Alkylation
To a suspension of 5 (5 mmol) and Cs2CO3 (9.8 mmol) in dry DMF (5 mL) was added the appropriate alkyl halide (5.4 mmol), and the mixture was stirred at room temperature overnight under argon. After addition of water (20 mL), the solution was extracted with ethyl acetate (3 × 20 mL). The organic layer was extracted with brine (3 × 10 mL). The combined organic layers were dried over MgSO4 and evaporated under reduce pressure. The residue was purified by HPLC.
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-methanesulfonyl-benzyl)-amide (69)
1H NMR (300 MHz, methanol-d4) δ 8.86 (s, 1H), 8.71 (d, J = 7.2 Hz, 1H), 8.10 (td, J = 1.5, 8.1 Hz, 1H), 7.99 (dt, J = 1.2, 8,1 Hz, 1H), 7.90 (d, J = 8.1 Hz, 2H), 7.69 (ddd, J = 1.2, 4.8, 7.5 Hz, 1H), 7.61 (d, J = 8.1 Hz, 2H), 7.33 (dd, J = 2.1, 9.0 Hz, 1H), 7.28 (s, 1H), 7.25 (d, J = 2.1 Hz, 1H), 6.65 (d, J = 8.7 Hz, 1H), 4.79 (d, J = 17.1 Hz, 1H), 4.62 (d, J = 17.1 Hz, 1H), 4.39–4.59 (m, 3H), 3.8 (s, 3H), 3.55–3.62 (m, 1H), 3.42–3.47 (m, 1H), 3.15 (s, 3H), 3.07–3.14 (m, 1H), 2.97 (dd, J = 4.5, 15.9 Hz, 1H). MS m/z 577.4 (M + H+).
6-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-pyridine-2-carboxylic Acid Methyl Ester (67)
1H NMR (300 MHz, CDCl3) δ 8.78 (s, 1H), 8.63 (d, J = 7.0 Hz, 1H), 7.78–8.04 (m, 5H), 7.51–7.76 (m, 1H), 7.30–7.34 (m, 2H), 7.18 (s, 1H), 6.56 (d, J = 8.7 Hz, 1H), 4.76 (d, J = 17.1 Hz, 1H), 4.65 (d, J = 17.1 Hz, 1H), 4.47–4.61 (m, 3H), 3.96 (s, 3H), 3.85 (s, 3H), 3.54–3.62 (m, 1H), 3.42–3.51 (m, 1H), 3.13 (dd, J = 11.1, 15 Hz, 1H), 2.91 (dd, J = 3.6, 15 Hz, 1H). MS m/z 558.3 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid tert-Butyl Ester (6)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 8.71 (d, J = 7.2 Hz, 1H), 8.05 (td, J = 1.8, 7.5 Hz, 1H), 8.01–8.02 (m, 1H), 7.66 (ddd, J = 1.2, 4.8, 7.5 Hz, 1H), 7.43 (s, 1H), 7.39 (dd, J = 1.5, 8.4 Hz, 1H), 7.33 (s, 1H), 6.80 (d, J = 9.7 Hz, 1H), 4.81 (d, J = 16.8 Hz, 1H), 4.68 (d, J = 16.8 Hz, 1H), 4.32–4.44 (m, 1H), 4.05–4.09 (m, 2H), 3.91 (s, 3H), 3.58–3.66 (m, 2H), 3.53–3.55 (m, 1H), 3.11–3.21 (m, 3H), 2.87–2.88 (m, 1H), 2.63–2.69 (m, 1H), 1.64–1.77 (m, 3H), 1.3 (s, 9H), 1.01–1.11 (m, 2H). MS m/z 606.6 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(2-pyrazol-1-yl-ethyl)-amide (56)
1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 8.73 (d, J = 6.9 Hz, 1H), 8.01–8.12 (m, 2H), 7.64–7.71 (m, 1H), 7.61 (d, J = 2.1 Hz, 1H), 7.50 (d, J = 1.5 Hz, 1H), 7.32–7.38 (m, 2H), 7.21 (s, 1H), 6.79 (d, J = 8.7 Hz, 1H), 6.30 (t, J = 2.1 Hz, 1H), 4.68 (d, J = 18.3 Hz, 1H), 4.60 (d, J = 18.3 Hz, 1H), 4.70 (t, J = 6.3 Hz, 2H), 4.25–4.35 (m, 1H), 3.83 (s, 3H), 3.74 (t, J = 6.3 Hz, 2H), 3.10–3.21 (m, 2H), 2.86 (dd, J = 12, 15.3 Hz, 1H), 2.58 (dd, J = 3.9, 15.3 Hz, 1H). MS m/z 503.4 (M + H+).
{3-[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-propyl}-methyl-carbamic Acid Methyl Ester (48)
1H NMR (300 MHz, methanol-d4) δ 8.94 (d, J = 0.9 Hz, 1H), 8.70 (d, J = 7.5 Hz, 1H), 8.08 (td, J = 1.8, 7.8 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.43 (d, J = 1.2 Hz, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.33 (s, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.82 (d, J = 17.5 Hz, 1H), 4.62 (d, J = 17.5 Hz, 1H), 4.40–4.52 (m, 1H), 3.92 (s, 3H), 3.65 (s, 3H), 3.55–3.47 (m, 4H), 3.28–3.05 (m, 4H), 2.93 (s, 3H), 1.78–1.88 (m, 2H). MS m/z 538.4 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-fluoro-benzyl)-amide (71)
1H NMR (300 MHz, methanol-d4) δ 8.89 (s, 1H), 8.74 (d, J = 6.5 Hz, 1H), 8.09 (td, J = 1.8, 7.8 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.67 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.39–7.43 (m, 2H), 7.35 (dd, J = 2.1, 8.7 Hz, 1H), 7.33 (s, 1H), 7.25 (s, 1H), 7.01–7.07 (m, 2H), 6.75 (d, J = 8.7 Hz, 1H), 4.43–4.69 (m, 5H), 3.82 (s, 3H), 3.55–3.47 (m, 1H), 3.17–3.25 (m, 1H), 3.07 (dd, J = 12, 15 Hz, 1H), 2.91 (dd, J = 3.9, 15 Hz, 1H). MS m/z 517.4 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[2-(2-trifluoromethyl-phenyl)-ethyl]-amide (76)
1H NMR (300 MHz, methanol-d4) δ 8.87 (s, 1H), 8.74 (d, J = 4.5 Hz, 1H), 8.12 (td, J = 1.8, 7.8 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.63–7.71 (m, 2H), 7.58–7.63 (m, 1H), 7.37–7.49 (m, 5H), 6.81 (d, J = 8.7 Hz, 1H), 4.92 (d, J = 17.7 Hz, 1H), 4.82 (d, J = 17.7 Hz, 1H), 4.50–4.56 (m, 1H), 3.98 (s, 3H), 3.55–3.47 (m, 4H), 3.18–3.31 (m, 3H), 3.02–3.08 (m, 1H). MS m/z 581.4 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[2-(2-fluoro-phenyl)-ethyl]-amide (113)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 7.76 (s, 1H), 7.73 (s, 1H), 7.51 (s, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.23–7.32 (m, 3H), 7.10–7.17 (m, 2H), 6.84 (d, J = 8.6 Hz, 1H), 4.92 (d, J = 17.7 Hz, 1H), 4.82 (d, J = 17.7 Hz, 1H), 4.48–4.50 (m, 1H), 4.07 (s, 3H), 3.82 (s, 3H), 3.35–3.57 (m, 4H), 3.16–3.29 (m, 3H), 2.92–3.00 (m, 1H). MS m/z 534.4 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[2-(4-fluoro-phenyl)-ethyl]-amide (112)
1H NMR (300 MHz, acetone-d6) δ 8.93 (s, 1H), 7.75 (s, 1H), 7.74 (s, 1H), 7.49 (s, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.32 (s, 1H) 7.22–7.25 (m, 2H), 7.01–7.07 (m, 2H), 6.93 (d, J = 8.6 Hz, 1H), 4.93 (d, J = 16.8 Hz, 1H), 4.80 (d, J = 16.8 Hz, 1H), 4.38–4.46 (m, 1H), 4.07 (s, 3H), 3.82 (s, 3H), 3.34–3.54 (m, 4H), 3.07 (dd, J = 12, 15 Hz, 1H), 2.98 (t, J = 8.1 Hz, 2H) 2.90 (dd, J = 3.7, 15.2 Hz, 1H). MS m/z 534.4 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-fluoro-benzyl)-amide (109)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.81 (s, 1H), 7.73 (s, 1H), 7.36–7.42 (m, 3H), 7.33 (s, 1H), 7.27 (s, 1H), 7.10–7.17 (m, 2H), 6.84 (d, J = 8.6 Hz, 1H), 4.28–4.65 (m, 5H), 3.87 (s, 3H), 3.80 (s, 3H), 3.34–3.39 (m, 1H), 3.18–3.26 (m, 1H), 3.07 (dd, J = 11.4, 15.3 Hz, 1H), 2.87 (dd, J = 3.7, 15.2 Hz, 1H). MS m/z 520.4 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-ethanesulfonyl-benzyl)-amide (132)
1H NMR (300 MHz, methanol-d4) δ 8.87 (s, 1H), 7.87 (d, J = 8.1 Hz, 2H), 7.85 (s, 1H), 7.77 (s, 1H), 7.62 (d, J) 8.1 Hz, 2H), 7.34 (dd, J = 1.8, 8.7 Hz, 1H), 7.25 (d, J = 1.8 Hz, 1H), 6.68 (d, J = 8.7 Hz), 4.75 (d, 2H), 4.53–4.55 (m, 1H), 4.51 (d, J = 17.7 Hz, 1H), 4.45 (d, J = 17.7 Hz, 1H), 3.81 (s, 3H), 3.80 (s, 3H), 3.10–3.25 (m, 6H), 2.95–3.05 (m, 1H), 1.21–1.25 (t, 3H). MS m/z 594.5 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid (4-Benzenesulfonyl-benzyl)-[6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (145)
1H NMR (300 MHz, methanol-d4) δ 8.89 (s, 1H), 7.89–8.10 (m, 4H), 7.87 (d, J = 8.1 Hz, 2H), 7.77 (s, 1H), 7.50–7.72 (m, 5H), 7.34 (dd, J = 1.8, 8.7 Hz, 1H), 7.25 (d, J = 1.8 Hz, 1H), 6.68 (d, J = 8.7 Hz, 1H), 4.75 (m, 2H), 4.53–4.55 (m, 1H), 4.51 (d, J = 17.7 Hz, 1H), 4.45 (d, J = 17.7 Hz, 1H), 3.81 (s, 3H), 3.80 (s, 3H), 3.40–3.45 (m, 2H), 2.95–3.05 (m, 1H). MS m/z 642.6 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4,4-dioxo-3,4-dihydro-2H-4λ6-benzo[1,4]oxathiin-7-ylmethyl)-amide (146)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.85 (s, 1H), 7.68 (d, J = 8.1 Hz, 1H), 7.45 (s, 1H), 7.30 (s, 1H), 7.28 (dd, J = 1.8, 8.7 Hz, 1H), 7.25 (d, J = 1.8 Hz, 1H), 7.15 (dd, J = 8.7, 1.8 Hz, 1H), 7.08 (d, J = 2.1 Hz, 1H), 6.68 (d, J = 8.7 Hz, 1H), 4.48–4.52 (m, 2H), 4.51 (d, J = 17.7 Hz, 1H), 4.45 (d, J = 17.7 Hz, 1H), 3.90 (s, 3H), 3.80 (s, 3H), 3.55–3.60 (m, 2H), 3.40–3.55 (m, 2H), 2.95–3.05 (m, 1H), 2.68–2.75 (m, 1H). MS m/z 608.4 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(1,5-dimethyl-1H-imidazole-4-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (234)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 7.73 (s, 1H), 7.40 (s, 1H), 7.37 (dd, J = 3.0, 9.0 Hz, 1H), 7.32 (d, J = 1.8 Hz, 1H), 6.79 (d, J) 9 Hz, 1H), 4.81 (d, J = 18.0 Hz, 1H), 4.68 (d, J = 18.0 Hz, 1H), 4.32–4.42 (m, 1H), 4.08–4.18 (m, 2H), 3.92 (s, 3H), 3.67 (s, 3H), 3.68 (s, 3H), 3.51–3.58 (m, 2H), 2.98–3.17 (m, 3H), 2.73–2.87 (m, 3H), 2.48 (s, 3H), 1.69–1.85, (m, 3H), 1.01–1.27 (m, 2H). MS m/z 581.6 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3yl]-(1,5-dimethyl-1H-imidazole-4-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid tert-Butyl Ester (235)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 7.73 (s, 1H), 7.43 (d, J = 1.8 Hz, 1H), 7.37 (dd, J = 3, 9 Hz, 1H), 7.32 (s, 1H), 6.79 (s, 1H), 4.81 (d, J = 18.0 Hz, 1H), 4.68 (d, J = 18.0 Hz, 1H), 4.33–4.40 (m, 1H), 4.06–4.11 (m, 2H), 3.92 (s, 3H), 3.67 (s, 3 H), 3.53–3.58 (m, 2H), 3.09–3.17 (m, 3 H), 2.64–2.81 (m, 3H), 2.48 (s, 3H), 1.63–1.81 (m, 3H), 1.4 (s, 9 H), 0.90–1.13 (m, 2H). MS m/z 623.6 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[2-(2-oxo-pyrrolidin-1-yl)-ethyl]-amide (117)
1H NMR (300 MHz, methanol-d4) δ 8.91 (s, 1H), 7.30–7.45 (m, 2H), 7.17–7.24 (m, 1H), 7.12 (s, 1H), 6.82 (d, J = 9.0 Hz, 1H), 6.52 (d, J = 9.0 Hz, 1H), 4.28–4.40 (m, 2H), 3.93 (s, 3H), 3.80 (s, 3H), 3.66–3.73 (m, 1H), 3.51–3.63 (m, 2H), 3.40–3.48 (m, 2H), 3.03–3.21 (m, 2H), 2.78–2.93 (m, 2H), 2.62–2.76 (m, 2H), 2.38 (t, J = 6.0 Hz, 2H), 2.07 (t, J = 6.0 Hz, 2H). MS m/z 523.3 (M + H+).
