Summary
Dendrite branching and spine formation determines the function of morphologically distinct and specialized neuronal subclasses. However, little is known about the programs instructing specific branching patterns in vertebrate neurons and whether such programs influence dendritic spines and synapses. Using knockout and knockdown studies combined with morphological, molecular and electrophysiological analysis we show that the homeobox Cux1 and Cux2 are intrinsic and complementary regulators of dendrite branching, spine development and synapse formation in layer II–III neurons of the cerebral cortex. Cux genes control the number and maturation of dendritic spines partly through direct regulation of the expression of Xlr3b and Xlr4b, chromatin remodeling genes previously implicated in cognitive defects. Accordingly, abnormal dendrites and synapses in Cux2−/− mice correlate with reduced synaptic function and defects in working memory. These demonstrate critical roles of Cux in dendritogenesis and highlight novel subclass-specific mechanisms of synapse regulation that contribute to the establishment of cognitive circuits.
Keywords: cerebral cortex, Cut, Cutl1, Cutl2, transcription factor