1,5-Dimethyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-methanesulfonyl-benzyl)-amide (236)
1H NMR (300 MHz, methanol-d4) δ 8.85 (s 1H), 7.91 (d, J = 9.0 Hz, 2H), 7.77 (s, 1H), 7.60 (d, J = 9.0 Hz, 2H), 7.35 (d, J = 9.0 Hz 2H), 7.29 (s, 1H), 6.68 (d, J = 9.0 Hz, 1H), 4.40–4.71 (m, 5H), 3.83 (s, 3H), 3.68 (s, 3H), 3.45–3.55 (m, 2H), 3.17 (s, 3H), 2.88–3.02 (m, 2H), 2.42 (s, 3H). MS m/z 594.5 (M + H+).
1,5-Dimethyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-ethanesulfonyl-benzyl)-amide (237)
1H NMR (300 MHz, methanol-d4) δ 8.88 (s 1H), 7.85 (d, J = 9.0 Hz, 2H), 7.78 (s, 1H), 7.60 (d, J = 9.0 Hz, 2H), 7.35 (d, J = 9.0 Hz 2H), 7.29 (s, 1H), 6.68 (d, J = 9.0 Hz, 1H), 4.40–4.76 (m, 5H), 3.97 (s, 3H), 3.82 (s, 3H), 3.45–3.55 (m, 2H), 3.15–3.30 (m, 2H), 2.88–3.05 (m, 2H), 2.48 (s, 3H), 1.22 (t, J = 9.6 Hz, 3H). MS m/z 608.4 (M + H+).
1,5-Dimethyl-1H-imidazole-4-sulfonic Acid (5-Bromo-2-fluorobenzyl)-[6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (238)
1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.76 (s, 1H), 7.53 (m, 1H), 7.31–7.45 (m, 3H), 7.28 (s, 1H), 6.99 (d, J = 9.0 Hz, 1H), 6.72 (d, J = 9.0 Hz, 1H), 4.42–4.75 (m, 5H), 3.89 (s, 3H), 3.67 (s, 3H), 3.50–3.62 (m, 2H), 2.95–3.10 (m, 2H), 2.40 (s, 3H). MS m/z 612.8 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-methanesulfonyl-benzyl)-amide (131)
1H NMR (300 MHz, methanol-d4) δ 9.0 (s 1H), 7.95–7.85 (m, 3H), 7.60 (d, J = 8.7 Hz, 2H), 7.35 (t, J = 7.8 Hz, 3H), 7.29 (s, 1H), 6.70 (d, J = 8.7 Hz, 1H), 4.60–4.40 (m, 4H), 4.40–4.30 (m, 1H), 3.85 (s, 3H), 3.80 (s, 3H), 3.40–3.30 (m, 2H), 3.0–2.90 (m, 1H), 2.83–2.75 (m, 1H), 2.50 (s, 3H). MS m/z 580.5 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-yl-methyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(3-methoxy-propyl)-amide (91)
1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.80 (s, 1H), 7.75 (s, 1H), 7.43 (s, 1H), 7.39 (d, J = 8.7 Hz, 1H), 7.32 (s, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.78 (d, J = 16.7 Hz, 1H), 4.69 (d, J = 16.7 Hz, 1H), 4.45–4.38 (m, 1H), 3.92 (s, 3H), 3.81 (s, 3H), 3.59–3.51 (m, 2H), 3.43–3.38 (m, 2H), 3.32 (s, 3H), 3.28–3.20 (m, 2H), 3.12–3.07 (m, 1H), 2.85 (dd, J = 3.3, 15.3 Hz, 1H), 1.96–1.85 (m, 4H). MS m/z 484.5 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(5-trifluoromethyl-furan-2-ylmethyl)-amide (108)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.72 (s, 1H), 7.60 (s, 1H), 7.42–7.36 (m, 2H), 7.40 (s, 1H), 6.90 (s, 1H), 6.70 (d, J = 8.5 Hz, 1H), 6.50 (s, 1H), 4.75–4.45 (m, 5H), 3.90 (s, 3H), 3.75 (s, 3H), 3.50–3.40 (m, 2H), 3.20–3.12 (m, 1H), 2.89 (dd, J = 3.2, 15.1 Hz, 1H). MS m/z 560.4 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[4-(1,3-dioxo-1,3-dihydro-isoindol-2-yloxy)-butyl]-amide (143)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s 1H), 7.78–7.65 (m, 6H), 7.60 (s, 1H), 7.40–7.30 (m, 2H), 6.80 (d, J = 8.7 Hz, 1H), 4.85 (d, J = 16.7 Hz, 1H), 4.75 (d, J = 16.7 Hz, 1H), 4.50–4.40 (m, 1H), 4.20 (t, 2H), 4.0 (s, 3H), 3.80 (s, 3H), 3.70–3.55 (m, 2H), 3.40–3.20 (m, 3H), 2.89 (dd, J = 3.2, 15.1 Hz, 1H), 1.96–1.70 (m, 4H). MS (EI) m/z 629.6 (M + H+).
General Procedure for the Synthesis of Compounds According to Scheme 2
Compound 6 (400 mg) was dissolved in dichloromethane (5 mL), and trifluoroacetic acid (1 mL) was added. The reaction mixture was stirred for 45 min. Solvent was removed in vacuo with care to be sure that all trifluoroacetic acid was removed. The residue was used without further purification.
The residue (0.28 mmol) was dissolved in dichloromethane (2.5 mL) together with diisopropylethyl amine (0.33 mmol). To this solution was added the appropriate chloroformate, isocyanate, or sulfonyl chloride (0.30 mmol) dropwise at 0 °C, and the mixture was stirred for 1 h. Aqueous NH4OH (1 mL) was added to the reaction mixture. After being stirred for an additional 15 min, the mixture was diluted with ethyl acetate (50 mL) and washed with brine. The organic layer was dried over MgSO4 and evaporated under reduced pressure to yield the crude product, which was purified by preparative HPLC. The appropriate fractions were combined and concentrated in vacuo to give the corresponding derivatives as the trifluoroacetate salt.
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-piperidin-4-ylmethyl-amide (54)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 8.71 (d, J = 7.2 Hz, 1H), 8.07 (td, J = 1.5, 7.5 Hz, 1H), 8.01 (dt, J = 0.9, 7.8 Hz, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.40 (s, 1H), 7.37 (d, J = 2.1 Hz, 1H), 7.33 (s, 1H), 6.79 (d, J = 8.7 Hz, 1H), 4.80 (d, J = 17.4 Hz, 1H), 4.65 (d, J = 17.4 Hz, 1H), 4.33–4.44 (m, 1H), 3.91 (s, 3H), 3.63 (t, J = 10.8 Hz, 1H), 3.40–3.54 (m, 4H), 3.14–3.23 (m, 2H), 2.82–3.02 (m, 3H), 1.97–2.09 (m, 3 H), 1.30–1.52 (m, 2H). MS m/z 506.5 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (162)
1H NMR (300 MHz, methanol-d4) δ 8.94 (d, J = 0.9 Hz, 1H), 8.70 (d, J = 7.5 Hz, 1H), 8.08 (td, J = 1.8, 7.8 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.43 (d, J = 1.2 Hz, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.33 (s, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.84 (d, J = 17.7 Hz, 1H), 4.68 (d, J = 17.7 Hz, 1H), 4.38–4.51 (m, 1H), 4.08–4.16 (m, 2H), 3.92 (s, 3H), 3.65 (s, 3H), 3.51–3.62 (m, 2H), 3.21–3.26 (m, 2H), 3.11–3.18 (m, 1H), 2.79–2.87 (m, 3H), 1.68–1.88 (m, 3H), 1.05–1.17 (m, 2H). MS m/z 564.4 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Isobutyl Ester (167)
1H NMR (300 MHz, methanol-d4) δ 8.94 (d, J = 0.9 Hz, 1H), 8.70 (d, J = 7.5 Hz, 1H), 8.08 (td, J = 1.8, 7.8 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.43 (d, J = 1.2 Hz, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.33 (s, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.84 (d, J = 17.7 Hz, 1H), 4.68 (d, J = 17.7 Hz, 1H), 4.38–4.51 (m, 1H), 4.08–4.16 (m, 2H), 3.92 (s, 3H), 3.85 (d, J = 6.6 Hz, 2H), 3.51–3.62 (m, 2H), 3.21–3.26 (m, 2H), 3.11–3.18 (m, 1H), 2.79–2.87 (m, 3H), 1.91–1.99 (m, 1H), 1.68–1.88, (m, 3H), 1.05–1.17 (m, 2H), 0.95 (d, J = 6.9 Hz, 6H). MS m/z 606.5 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[1-(2,2-dimethyl-propionyl)-piperidin-4-ylmethyl]-amide (150)
1H NMR (500 MHz, methanol-d4) δ 8.94 (s, 1H), 8.71 (d, J = 6 Hz, 1H), 8.09 (td, J = 1, 8 Hz, 1H), 8.01 (dt, J = 1, 8 Hz, 1H), 7.66 (ddd, J = 1.5, 5, 7.5 Hz, 1H), 7.43 (s, 1H), 7.38 (dd, J = 2, 8 Hz, 1H), 7.33 (s, 1H), 6.82 (d, J = 9 Hz, 1H), 4.82 (d, J = 18.5 Hz, 1H), 4.69 (d, J = 18.5 Hz, 1H), 4.38–4.45 (m, 3 H), 3.92 (s, 3H), 3.61–3.66 (m, 1H), 3.52–3.55 (m, 1H), 3.15–3.30 (m, 3H), 2.76–2.88 (m, 3H), 1.93–1.97 (m, 1H), 1.77–1.85 (m, 2H), 1.27 (s, 9H), 1.07–1.15 (m, 2H). MS m/z 590.6 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-[1-(3,3,3-trifluoro-propionyl)-piperidin-4-ylmethyl]-amide (155)
1H NMR (300 MHz, methanol-d4) δ 8.85 (s, 1H), 8.67 (d, J = 6 Hz, 1H), 8.09 (td, J = 1, 8 Hz, 1H), 8.01 (dt, J = 1, 8 Hz, 1H), 7.60 (m, 1H), 7.43 (s, 1H), 7.38 (d, J = 8 Hz, 1H), 7.23 (s, 1H), 6.82 (d, J = 9 Hz, 1H), 4.82 (d, J = 18.5 Hz, 1H), 4.69 (d, J = 18.5 Hz, 1H), 4.38–4.45 (m, 3 H), 3.92 (s, 3H), 3.61–3.66 (m, 1H), 3.50–3.52 (m, 1H), 3.12–3.28 (m, 5H), 2.71–2.81 (m, 3H), 1.93–1.97 (m, 1H), 1.72–1.80 (m, 2H), 1.03–1.12 (m, 2H). MS m/z 616.4 (M + H+).
Pyridine-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(1-methanesulfonyl-piperidin-4-ylmethyl)-amide (177)
1H NMR (300 MHz, methanol-d4) δ 8.93 (d, J = 1.2 Hz, 1H), 8.71 (d, J = 7.5 Hz, 1H), 8.08 (dt, J = 1.8, 7.8 Hz, 1H), 8.02 (td, J = 1.2, 8.1 Hz, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.42 (d, J = 1.5 Hz, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.33 (s, 1H), 6.80 (d, J = 8.7 Hz, 1H), 4.8 (d, J = 17.7 Hz, 1H), 4.68 (d, J = 17.7 Hz, 1H), 4.37–4.48 (m, 1H), 3.91 (s, 3H), 3.69–3.72 (m, 2H), 3.58–3.65 (m, 1H), 3.49–3.55 (m, 1H), 3.22–3.28 (m, 2H), 3.13–3.20 (m, 1H), 2.85–2.89 (m, 1H), 2.82 (s, 3H), 2.64–2.74 (m, 2H), 1.75–1.93 (m, 3H), 1.17–1.35 (m, 2H). MS m/z 584.4 (M + H+).
Pyridine-2-sulfonic Acid (1-Benzenesulfonyl-piperidin-4-yl-methyl)-[6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (180)
1H NMR (300 MHz, methanol-d4) δ 8.93 (d, J = 1.2 Hz, 1H), 8.71 (d, J = 7.5 Hz, 1H), 8.08 (dt, J = 1.8, 7.8 Hz, 1H), 8.02 (td, J = 1.2, 8.1 Hz, 1H), 7.56–7.78 (m, 6H), 7.32 (s, 1H), 7.29 (d, J = 8.7 Hz, 1H), 7.21 (s, 1H), 6.80 (d, J = 8.7 Hz, 1H), 4.75 (d, J = 17.7 Hz, 1H), 4.60 (d, J = 17.7 Hz, 1H), 4.37–4.48 (m, 1H), 3.91 (s, 3H), 3.69–3.72 (m, 2H), 3.58–3.65 (m, 1H), 3.49–3.55 (m, 1H), 3.22–3.28 (m, 2H), 3.13–3.20 (m, 1H), 2.85–2.89 (m, 1H), 2.64–2.74 (m, 2H), 1.75–1.93 (m, 3H), 1.17–1.35 (m, 2H). MS m/z 646.5 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Ethylamide (170)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 8.70 (d, J = 7.2 Hz, 1H), 8.04–8.10 (m, 1H), 8.99–8.02 (m, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.42 (s, 1H), 7.39 (dd, J = 2.1, 8.4 Hz, 1H), 7.33 (s, 1H), 6.82 (d, J) 8.7 Hz, 1H), 4.8 (d, J = 17.7 Hz, 1H), 4.69 (d, J = 17.7 Hz, 1H), 4.38–4.47 (m, 1H), 3.98–4.02 (m, 2H), 3.91 (s, 3H), 3.62 (t, J = 10.8 Hz, 1H), 3.54–3.56 (m, 1H), 3.12–3.29 (m, 5H), 2.81–2.90 (m, 1H), 2.66–2.74 (m, 2H), 1.65–1.87 (m, 3H), 1.17–1.18 (m, 2H), 1.10 (t, J = 7.2 Hz, 3H). MS m/z 577.5 (M + H+).
[2-(4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-piperidin-1-yl)-2-oxo-ethyl]-carbamic Acid tert-Butyl Ester (157)
A mixture of 54 (22 mg, 0.044 mmol), Boc-Gly-OH (9.2 mg, 0.051 mmol), dicyclohexylcarbodiimide (10 mg, 0.051 mmol), 4-dimethylamino-pyridine (2 mg), and dichloromethane (5 mL) was stirred at room temperature for 18 h. Upon completion of reaction, the mixture was filtered to remove dicyclohexylurea, and the residue was washed with dichloromethane (5 mL). The filtrate and washings were combined and evaporated to dryness in Vacuo. The residue was purified by HPLC to afford 15 mg (54%) of 157 as the trifluoroacetate salt. 1H NMR (500 MHz, methanol-d4) δ 8.94 (s, 1H), 8.71 (d, J = 7.5 Hz, 1H), 8.06–8.12 (m, 1H), 7.88–8.01 (m, 1H), 7.65–7.68 (m, 1H), 7.42 (s, 1H), 7.38 (dd, J = 1.5, 8.5 Hz, 1H), 7.33 (s, 1H), 6.81 (d, J = 8 Hz, 1H), 4.81 (d, J = 18 Hz, 1H), 4.69 (d, J = 18 Hz, 1H), 4.35–4.50 (m, 3H), 3.92 (s, 3H), 3.80–3.92 (m, 2H), 3.59–3.63 (m, 1H), 3.52–3.54 (m, 1H), 3.13–3.33 (m, 3H), 3.00–3.12 (m, 1H), 2.82–2.88 (m, 1H), 2.57–2.65 (m, 1H), 1.72–1.95 (m, 3H), 1.46 (s, 9 H), 1.06–1.25 (m, 2H). MS m/z 663.6 (M + H+).
General Procedure for the Synthesis of Compounds According to Scheme 3
To a solution of 6-cyano-1,2,3,4-tetrahydro-quinolin-3-ylamine hydrochloride 3 (4.18 g, 20 mmol) and N,N-diisopropylethyl amine (5.1 g, 40 mmol) in 25 mL of anhydrous dichloromethane was added benzyl chloroformate (5.1 g, 30 mmol) at 0 °C. After stirring at ambient temperature for 5 h, the reaction mixture was quenched with water. The mixture was partitioned between water and ethyl acetate. The organic layer was washed with brine, dried over sodium sulfate, and evaporated under reduce pressure. Purification by flash chromatography gave 9 (5.49 g 89%) as a white foam.
To a solution of 9 (5 g, 16.2 mmol) and trifluoroacetic acid (14 mL) in 20 mL of dichloroethane (14 mL) at room temperature under nitrogen was added 3-methyl-3H-imidazole-4-carboxaldehyde (5.37 g, 48.6 mmol). The mixture was stirred for 1 h at room temperature, and then triethylsilane (7.75 mL, 48.6 mmol) was added dropwise. The mixture was heated in an oil bath at 45 °C for 15 h. The volatile materials were removed under vacuum. The reaction mixture was diluted with ethyl acetate and washed with aqueous NaHCO3, water, and brine solution. The organic layer was dried over MgSO4, filtered, and concentrated. The crude residue was purified on a flash silica column to afford 10 (3.1 g, 47%).
A solution of [6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-carbamic acid benzyl ester 10 (1.3 g, 3 mmol), 15 mL methanol, and 10% Pd/C catalyst (0.5 g) was stirred under atmospheric pressure of hydrogen for 4 h. The catalyst was filtered off, and the filtrate was concentrated to give 11 as an off-white foam (790 mg, 92%).
To a suspension of 11 (0.55 g, 0.2 mmol) and Cs2CO3 (1.34 g, 0.4 mmol) in dry DMF (3 mL) was added the appropriate alkyl halide (0.2 mmol), and the mixture was stirred at room temperature overnight under argon. After addition of water (10 mL), the solution was extracted with ethyl acetate (3 × 10 mL). The organic layer was extracted with brine (3 × 10 mL). The combined organic layers were dried over MgSO4 and evaporated under reduce pressure. The residue was purified by HPLC. Appropriate fractions were collected, and the pure product 12 was obtained as the TFA salt.
Compound 12 (0.1 mmol) dissolved in dichloromethane (1.5 mL) together with N,N-diisopropylethyl amine (0.2 mmol) was cooled to 0 °C. Acetyl chloride or sulfonyl chloride (0.1.2 mmol) dissolved in dichloromethane (0.5 mL) was added dropwise, and the mixture was stirred for 1 h. Aqueous NH4OH (1 mL) was added to the reaction mixture. After being stirred for an additional 15 min, the reaction mixture was diluted with ethyl acetate (50 mL) and washed with brine solution. The organic layer was dried over MgSO4 and evaporated under reduced pressure, yielding the crude amide 13, which was purified by HPLC. The appropriate fractions were combined, concentrated in vacuo, and lyophilized to give the corresponding compounds as the trifluoroacetate salt.
(6-Cyano-1,2,3,4-tetrahydro-quinolin-3-yl)-carbamic Acid Benzyl Ester (9)
1H NMR (300 MHz, methanol-d4) δ 7.02–7.15 (m, 7H), 6.58 (d, J = 8 Hz, 1H), 5.09 (s, 2H), 3.94–3.98 (m, 1H), 3.45 (m, 1H), 3.20 (m, 1H), 2.99 (dd J = 4.2, 16.5 Hz, 1H), 2.74 (ddd, J = 2.1, 4.8, 16.5 Hz, 1H). MS m/z 308.2 (M + H+).
[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-carbamic Acid Benzyl Ester (10)
1H NMR (300 MHz, CD3OD) δ 7.61 (s, 1H), 7.31–7.38 (m, 7H) 6.87 (d, J = 8.7 Hz, 1H), 6.83 (s, 1H), 5.07 (s, 2H), 4.59 (s, 2H), 4.00–4.02 (m, 1H), 3.64 (s, 3H), 3.36 (dd, J = 2.7, 12 Hz, 1H), 3.23 (dd, J = 4.5, 12.1 Hz, 1H), 2.03 (dd J = 5.1, 15.9 Hz, 1H), 2.71 (dd, J = 4.8, 16.2 Hz, 1H). MS m/z 402.2 (M + H+).
3-Amino-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinoline-6-carbonitrile (11)
1H NMR (300 MHz, methanol-d4) δ 7.61 (s, 1H), 7.32 (dd, J = 2.1, 8.4 Hz, 1H), 7.29 (s, 1H), 6.83 (d, J = 8.4 Hz, 1H), 6.80 (s, 1H), 4.64 (d, J = 16.5 Hz, 1H), 4.58 (d, J = 16.5 Hz, 1H), 3.57 (s, 3H), 3.42 (dd, J = 2.4, 11.7 Hz, 1H), 3.35 (m, 1H), 3.21 (m, 1H), 2.98 (dd, J = 4.1, 16.5 Hz, 1H), 2.67 (dd, J = 4.8, 16.4 Hz, 1H). MS m/z 268.3 (M + H+).
3-(2-Fluoro-benzylamino)-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinoline-6-carbonitrile Trifluoroacetate Salt (12)
1H NMR (300 MHz, methanol-d4) δ 8.83 (s, 1H), 7.20–7.48 (m, 5H), 7.12–7.17 (m, 2H), 6.82 (d, J = 8.5 Hz, 1H), 4.74 (d, J = 17.4 Hz, 1H), 4.60 (d, J = 17.4 Hz, 1H), 4.35 (s, 2H), 3.88 (m, 1H), 3.75 (s, 3H), 3.74 (dd, J = 2.1, 14.1 Hz, 1H), 3.49 (dd, J = 4.8, 14.5 Hz, 1H), 3.31 (dd, J = 4.5, 16.5 Hz, 1H), 2.67 (dd, J = 6.6, 16.5 Hz, 1H). MS m/z 268.3 (M + H+).
4-{[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-ylamino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (12)
1H NMR (300 MHz, methanol-d4) δ 8.87 (s, 1H), 7.43 (s, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 7.33 (s, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.84 (d, J = 17.7 Hz, 1H), 4.68 (d, J = 17.7 Hz, 1H), 3.89–4.08 (m, 3H), 3.82 (s, 3H), 3.65 (s, 3H), 3.51–3.62 (m, 2H), 3.21–3.26 (m, 2H), 3.11–3.18 (m, 1H), 2.79–2.87 (m, 3H), 1.68–1.88, (m, 3H), 1.05–1.17 (m, 2H). MS m/z 423.4 (M + H+).
N-[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-N-(2-fluoro-benzyl)-4-methoxy-benzene-sulfonamide (215)
1H NMR (300 MHz, methanol-d4) δ 8.91 (s, 1H), 7.83 (d, J = 9.1 Hz, 2H), 7.49–7.52 (m, 1H), 7.28–7.33 (m, 3H), 7.16–7.19 (m, 2H), 7.1 (d, J = 9.0 Hz, 2H), 6.96–7.03 (m, 1H), 6.71 (d, J = 9.0 Hz, 1H), 4.30–4.61 (m, 5H), 3.91 (s, 3H), 3.81 (s, 3H), 3.30–3.39 (m, 2H), 2.94–3.04 (m, 1H), 2.75 (dd, J = 4.8, 15.6 Hz, 1H). MS m/z 546.5 (M + H+).
1-Methyl-1H-pyrazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(2-fluoro-benzyl)-amide (213)
1H NMR (300 MHz, methanol-d4) δ 8.91 (s, 1H), 8.19 (s, 1H), 7.82 (s, 1H), 7.56–7.61 (m, 1H), 7.27–7.36 (m, 4H), 7.14–7.22 (m, 1H), 7.01–7.09 (m, 1H), 6.72 (d, J = 8.4 Hz, 1H), 4.32–4.59 (m, 5H), 3.96 (s, 3H), 3.82 (s, 3H), 3.30–3.39 (m, 2H), 3.03–3.12 (m, 1H), 2.86 (dd, J = 4.8, 15.6 Hz, 1H). MS m/z 520.4 (M + H+).
3-Methyl-3H-imidazole-4-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-methanesulfonyl-benzyl)-amide (221)
1H NMR (300 MHz, methanol-d4) δ 8.89 (s, 1H), 7.93 (d, J = 8.2 Hz, 2H), 7.83 (s, 1H), 7.76 (s, 1H), 7.62 (d, J = 8.4 Hz, 2H), 7.31–7.35 (m, 2H), 7.26 (s, 1H), 6.68 (d, J = 8.7 Hz, 1H), 4.39–4.68 (m, 5H), 3.81 (s, 6H), 3.40–3.46 (m, 2H), 3.15 (s, 3H), 3.03–3.11 (m, 1H), 2.86 (m, 1H). MS m/z 580.5 (M + H+).
N-[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-N-(4-methanesulfonyl-benzyl)-benzene-sulfonamide (218)
1H NMR (300 MHz, methanol-d4) δ 8.91 (s 1H), 7.92–7.96 (m, 4H), 7.67–7.72 (m, 5H), 7.31 (dd, J = 2.1, 8.4 Hz, 1H), 7.27 (s, 1H), 7.16 (s, 1H), 6.63 (d, J = 8.4 Hz, 1H), 4.35–4.70 (m, 5H), 3.82 (s, 3H), 3.41–3.46 (m, 2H), 3.17 (s, 3H), 2.95–3.05 (m, 1H), 2.79–2.82 (m, 1H). MS m/z 576.6 (M + H+).
4-({[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-ethanesulfonyl-amino}-methyl)-piperidine-1-carboxylic Acid Methyl Ester (200)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 7.42 (s, 1H), 7.42 (s, 1H), 7.33 (dd, J = 1.8, 8.4 Hz, 1H), 6.82 (d, J = 8.7 Hz, 1H), 4.79 (d, J = 18 Hz, 1H), 4.70 (d, J = 18 Hz, 1H), 4.05–4.18 (m, 3H), 3.92 (s, 3H), 3.65 (s, 3H), 3.51–3.56 (m, 2H), 3.12–3.24 (m, 6H), 2.71–2.78 (m, 2H), 1.72–1.84 (m, 3H), 1.34 (t, J = 7.5 Hz, 3H), 1.05–1.14 (m, 2H). MS m/z 515.4 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(3-methyl-3H-imidazole-4-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (191)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s 1H), 7.76 (s, 1H), 7.74 (s, 1H), 7.45 (s, 1H), 7.33 (dd, J = 1.8, 8.4 Hz, 1H), 7.32 (s, 1H), 6.82 (d, J = 8.7 Hz, 1H), 4.78 (d, J = 17.4 Hz, 1H), 4.70 (d, J = 17.4 Hz, 1H), 4.32–4.41 (m, 1H), 3.99–4.1 (m, 2H), 3.92 (s, 3H), 3.80 (s, 3H), 3.65 (s, 3H), 3.51–3.56 (2H), 2.96–3.14 (m, 4H), 2.71–2.78 (m, 2H), 1.71–1.88 (m, 3 H), 1.00–1.13 (m, 2H). MS m/z 567.5 (M + H+).
4-({(Benzo[1,2,5]thiadiazole-4-sulfonyl)-[6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amino}-methyl)-piperidine-1-carboxylic Acid Methyl Ester (188)
1H NMR (300 MHz, methanol-d4) δ 8.94 (s 1H), 8.32–8.35 (m, 1H), 8.32 (s, 1H), 7.08–7.85 (m, 1H), 7.42 (s, 1H), 7.35 (dd, J = 1.8, 8.7 Hz, 1H), 7.24 (s, 1H), 6.82 (d, J = 8.7 Hz, 1H), 4.78 (d, J = 18 Hz, 1H), 4.62 (d, J = 18 Hz, 1H), 4.04–4.08 (m, 3H), 3.91 (s, 3H), 3.66 (s, 3H), 3.49–3.51 (m, 3H), 3.03–3.10 (m, 2H), 2.61–2.73 (m, 3H), 1.71–1.88 (m, 3 H), 1.02–1.14 (m, 2H). MS m/z 567.5 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(3,4-dihydro-2H-benzo[b][1,4]dioxepine-7-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (204)
1H NMR (300 MHz, methanol-d4) δ 8.94 (s, 1H), 7.45 (s, 1H), 7.39 (dd, J = 2.1, 8.7 Hz, 1H), 7.36 (dd, J = 2.1, 8.7 Hz, 1H), 7.35 (d, J = 2.1 Hz, 1H), 7.29 (s, 1H), 7.25 (d, J = 8.7 Hz, 1H), 7.10 (d, J = 8.7 Hz, 1H), 6.88 (d, J = 8.7 Hz, 1H), 4.64 (d, J = 16.7 Hz, 1H), 4.51 (d, J = 16.7 Hz, 1H), 4.24–4.33 (m, 4H), 4.04–4.14 (m, 3H), 3.85 (s, 3H), 3.64 (s, 3H), 3.16–3.19 (m, 1H), 2.99–3.03 (m, 3H), 2.63–2.80 (m, 4H), 2.21–2.29 (m, 2H), 1.68–1.78 (m, 3H), 1.00–1.06 (m, 2H). MS m/z 635.7 (M + H+).
4-({(Benzo[1,2,5]oxadiazole-4-sulfonyl)-[6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amino}-methyl)-piperidine-1-carboxylic Acid Methyl Ester (205)
1H NMR (300 MHz, methanol-d4) δ 8.94 (s, 1H), 8.27–8.29 (m, 1H), 8.19–8.22 (m, 1H), 7.68 (s, 1H), 7.40–7.49 (m, 1H), 7.35 (dd, J = 1.8, 8.7 Hz, 1H), 7.29 (s, 1H), 6.82 (d, J = 8.7 Hz, 1H), 4.78 (d, J = 16.7 Hz, 1H), 4.62 (d, J = 16.7 Hz, 1H), 4.04–4.08 (m, 3H), 3.91 (s, 3H), 3.66 (s, 3H), 3.49–3.51 (m, 3H), 3.03–3.10 (m, 2H), 2.61–2.73 (m, 3H), 1.71–1.88 (m, 3H), 1.02–1.14 (m, 2H). MS m/z 605.4 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(2-oxo-2H-chromene-6-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (206)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 8.22 (d, J = 2.1 Hz, 1H), 8.07 (dd, J = 2.1, 8.7 Hz, 1H), 8.04 (d, J = 9.9 Hz, 1H), 7.53 (d, J = 8.7 Hz, 1H), 7.41 (d, J = 2.1 Hz, 1H), 7.37 (dd, J = 2.1, 8.7 Hz, 1H), 7.26 (s, 1H), 6.78 (d, J = 8.7 Hz, 1H), 6.58 (d, J = 9.9 Hz, 1H), 4.75 (d, J = 16.7 Hz, 1H), 4.68 (d, J = 16.7 Hz, 1H), 4.04–4.08 (m, 3H), 3.90 (s, 3H), 3.66 (s, 3H), 3.49–3.51 (m, 1H), 3.03–3.15 (m, 3H), 2.61–2.80 (m, 3H), 1.60–1.71 (m, 3H), 1.02–1.16 (m, 2H). MS m/z 631.4 (M + H+).
4-({(4-Carboxy-furan-3-sulfonyl)-[6-cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amino}-methyl)-piperidine-1-carboxylic Acid Methyl Ester (197)
1H NMR (300 MHz, methanol-d4) δ 8.93 (s, 1H), 7.43(dd, J = 2.1 Hz, 1H), 7.30 (d, J = 8.7 Hz, 1H), 7.38 (d, J = 6.0 Hz, 1H), 7.36 (s, 1H), 7.25 (d, J = 6.0 Hz, 1H), 6.83 (d, J = 8.7 Hz, 1H), 6.88 (d, J = 8.7 Hz, 1H), 4.78 (d, J = 16.7 Hz, 1H), 4.62 (d, J = 16.7 Hz, 1H), 4.08–4.04 (m, 3H), 3.91 (s, 3H), 3.66 (s, 3H), 3.49–3.51 (m, 3H), 3.03–3.10 (m, 2H), 2.61–2.73 (m, 3H), 1.71–1.88 (m, 3H), 1.02–1.14 (m, 2H). MS m/z 597.4 (M + H+).
4-({[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-methanesulfonylmethanesulfonyl-amino}-methyl)-piperidine-1-carboxylic Acid Methyl Ester (207)
1H NMR (300 MHz, methanol-d4) δ 8.96 (s, 1H), 7.49 (dd, J = 2.1, 8.7 Hz, 1H), 7.47 (d, J = 2.1 Hz, 1H), 6.88 (d, J = 8.7 Hz, 1H), 5.15 (s, 2H), 4.68 (d, J = 16.7 Hz, 1H), 4.62 (d, J = 16.7 Hz, 1H), 4.40–4.25 (m, 1H), 4.04–4.14 (m, 2H), 3.92 (s, 3H), 3.73 (s, 3H), 3.55–3.61 (m, 3H), 3.25 (s, 3H), 3.16–3.19 (m, 1H), 2.99–3.03 (m, 2H), 2.63–2.80 (m, 2H), 2.21–2.29 (m, 1H), 1.68–1.78 (m, 2H), 1.05–1.16 (m, 2H). MS m/z 579.2 (M + H+).
4-({[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-cyclopropanesulfonyl-amino}-methyl)-piperidine-1-carboxylic Acid Methyl Ester (202)
1H NMR (300 MHz, methanol-d4) δ 8.94 (s, 1H), 7.52 (dd, J = 1.8, 8.7 Hz, 1H), 7.48 (d, J = 2.0 Hz), 7.42 (s, 1H) 6.85 (d, J = 8.7 Hz, 1H), 4.78 (d, J = 16.7 Hz, 1H), 4.62 (d, J = 16.7 Hz, 1H), 4.08–4.30 (m, 3H), 3.95 (s, 3H), 3.75 (s, 3H), 3.51–3.72 (m, 4H), 3.10–3.40 (m, 4H), 2.80–3.02 (m, 2H), 2.68–2.73 (m, 1H), 1.75–2.01 (m, 3H), 1.10–1.30 (brm, 6H). MS m/z 527.4 (M + H+).
3,4-Dihydro-2H-benzo[b][1,4]dioxepine-7-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-methanesulfonyl-benzyl)-amide (223)
1H NMR (300 MHz, methanol-d4) δ 8.94 (s, 1H), 7.87 (d, J = 8.1 Hz, 2H), 7.62 (d, J = 8.1 Hz, 2H), 7.45 (s, 1H), 7.39 (dd, J = 2.1, 8.7 Hz, 1H), 7.36 (dd, J = 2.1, 8.7 Hz, 1H), 7.35 (d, J = 2.1 Hz, 1H), 7.29 (s, 1H), 7.25 (d, J = 8.7 Hz, 1H), 7.10 (d, J = 8.7 Hz, 1H), 6.68 (d, J = 8.7 Hz, 1H), 4.64 (d, J = 16.7 Hz, 1H), 4.51 (d, J = 16.7 Hz, 1H), 4.24–4.33 (m, 4H), 4.04–4.14 (m, 2H), 3.85 (s, 3H), 3.20 (s, 3H), 2.95–3.05 (m, 1H), 1.12–1.25 (m, 2H). MS m/z 648.5 (M + H+).
3-Methyl-thiophene-2-sulfonic Acid [6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(4-methanesulfonyl-benzyl)-amide (224)
1H NMR (300 MHz, methanol-d4) δ 8.87 (s, 1H), 7.98 (d, J = 8.1 Hz, 2H), 7.72 (d, J = 5.1 Hz, 1H), 7.61 (dd, J = 1.8, 8.7 Hz, 1H), 7.35 (d, J = 8.7 Hz, 1H), 7.25 (m, 2H), 7.07 (d, J = 5.1 Hz, 1H), 6.70 (d, J = 8.7 Hz, 1H) 4.75 (d, 2H), 4.53–4.55 (m, 1H), 4.51 (d, J = 17.7 Hz, 1H), 4.48 (d, J = 17.7 Hz, 1H), 3.79 (s, 3H), 3.10–3.25 (m, 7H), 2.95–3.05 (m, 1H), 2.49 (s, 3H). MS m/z 596.6 (M + H+).
N-[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-2-dimethylamino-N-(2-fluoro-benzyl)-acetamide (248)
1H NMR (300 MHz, methanol-d4) δ 8.78 (s, 1H), 7.01–7.34 (m,7 H), 6.68 (d, J = 8.7 Hz, 1H), 4.42–4.61 (m, 3H), 4.25 (d, J = 15.9 Hz, 1H), 4.18 (d, J = 15.9 Hz, 1H), 3.74 (s, 3H), 3.41–3.59 (m, 2H), 3.19 (s, 2H), 2.94–3.04 (m, 1H), 2.86 (m, 7 H). MS m/z 461.3 (M + H+).
N-[6-Cyano-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-N-(2-fluoro-benzyl)-3-methylsulfanyl-propionamide (247)
1H NMR (300 MHz, methanol-d4) δ 8.84 (s, 1H), 7.01–7.34 (m, 7H), 6.78 (d, J = 7.6 Hz, 1H), 4.42–4.77 (m, 5H), 3.84 (s, 3H), 3.41–3.59 (m, 2H), 2.94–3.12 (m, 6H), 2.06 (s, 3H). MS m/z 478.4 (M + H+).
General Procedure for the Synthesis of Compounds According to Scheme 4. 1-Methyl-1H-imidazole-4-sulfonic Acid [6-Cyano-1-(1-trityl-1H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (15)
A solution of 14 (0.397 g, 1 mmol), N,N-diisopropylethyl amine (2.37 mL, 2 mmol), and 3 mL of dry DMF was stirred at room temperature for 30 min. To this solution was added dropwise a solution of triphenylmethyl chloride (0.278 g, 1 mmol) dissolved in dry DMF (3 mL). The reaction mixture was stirred at room temperature for 20 h. After addition of water (10 mL), the solution was extracted with dichloromethane (3 × 15 mL). The combined organic layer was washed with brine (20 mL) and the organic layer was dried over MgSO4 and evaporated under reduce pressure, yielding the crude product, which was purified on a silica gel column eluting with 10% methanol in ethyl acetate to afford 15 (0.562 g, 88%). 1H NMR (300 MHz, CDCl3) δ 8.0 (s, 1H), 7.9 (s, 1H), 7.4 (s, 1H), 7.25–7.37 (m, 18 H), 6.48 (d, J = 8.7 Hz, 1H), 4.5 (d, J = 16.2 Hz, 1H), 4.23 (d, J = 16.2 Hz, 1H), 4.00–4.09 (m, 1H), 3.70 (s, 3H), 3.51–3.61 (m, 1H), 3.31–3.34 (m, 1H), 3.09–3.12 (m, 1H), 2.86–3.07 (m, 1H). MS m/z 640.3 (M + H+).
A solution of 16 (0.21 mmol), dichloromethane (3 mL), and trifluoroacetic acid (0.5 mL) was stirred for 30 min. The reaction mixture was filtered to remove trityl alcohol, and the residue was washed with dichloromethane. The filtrate and washings were combined and then evaporated to dryness via rotary evaporation. The residue was purified by HPLC.
1-Methyl-1H-imidazole-4-sulfonic Acid Benzyl-[6-cyano-1-(3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (253)
1H NMR (300 MHz, methanol-d4) δ 8.86 (s, 1H), 7.83 (s, 1H), 7.71 (s, 1H), 7.18–7.39 (m, 8 H), 6.71 (d, J = 9.7 Hz, 1H), 4.29–4.73 (m, 5H), 3.80 (s, 3H), 3.36–3.42 (m, 1H), 3.18–3.26 (m, 1H), 3.12 (dd, J = 8.7, 15.6 Hz, 1H), 2.89 (dd, J = 3.3, 15.3 Hz, 1H). MS m/z 488.1 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid (2-Bromo-allyl)-[6-cyano-1-(3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (252)
1H NMR (300 MHz, methanol-d4) δ 8.86 (s, 1H), 7.83 (s, 1H), 7.79 (s, 1H), 7.28 (s, 1H), 7.33 (dd, J = 2.1, 9.0 Hz, 1H), 7.28 (s, 1H), 6.65 (d, J = 8.7 Hz, 1H), 6.04 (s, 1H), 5.65 (s, 1H), 4.79 (d, J = 17.1 Hz, 1H), 4.62 (d, J = 17.1 Hz, 1H), 4.39–4.59 (m, 1H), 4.19 (d, J = 17.1 Hz, 1H) 4.0 (d, J = 17.1 Hz, 1H), 3.81 (s, 3H), 3.55–3.62 (m, 1H), 3.42–3.47 (m, 1H), 3.07–3.14 (m, 1H), 2.97(dd, J = 4.5, 15.9 Hz, 1H). MS m/z 518.2 (M + H+).
Synthesis of 255 According to Scheme 5. 4-{[(6-cyano-1,2,3,4-tetrahydro-quinolin-3-yl)-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (18)
1H NMR (300 MHz, methanol-d4) δ 8.70 (d, J = 7.5 Hz, 1H), 8.08 (td, J = 1.8, 7.8 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.66 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.43 (d, J = 1.2 Hz, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.38–4.51 (m, 1H), 4.08–4.16 (m, 2H), 3.65 (s, 3H), 3.51–3.62 (m, 2H), 3.21–3.26 (m, 2H), 3.11–3.18 (m, 1H), 2.79–2.87 (m, 3H), 1.68–1.88, (m, 3H), 1.05–1.17 (m, 2H). MS m/z 470.2 (M + H+).
4-{[[6-Cyano-1-(3-methyl-3H-imidazole-4-sulfonyl)-1,2,3,4-tetrahydro-quinolin-3-yl]-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (255)
A solution of 18 (47 mg, 0.1 mmol), 3-methyl-3H-imidazole-4-sulfonyl chloride hydrochloride salt (26 mg, 0.12 mmol, Apollo Scientific Intermediates), N,N-dimethyl pyridine (15 mg, 0.12 mmol), and 3 mL of dry acetonitrile was refluxed overnight under nitrogen. Then the reaction mixture was quenched with water and extracted with dichloromethane (3 × 10 mL). The combine organic layer was washed with brine (10 mL), and the organic layer was dried over MgSO4 and evaporated under reduced pressure, yielding the crude product, which was purified by HPLC. 1H NMR (300 MHz, methanol-d4) δ 8.76 (d, J = 4.8 Hz, 1H), 8.08 (td, J = 1.8, 8.1 Hz, 1H), 8.0 (dt, J = 1.2, 7.8 Hz, 1H), 7.89 (d, J = 8.4 Hz, 1H), 7.81 (s, 1H), 7.66 (m, 2H), 7.48 (dd, J = 2.1, 8.7 Hz, 1H), 7.43 (s, 1H), 4.32–4.38 (m, 1H), 4.14–4.25 (m, 1H), 4.09–4.13 (m, 2H), 3.72 (s, 3H), 3.68 (s, 3H), 3.51–3.62 (m, 1H), 3.21–3.26 (m, 2H), 3.11–3.18 (m, 1H), 2.79–2.87 (m, 3H), 1.92–1.99 (m, 1H), 1.68–1.88 (m, 2H), 1.05–1.17 (m, 2H). MS m/z 614.4 (M + H+).
[[1-(3-Benzyl-3H-imidazol-4-ylmethyl)-6-cyano-1,2,3,4-tetrahydro-quinolin-3-yl]-(1-methyl-1H-imidazole-4-sulfonyl)-amino]-acetic Acid tert-Butyl Ester (254)
1H NMR (300 MHz, methanol-d4) δ 8.86 (s, 1H), 7.11–7.50 (m, 10 H), 6.55 (d, J) 8.5 Hz, 1H), 5.12 (s, 2H), 4.62–4.78 (m, 2H), 4.39–4.59 (m, 1H), 4.19 (d, J) 17.1 Hz, 1H) 4.0 (d, J) 17.1 Hz, 1H), 3.81 (s, 3H), 3.55–3.62 (m, 1H), 3.42–3.47 (m, 1H), 3.07–3.14 (m, 1H), 2.97 (m, 1H), 1.42 (s, 9H). MS m/z 602.2 (M + H+).
4-{[(6-Cyano-1-pyridin-3-ylmethyl-1,2,3,4-tetrahydro-quinolin-3-yl)-(pyridine-2-sulfonyl)-amino]-methyl}-piperidine-1-carboxylic Acid Methyl Ester (256)
1H NMR (300 MHz, methanol-d4) δ 8.84–8.89 (m, 2H), 8.70–8.75 (m, 1H), 8.49–8.51 (m, 1H), 8.03–8.19 (m, 3H), 7.68 (ddd, J = 1.5, 4.8, 7.5 Hz, 1H), 7.43 (d, J = 1.2 Hz, 1H), 7.38 (dd, J = 2.1, 8.7 Hz, 1H), 6.81 (d, J = 8.7 Hz, 1H), 4.68–4.83 (m, 2H), 4.38–4.51 (m, 1H), 4.08–4.16 (m, 2H), 3.65 (s, 3H), 3.51–3.62 (m, 2H), 3.21–3.26 (m, 2H), 3.11–3.18 (m, 1H), 2.79–2.87 (m, 3H), 1.68–1.88, (m, 3H), 1.05–1.17 (m, 2H). MS m/z 561.4 (M + H+).
General Procedure for the Synthesis of Compounds According to Scheme 6
A suspension of the aryl bromide 21 (500 mg, 1.53 mmol), phenyl boronic acid (559 mg, 4.58 mmol), and tetrakis triphenylphosphine palladium (177 mg, 10 mol %) in dimethoxyethane (15 mL) was thoroughly degassed and stirred under argon. Deionized water (3 mL) and barium hydroxide octahydrate (1.45 gm, 4.58 mmol) were added, and the reaction mixture was heated at reflux for 2 h. The reaction mixture was diluted with ethyl acetate, washed with water and brine, dried (Na2SO4), and concentrated under reduced pressure. The residue was purified by flash chromatography over a silica gel column to give 22 (120 mg, 45%) as a colorless solid. 1H NMR (CDCl3) δ 7.49 (d, J = 8.1 Hz, 1H), 7.36 (t, J = 7.8 Hz, 2H), 7.23 (t, J = 7.4 Hz, 4H), 6.51 (d, J = 8.1 Hz, 1H), 5.04 (d, J = 7.5 Hz, 1H), 4.11 (br s, 1H), 3.95 (s, 1H), 3.37–3.33 (m, 1H), 3.19–3.16 (m, 1H), 3.09–3.02 (m, 1H), 2.77–2.72 (m, 1H), 1.43 (s, 9H).
To a stirred solution of the phenyl derivative 22 (120 mg, 0.37 mmol) in CH2Cl2 (1.6 mL) at room temperature, trifluoroacetic acid (0.4 mL) was added, and the reaction mixture was stirred for 2 h. The solution was concentrated under reduced pressure, and the crude amine (trifluoroacetate salt) 23 was taken to the next step without further purification.
To a mixture of the amine 23 in CH2Cl2 (2 mL), diisopropylethylamine (0.5 mL) was added and the reaction mixture was stirred at room temperature for 0.5 h. The appropriate sulfonyl chloride (0.37 mmol) was added, and the reaction mixture was stirred for an additional 2 h. The reaction mixture was partitioned between water and ethyl acetate. The organic layer was washed with brine, dried (Na2SO4), and evaporated under reduced pressure. Purification by flash chromatography gave 24 (89%) as a pale yellow solid. 1H NMR (methanol-d4) δ 7.93 (s, 1H), 7.76 (s, 1H), 7.69 (d, J = 1.2 Hz, 1H), 7.49 (dd, J = 1.2, 8.4 Hz, 2H), 7.35 (t, J = 7.5 Hz, 2H), 7.22 (d, J = 7.5 Hz, 1H), 7.11 (s, 1H), 6.60 (d, J = 8.4 Hz, 1H), 3.77 (s, 3H), 3.74–3.69 (m, 1H), 3.39–3.36 (m, 1H), 3.09 (dd, J = 8.1, 11.4 Hz, 1H), 2.93 (dd, J = 4.5, 15.6 Hz, 1H), 2.76 (dd, J = 8.1, 15.6 Hz, 1H).
To a stirred solution of the sulfonamide 24 (0.33 mmol) in dichloroethane (1 mL) and trifluoroacetic acid (1 mL) was added 3-methyl-3H-imidazole-4-carbaldehyde (91 mg, 0.83 mmol) at room temperature, and the reaction mixture was heated to 50 °C for 1 h. Triethylsilane (0.13 mL, 0.83 mmol) was added and stirring was continued for an additional 48 h. Volatile materials were removed under vacuum, and the reaction mixture was diluted with ethyl acetate, washed with aqueous NaHCO3 solution, water, and brine, dried (Na2SO4), filtered, and concentrated in vacuum. Purification by flash chromatography afforded 25 (50 mg, 33%) as a pale yellow foam. 1H NMR (methanol-d4) δ 8.89 (s, 1H), 8.06 (s, 1H), 7.67 (s, 1H), 7.49 (t, J = 7.8 Hz, 3H), 7.37–7.33 (m, 3H), 7.24–7.20 (m, 2H), 6.85 (d, J = 7.8 Hz, 1H), 4.74 (d, J = 17.1 Hz, 1H), 4.57 (d, J = 17.1 Hz, 1H), 3.93–3.71 (m, 8H), 3.35–3.31 (m, 1H), 3.04 (dd, J = 4.5, 15.6 Hz, 1H), 2.76 (dd, J = 6.3, 15.9 Hz, 1H).
A mixture of 25 (1 equiv), Cs2CO3 (2 equiv), and the appropriate bromide (1 equiv) in anhydrous DMF was stirred overnight at room temperature. The reaction mixture was partitioned between water and ethyl acetate. The organic layer was washed with brine, dried (Na2SO4), and evaporated under reduced pressure. The crude residue was purified by HPLC.
1-Methyl-1H-imidazole-4-sulfonic Acid [1-(3-Methyl-3H-imi-dazol-4-ylmethyl)-6-phenyl-1,2,3,4-tetrahydro-quinolin-3-yl]-(2-pyrrol-1-yl-ethyl)-amide (272)
1H NMR (methanol-d4) δ 7.74 (d, J = 1.0 Hz, 1H), 7.70 (d, J = 1.5 Hz, 1H), 7.53 (br s, 1H), 7.52–7.50 (m, 2H), 7.39–7.33 (m, 3H), 7.26–7.22 (m, 1H), 7.17–7.16 (m, 1H), 6.90 (d, J = 8.5 Hz, 1H), 6.80 (br s, 1H), 6.63 (t, J = 2.0 Hz, 2H), 6.04 (t, J = 2.0 Hz, 2H), 4.44 (d, J = 15.5 Hz, 1H), 4.27 (d, J= 15.5 Hz, 1H), 4.22–4.17 (m, 4H), 3.75 (s, 3H), 3.63 (s, 3H), 3.48–3.44 (m, 2H), 2.91 (dd, J = 4.5, 11.5 Hz, 1H), 2.76–2.66 (m, 2H). MS m/z 556.3 (M + H+).
N-tert-Butyl-2-{(1-methyl-1H-imidazole-4-sulfonyl)-[1-(3-methyl-3H-imidazol-4-ylmethyl)-6-phenyl-1,2,3,4-tetrahydro-quinolin-3-yl]-amino}-acetamide (267)
1H NMR (methanol-d4) δ 8.87 (s, 1H), 7.78 (d, J = 1.2 Hz, 2H), 7.45 (d, J = 7.2 Hz, 2H), 7.41 (s, 1H), 7.32 (t, J= 7.5 Hz, 3H), 7.19 (t, J = 7.2 Hz, 2H), 6.82 (d, J = 7.8 Hz, 1H), 4.65 (d, J = 15.6 Hz, 1H), 4.52 (d, J = 15.6 Hz, 1H), 4.36–4.18 (m, 1H), 3.94–3.75 (m, 9H), 3.31–3.40 (m, 1H), 3.02–2.85 (m, 2H), 1.31 (m, 9H). MS m/z 576.4 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid (4-Methanesulfonyl-benzyl)-[1-(3-methyl-3H-imidazol-4-ylmethyl)-6-phenyl-1,2,3,4-tetrahydroquinolin-3-yl]-amide (278)
1H NMR (300 MHz, methanol-d4) δ 8.87 (s, 1H), 7.92 (d, J = 8.4 Hz, 2H), 7.85 (s, 1H), 7.80 (s, 1H), 7.62 (d, J = 8.4 Hz, 2H), 7.52 (d, J = 8.4 Hz, 2H), 7.40–7.30 (m, 4H), 7.28–7.22 (m, 2H), 6.72 (d, J = 8.4 Hz, 1H), 4.75 (d, J = 16.5 Hz, 2H), 4.55–4.50 (m, 1H), 4.48 (d, J = 17.4 Hz, 1H), 4.35 (d, J = 16.5 Hz, 1H), 3.85 (s, 3H), 3.80 (s, 3H), 3.42–3.36 (m, 2H), 3.12 (s, 3H), 3.08–2.95 (m, 2H). MS m/z 631.6 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid (2-Bromo-allyl)-[1-(3-methyl-3H-imidazol-4-ylmethyl)-6-phenyl-1,2,3,4-tetrahydro-quinolin-3-yl]-amide (264)
1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.80 (d, J = 3.9 Hz, 2H), 7.55 (dd, J = 1.2, 8.4 Hz, 2H), 7.45 (s, 1H), 7.40 (t, J = 8.7 Hz, 3H), 7.32–7.20 (m, 2H), 6.90 (d, J = 8.7 Hz, 1H), 6.10 (d, J = 2.1 Hz, 1H), 5.65 (d, J = 2.1 Hz, 1H), 4.75 (d,J = 16.8 Hz, 1H), 4.60 (d, J = 17.1 Hz, 1H), 4.50–4.40 (m, 1H), 4.20 (d, J = 18.0 Hz, 1H), 4.10 (d, J = 18.0 Hz, 1H), 3.95 (s, 3H), 3.80 (s, 3H), 3.49–3.40 (m, 2H), 3.15–3.05 (m, 1H), 2.87 (dd, = 5.1, 15.6 Hz, 1H). MS m/z 583.0 (M + H+).
1-Methyl-1H-imidazole-4-sulfonic Acid Ethyl-[1-(3-methyl-3H-imidazol-4-ylmethyl)-6-phenyl-1,2,3,4-tetrahydro-quinolin-3-yl]amide (259)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.75 (s, 1H), 7.72 (s, 1H), 7.52 (dd, J = 1.2, 8.7 Hz, 2H), 7.45 (s, 1H), 7.35 (t, J = 7.2 Hz, 3H), 7.30–7.20 (m, 2H), 6.87 (d, J = 8.7 Hz, 1H), 4.75 (d, J = 16.8 Hz, 1H), 4.57 (d, J = 16.8 Hz, 1H), 4.47–4.37 (m, 1H), 3.95 (s, 3H), 3.80 (s, 3H), 3.45–3.25 (m, 4H), 3.20–3.10 (m, 1H), 2.90 (m, 1H), 1.25 (t, J = 6.9 Hz, 3H). MS m/z 491.5 (M + H+).
{(1-Methyl-1H-imidazole-4-sulfonyl)-[1-(3-methyl-3H-imidazol-4-ylmethyl)-6-phenyl-1,2,3,4-tetrahydro-quinolin-3-yl]-amino}-acetic Acid Methyl Ester (266)
1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.80 (s, 1H), 7.75 (s, 1H), 7.52 (dd, J = 1.2, 8.4 Hz, 2H), 7.44 (s, 1H), 7.36 (t, J = 7.5 Hz, 3H), 7.30–7.25 (m, 2H), 6.85 (d, J = 8.7 Hz, 1H), 4.75 (d, J = 16.8 Hz, 1H), 4.55 (d, J = 16.8 Hz, 1H), 4.50–4.40 (m, 1H), 4.15 (d, J = 18.3 Hz, 1H), 4.08 (d, J = 18.6 Hz, 1H), 3.95 (s, 3H), 3.80 (s, 3H), 3.70 (s, 3H), 3.55–3.45 (m, 2H), 3.05–2.90 (m, 2H). MS m/z 535.4 (M + H+).
General Procedure for the Synthesis of Compounds According to Scheme 7. General Procedure for the Conversion of Nitrile (5) to Amide (28)
To a stirred solution of nitrile 5 (441 mg, 1 mmol) in alcohol (8 mL) was slowly added concentrated sulfuric acid (0.05 mL) at room temperature. The resulting solution was stirred at 40 °C for 2 h. The mixture was poured into cold aqueous 20% KHCO3 solution to neutralize the acid and precipitate the product. The product was filtered, washed with cold water, and dried under vacuum to give amides 28 (40–45%).
3-(1-Methyl-1H-imidazole-4-sulfonylamino)-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydroquinoline-6-carboxylic Acid tert-Butylamide (28)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.80 (s, 1H), 7.70 (s, 1H), 7.51 (dd, J = 2.1, 8.7 Hz, 1H), 7.45 (s, 1H), 7.40 (d, J = 2.1 Hz, 1H), 6.80 (d, J = 8.7 Hz, 1H), 4.80 (d, J = 16.8 Hz, 1H), 4.60 (d, J = 16.8 Hz, 1H), 3.92 (s, 3H), 3.86–3.83 (m, 1H), 3.80 (s, 1H), 3.52–3.45 (m, 1H), 3.42–3.35 (m, 1H), 3.0 (dd, J = 4.8, 16.5 Hz, 1H), 2.74 (dd, J = 6.3, 16.2 Hz, 1H), 1.45 (s, 9H) MS m/z 486.4 (M + H+).
3-[(tert-Butylcarbamoyl-methyl)-(1-methyl-1H-imidazol-4-sulfonyl)-amino]-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-6-carboxylic Acid tert-Butylamide (285)
The compound was prepared using the general N-alkylation procedure (5 to 2, Scheme 1). 1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.85 (s, 1H),7.80 (s, 1H), 7.52 (dd, J = 2.1, 8.7 Hz, 1H), 7.43 (s, 1H), 7.40 (d, J = 2.1 Hz, 1H), 6.79 (d, J = 8.7 Hz, 1H), 4.75 (d, J = 16.8 Hz, 1H), 4.63 (d, J = 16.8 Hz, 1H), 4.33–4.25 (m, 1H), 3.92 (s, 3H), 3.87 (s, 2H), 3.81 (s, 3H), 3.57–3.51 (m, 1H), 3.44–3.40 (m, 1H), 3.03 (dd, J = 4.5, 15.9 Hz, 1H), 2.80 (dd, J = 4.5, 15.9 Hz, 1H), 1.43 (s, 9H), 1.35 (s, 9H). MS m/z 599.5 (M + H+).
General Procedure for the Conversion of Nitrile (5) to Acid (27)
A solution of 5 (441 mg, 1.0 m mol) in concentrated HCl (20 mL) was stirred at 80 °C for 2 h. The reaction mixture was cooled to room temperature and then evaporated to dryness under reduced pressure. The residue was dissolved in saturated LiOH solution (pH 9) and then evaporated to dryness under reduced pressure. The residue was dissolved in 10% aqueous HCl solution (pH 2), evaporated, and dried under vacuum to give the corresponding acid 27, which was used in the next step without further purification. 1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.80 (s, 1H), 7.70 (s, 1H), 7.62 (dd, J = 2.1, 8.7 Hz, 1H), 7.52 (d, J = 2.1 Hz, 1H), 7.42 (s, 1H), 6.80 (d, J = 8.7 Hz, 1H), 4.80 (d, J = 16.8 Hz, 1H), 4.62 (d, J = 16.8 Hz, 1H), 3.92 (s, 3H), 3.90–3.82 (m, 1H), 3.80 (s, 3H), 3.55–3.45 (m, 1H), 3.43–3.38 (m, 1H), 3.03 (dd, J = 4.8, 16.5 Hz, 1H), 2.76 (dd, J = 6.3, 16.2 Hz, 1H). MS m/z 486.4 (M + H+).
To a mixture of the acid 27 (430 mg, 1.0 m mol) above, benzyl amine (107 mg, 1 mmol), and N,N-dimethylamino pyridine (18 mg, 0.15 mmol) in DMF was added EDC (287 mg, 1.5 mmol), and the mixture was stirred at room temperature for 20 h. The mixture was concentrated under reduced pressure, and aqueous NaHCO3 solution (20 mL) was added. The mixture was extracted with 10% MeOH in CHCl3 (3 × 30 mL), and the organic layer was dried (Na2SO4) and evaporated under reduced pressure. The crude product was purified by flash chromatography to afford 28 (220 mg, 38%).
3-(1-Methyl-1H-imidazole-4-sulfonylamino)-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydroquinoline-6-carboxylic Acid Benzylamide (28)
1H NMR (300 MHz, methanol-d4) δ 8.90 (s, 1H), 7.76 (s, 1H), 7.70 (s, 1H), 7.62 (dd, J = 2.1, 8.7 Hz, 1H), 7.50 (d, J = 2.1 Hz, 1H), 7.46 (s, 1H), 7.36–7.30 (m, 5H), 6.82 (d, J = 8.7 Hz, 1H), 4.75 (d, J = 16.8 Hz, 1H), 4.62 (d, J = 16.8 Hz, 1H), 4.55 (s, 2H), 3.92 (s, 3H), 3.90–3.82 (m, 1H), 3.80 (s, 3H), 3.53–3.48 (m, 1H), 3.43–3.39 (m, 1H), 3.03 (dd, J = 4.8, 16.5 Hz, 1H), 2.75 (dd, J = 6.3, 16.2 Hz, 1H).
3-[(tert-Butylcarbamoyl-methyl)-(1-methyl-1H-imidazol-4-sulfonyl)-amino]-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-6-carboxylic Acid Benzylamide (291)
Compound 291 was prepared as described from the general procedure for N-alkylation. 1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.80 (s, 1H), 7.78 (s, 1H), 7.63 (dd, J = 2.1, 8.7 Hz, 1H), 7.53 (d, J = 2.1 Hz, 1H), 7.42 (s, 1H), 7.36–7.30 (m, 5H), 6.82 (d, J = 8.7 Hz, 1H), 4.77 (d, J = 16.8 Hz, 1H), 4.65 (d, J = 17.4 Hz, 1H), 4.55 (s, 2H), 4.33–4.22 (m, 1H), 3.91 (s, 3H), 3.85 (s, 2H), 3.80 (s, 3H), 3.56–3.50 (m, 1H), 3.46–3.39 (m, 1H), 3.03 (dd, J = 5.1, 15.3 Hz, 1H), 2.86 (dd, J = 5.1, 15.3 Hz, 1H), 1.4 (s, 9H). MS m/z 633.8 (M + H+).
3-[(tert-Butylcarbamoyl-methyl)-(1-methyl-1H-imidazol-4-sulfonyl)-amino]-1-(3-methyl-3H-imidazol-4-ylmethyl)-1,2,3,4-tetrahydro-quinolin-6-carboxylic Acid (289)
Compound 289 was prepared as described from general procedure for N-alkylation. 1H NMR (300 MHz, methanol-d4) δ 8.92 (s, 1H), 7.82 (s, 1H), 7.79 (s, 1H), 7.62 (dd, J = 2.1, 8.7 Hz, 1H), 7.52 (d, J = 2.1 Hz, 1H), 7.42 (s, 1H), 6.80 (d, J = 8.7 Hz, 1H), 4.75 (d, J = 16.8 Hz, 1H), 4.65 (d, J = 16.8 Hz, 1H), 4.35–4.25 (m, 1H), 3.92 (s, 3H), 3.85 (s, 2H), 3.80 (s, 3H), 3.60–3.51 (m, 1H), 3.50–3.40 (m, 1H), 3.03 (dd, J = 4.8, 16.5 Hz, 1H), 2.80 (dd, J = 6.3, 16.2 Hz, 1H), 1.40 (s, 9H). MS m/z 543.5 (M + H+).
Synthesis of 34 According to Scheme 8. Preparation of 1-[4-(N-triphenylmethyl)imidazolyl]ethanol
To an ice-bath cooled solution of 3.35 mL (10.0 mmol) of 3 M methyl magnesium bromide in 40 mL dry ether was added a solution of 4-(N-triphenylmethyl)-imidazole carboxaldehyde (1.69 g, 10 mmol) in THF. After 1.5 h at ambient temperature, a concentrated solution of 0.67 g (25 mmol) NH4Cl in water was added to the reaction mixture. The mixture was stirred for 1 h and filtered, and the solids were washed with THF. The combined filtrate and washes were washed with water and brine, dried over NaSO4, and concentrated in vacuo to afford the required compound.17 1H NMR (300 MHz CDCl3) 7.42 (s, 1H), 7.30–7.39 (m, 9H), 7.10–7.21 (m, 6H), 6.85 (s, 1H), 4.85 (q, 1H), 1.5 (d, 3H). MS m/z 355.2 (M + H+).
A mixture of 33 (47 mg, 0.1 mmol) and 1-(4-(N-triphenylmethyl)imidazolyl)ethanol (17 mg, 0.05 mmol) in 5 mL of dry acetonitrile was added methanesulfonyl chloride (34 mg, 0.3 mmol), and the mixture was heated at 60 °C overnight under nitrogen. Aqueous NaHCO3 and ethyl acetate were added. The organic layer was washed with water and brine and dried over MgSO4. The crude product was purified by HPLC to give 34 as a mixture of isomers.18 1H NMR (300 MHz, methanol-d4) δ 8.95 (s, 1H), 8.94 (s, 1H), 8.72 (d, J = 7.5 Hz, 1H), 8.68 (d, J = 7.5 Hz, 1H), 7.90–8.10 (m, 4H), 7.59–7.70 (m, 4H), 7.31–7.48 (m, 4H), 6.95 (d, J = 8.7 Hz, 1H), 6.91 (d, J = 8.7 Hz, 1H), 5.50 (q, 1H), 5.40 (q, 1H), 4.38–4.51 (m, 2H), 4.08–4.16 (m, 4H), 3.70 (s, 6H), 3.32–3.45 (m, 4H), 3.01–3.22 (m, 6H), 2.65–2.90 (m, 6H), 1.68–1.88 (m, 6H), 1.60 (d, 6H), 1.05–1.17 (m, 4H). MS m/z 564.6 (M + H+).
Resolution of Enantiomers (163 and 164)
Enantiomerically pure amines derived from racemic 6-cyano-1,2,3,4-tetrahydro-quinolin-3-ylamine hydrochloride were obtained by coupling with S-mandelic acid to form diastereomeric amides as described previously.5,10 After resolution, the amines were converted to 163 and 164, as shown in Scheme 1.
Modeling Studies
We published a detailed procedure for building a homology model of Pf-PFT.7 Briefly, the model was generated with MODELLER19 by using the crystal structure of rat-PFT complexed with the nonsubstrate tetrapeptide inhibitor CVFM and farnesyl-pyrophosphate as the template structure (PDB entry 1JCR). The sequences of the two subunits (α and β) of Pf-PFT were obtained from the PlasmoDB20 (α: PFL2050w and β: chr11.glm_528) and aligned with the template. Only regions with reasonable reliability in the alignment were included. The model of Pf-PFT comprises the following sequence segments (the residue numbers of the corresponding segments of the rat-PFT subunits are given in parentheses): α: 72–164 (87–179), 300–411 (184–283); β: 421–677 (71–315), 806–896 (330–417). Note that the Pf-PFT sequences (α: 497; β: 967) are much longer than the rat ones (α: 377; β: 437)
A small molecule crystal structure of 7-nitro-1,2,3,4-THQ,21 with R = 0.034 at 0.77 Å resolution, shows that the tetrahydropyridine ring adopts an envelope conformation, with atom C3 deviating by 0.66 Å from the mean plane through the other five atoms. Upon substitution with an equatorial amino group, the deviation of the methyl from the plane is 0.40 Å, implying that when the two enantiomers are superimposed the amines are only 0.40 Å apart. Both conformers of the molecule were built with Insight (Accelrys) starting from the 7-nitro-1,2,3,4-THQ crystal structure, and the energies were minimized with the CFF force-field; a 4rε electrostatics model was used. The energies were 54.28 kcal/mol (equatorial) and 56.96 kcal/mol (axial).
All molecular docking calculations of the THQ compounds were carried out using the FLO/QXP v.6.02 package24 with as start model 162, as observed in the rat-PFT crystal structure transferred by superposition into the comparative model of Pf-PFT. THQ compounds were built with the FLO BUILDER module, specifically taking care to assign the mixed sp2/sp3 N atom type of the sulfonamide and allowing for mild pyramidalization. Monte Carlo searches were carried out for the THQ substituents, using 50 cycles per rotatable bond, while keeping the THQ scaffold fixed. Subsequently, the best solutions were minimized, allowing also for spatial freedom to the scaffold. Because the force-field has not been parametrized for transition metal complexes, the inhibitor N atom that coordinates the Zn2+ atom was spatially constrained as well as all other atoms of the Zn2+-binding imidazole ring.
Plasmodium Strains
The P. falciparum strains used in this study were 3D7 (Netherlands [airport-associated malaria], chloroquine sensitive) and K1 (Thailand, chloroquine resistant, pyrimethamine resistant). Strain 3D7 was provided by Dr. Pradipsinh Rathod from the University of Washington. P. falciparum strain K1 and P. berghei isolate NK65 (used for rodent malaria experiments) were obtained from the MR4 unit of the American Type Culture Collection (ATCC, Manassas, VA).
P. falciparum Culture
Strains of P. falciparum were cultured in vitro using experimental techniques described by Trager and Jensen.22 Cultures were maintained in RPMI-1640 (Sigma, St. Louis, MI) with 2 mM L-glutamine, 25 mM HEPES, 33 mM NaHCO3, 20 µg/mL gentamicin sulfate, and 20% (v/v) heat-inactivated human plasma type A+ (RP-20P). Type A+ erythrocytes were obtained from lab donors, washed three times with RPMI 1640, resuspended in 50% RPMI-1640, and stored at 4 °C. Parasites were grown in 10 mL of a 2% hematocrit/RP-20P (v/v) in 50 mL flasks under a 5% CO2, 5% O2, and 90% N2 atmosphere.
P. falciparum ED50 Determination
A total of 10 µL of PFTI in solution was added to each well of a 96-well plate followed by the addition of 190 µL of an asynchronous P. falciparum culture at parasitemia and hematocrit of 0.5%. PFTI solutions were prepared by diluting 20 mM THQ PFTI in dimethylsulfoxide by 200-fold with RP-20P for the highest concentration (100 µM stock gives final assay concentration of 5 µM), then performing further serial dilutions in RP-20P. Plates were flushed with 5% CO2, 5% O2, and 90% N2 then incubated at 37 °C for 48 h. 8[3H]-Hypoxanthine (0.3 µCi, 20 Ci/mmol, American Radiolabeled Chemicals) in 30 µL of RP-20P was added to cultures and incubated for an additional 24 h. Cells were harvested onto glass fiber filters by a cell harvester (Inotech Biosystems International, Inc Rockville, MD), and the radioactivity incorporated into the parasites was counted on a Chameleon 425–104 multilabel plate counter (Hidex Oy Turku, Finland). The background level detected with uninfected erythrocytes was subtracted from the data. The 3H-incorporation into infected erythrocytes with 1 µL of DMSO vehicle alone represents 100% malaria growth. ED50 values, the effective dose that reduces growth by 50%, were determined by linear regression analysis of the plots of 3H-hypoxanthine incorporation versus concentration of compound. Each compound was tested in duplicate, and the mean value is shown; individual measurements differed by less than 3-fold.
Pf-PFT IC50 Determination
The PFT assay used to determine the IC50s (inhibitor concentration that causes 50% enzyme inhibition) of the compounds is based on a PFT [3H] scintillation proximity assay (SPA; TRKQ7010 Amersham Biosciences Corp Piscataway, NJ).3 Assays were carried out in assay buffer (pH 7.5, 50 mM HEPES, 30 mM MgCl2, 20 mM KCl, 5 mM DTT, 0.01% Triton X-100), 1 µM human lamin-B carboxy-terminus sequence peptide (biotin-YRASNRSCAIM), and 1 µCi 3H-farnesylpyrophos-phate (Amersham specific activity 15 to 20 Ci/mM) in a total volume of 50 µL, which included 1 µL of Pf-PFT inhibitor solution in DMSO and 5 µL of partially purified Pf-PFT. Assays in the absence of Pf-PFT inhibitor and Pf-PFT were included as positive and negative controls, respectively. Reaction mixtures were incubated at 37 °C for 60 min and terminated by the addition of 70 µL of assay STOP solution and 5 µL of SPA beads. The assay mixture was incubated at room temperature for 30 min. The assay was counted on a plate Chameleon 425-104 multilabel counter (Hidex Oy Turku, Finland). IC50 values were calculated using linear regression analysis of the plots of the amount of radio-prenylation versus the concentration of compound.
Mammalian-PFT IC50 Determination
The PFT assay used to determine the IC50s of the compounds is based on a farnesyl transferase [3H] SPA enzyme assay (TRKQ7010 Amersham Biosciences Corp Piscataway, NJ). Rat PFT was prepared as described.23 Assays were carried out in assay buffer pH 7.5 (50 mM HEPES, 30 mM MgCl2, 20 mM KCl, 5 mM DTT, 0.01% Triton X-100) and 1 µM human lamin-B carboxy-terminus sequence peptide (biotin-YRASNRSCAIM) in a total volume of 50 µL, which included 1 µL of PFT inhibitor solution in DMSO and 5 µL of partially purified rat-PFT. Assays in the absence of PFT inhibitor and rat-PFT were included as positive and negative controls, respectively. Reaction mixtures were incubated at 37 °C for 15 min and terminated by the addition of 70 µL of assay STOP solution and 5 µL of SPA beads. The assay is incubated at room temperature for 30 min. The assay was counted on a plate Chameleon 425-104 multilabel counter (Hidex Oy Turku, Finland). IC50 values were calculated using linear regression analysis of the plots of 3H-FPP prenylation versus concentration of compounds.
Footnotes
Abbreviations: ED50, effective dose of inhibitor that reduces parasite growth in vitro by 50%; IC50, inhibitor concentration that reduces the rate of PFT in vitro by 50%; PFT, protein farnesyltransferase, PFTI, PFT inhibitor, THQ, tetrahydroquinoline.
Note Added after ASAP Publication. This manuscript was released ASAP on August 28, 2007, with errors in the compound numbers in Schemes 5 and 7 and with an error in the structure for compound 119 in Table 2. The correct version was posted on September 13, 2007.
Supporting Information Available: HPLC traces of key compounds 6, 162, 110, 191, and 234. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chakrabarti D, Azam T, DelVecchio C, Qiu L, Park Y, Allen CM. Protein prenyl transferase activities of Plasmodium falciparum. Mol. Biochem. Parasitol. 1998;94:175–184. doi: 10.1016/s0166-6851(98)00065-6. [DOI] [PubMed] [Google Scholar]
- 2.Chakrabarti D, Da Silva T, Barger J, Paquette S, Patel H, Patterson S, Allen CM. Protein farnesyltransferase and protein prenylation in Plasmodium falciparum. J. Biol. Chem. 2002;277:42066–42073. doi: 10.1074/jbc.M202860200. [DOI] [PubMed] [Google Scholar]
- 3.Nallan L, Bauer KD, Bendale P, Rivas K, Yokoyama K, Horney CP, Pendyala PR, Floyd D, Lombardo LJ, Williams DK, Hamilton A, Sebti S, Windsor WT, Weber PC, Buckner FS, Chakrabarti D, Gelb MH, Van Voorhis WC. Protein farnesyltransferase inhibitors exhibit potent antimalarial activity. J. Med. Chem. 2005;48:3704–3713. doi: 10.1021/jm0491039. [DOI] [PubMed] [Google Scholar]
- 4.Bell IM. Inhibitors of farnesyltransferase: A rational approach to cancer chemotherapy? J. Med. Chem. 2004;47:1869–1878. doi: 10.1021/jm0305467. [DOI] [PubMed] [Google Scholar]
- 5.Lombardo LJ, Camuso A, Clark J, Fager K, Gullo-Brown J, Hunt JT, Inigo I, Kan D, Koplowitz B, Lee F, McGlinchey K, Qian L, Ricca C, Rovnyak G, Traeger S, Tokarski J, Williams DK, Wu LI, Zhao Y, Manne V, Bhide RS. Design, synthesis, and structure-activity relationships of tetrahyroquinoline-based farnesyltransferase inhibitors. Bioorg. Med. Chem. Lett. 2005;15:1895–1899. doi: 10.1016/j.bmcl.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Hunt JT, Ding CZ, Batorsky R, Bednarz M, Bhide R, Cho Y, Chong S, Chao S, Gullo-Brown J, Guo P, Kim SH, Lee FY, Leftheris K, Miller A, Mitt T, Patel M, Penhallow BA, Ricca C, Rose WC, Schmidt R, Slusarchyk WA, Vite G, Manne V. Discovery of (R)-7-cyano-2,3,4,5-tetrahydro-1-(1H-imidazol-4-ylmethyl)-3-phenylmethyl)-4-(2-thienylsulfonyl)-1H-1,4-benzodiazepine (BMS-214662), a farnesyltransferase inhibitor with potent preclinical antitumor activity. J. Med. Chem. 2000;43:3587–3595. doi: 10.1021/jm000248z. [DOI] [PubMed] [Google Scholar]
- 7.Eastman RT, White J, Hucke O, Bauer K, Yokoyama K, Nallan L, Chakrabarti D, Verlinde CL, Gelb MH, Rathod PK, Van Voorhis WC. Resistance to a protein farnesyltransferase inhibitor in Plasmodium falciparum. J. Biol. Chem. 2005;280:13554–13559. doi: 10.1074/jbc.M413556200. [DOI] [PubMed] [Google Scholar]
- 8.Eastman RT, White J, Hucke O, Yokoyama K, Verlinde CL, Hast MA, Beese LS, Gelb MH, Rathod PK, Van Voorhis WC. Resistance mutations at the lipid substrate binding site of Plasmodium falciparum protein farnesyltransferase. Mol. Biochem. Parasitol. 2007;152(1):66–71. doi: 10.1016/j.molbiopara.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Voorhis WC, Rivas K, Bendale P, Nallan L, Hornéy C, Barrett LK, Ankala S, Hucke O, Verlinde CLMJ, Chakrabarti D, Strickland C, Yokoyama K, Buckner FS, Hamilton A, Williams DK, Lombardo LJ, Floyd D, Gelb MH. Tetrahydroquinoline Plasmodium falciparum protein farnesyltransferase inhibitors: Efficacy, pharmacokinetics, and metabolism. Antimicrob. Agents Chemother. 2007 doi: 10.1128/AAC.00246-07. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhide R. Inhibitors of farnesyl protein transferase. 1999 WO 99/01434. [Google Scholar]
- 11.Reid TS, Beese LS. Crystal structures of the anticancer clinical candidates R115777 (Tipifarnib) and BMS-214662 complexed with protein farnesyltransferase suggest a mechanism of FTI selectivity. Biochemistry. 2004;43:6877–6884. doi: 10.1021/bi049723b. [DOI] [PubMed] [Google Scholar]
- 12.Nicholas JB, Vance REM, Burke BJ, Hopfinger AJ. A molecular mechanics valence force field for sulfonamides derived by ab initio methods. J. Chem. Phys. 1991;95:9803–9811. [Google Scholar]
- 13.Liang GY, Bays JP, Bowen JP. Ab initio calculations and molecular mechanics (MM3) force field development for sulfonamide and its alkyl derivatives. Theochem. 1997;40(1/2):165–179. [Google Scholar]
- 14.Hillal SH, Karickhoff SW, Carreira LA. A rigorous test for SPARC’s chemical reactivity models: Estimation of more than 4300 ionization pK(a)s. Quant. Struct.-Act. Relat. 1995;14:348. [Google Scholar]
- 15.Hillal SH, El-Shabrawy Y, Carreira LA, Karickhoff WW, Toubar SS, Rizk M. Estimation of the ionization pKa of pharmaceutical substances using the computer program sparc. Talanta. 1996;43:607–619. doi: 10.1016/0039-9140(95)01789-5. [DOI] [PubMed] [Google Scholar]
- 16.Chen BC, Skoumbourdis AP, Sundenen JE, Rovnyak GC, Traeger SC. Efficient molar-scale synthesis of 1-methyl-5-acylimidazole triflic acid salts. Org. Process Res. Dev. 2000;4:513–614. [Google Scholar]
- 17.Kelley JL, Miller CA, McLean EW. Attempted inhibition of histidine decarboxylase with β-alkyl analogues of histidine. J. Med. Chem. 1977;20:721–723. doi: 10.1021/jm00215a021. [DOI] [PubMed] [Google Scholar]
- 18.Venishi J, Hamada M, Aburatani S, Matsui K, Yonemitsu O, Tsukube H. Synthesis of chiral nonracemic 1-(2-pyridinyl)ethylamines: Stereospecific introduction of amino function onto the 2-pyridinylmethyl carbon center. J. Org. Chem. 2004;69:6781–6789. doi: 10.1021/jo0491758. [DOI] [PubMed] [Google Scholar]
- 19.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 20.Bahl A, Brunk B, Crabtree J, Fraunholz MJ, Gajria B, Grant GR, Ginsburg H, Gupta D, Kissinger JC, Labo P, Li L, Mailman MD, Milgram AJ, Pearson DS, Roos DS, Schug J, Stoeckert CJ, Jr, Whetzel P. PlasmoDB: The Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu JM, Hu XR, Xu WM. 7-Nitro-1,2,3,4-tetrahydroquinoline. Acta Crystallogr. 2006;E62:o62–o63. [Google Scholar]
- 22.Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 23.Yokoyama K, Zimmerman K, Scholten J, Gelb MH. Differential prenyl pyrophosphate binding to mammalian protein geranylgeranyltransferase-I and protein farnesyltransferase and its consequence on the specificity of protein prenylation. J. Biol. Chem. 1997;272:3944–3952. doi: 10.1074/jbc.272.7.3944. [DOI] [PubMed] [Google Scholar]
- 24.McMartin C, Bohacek RS. QXP: Powerful, rapid computer algorithms for structure-based drug design. J. Comput.-Aided Mol. Des. 1997;11:333–344. doi: 10.1023/a:1007907728892. [DOI] [PubMed] [Google Scholar]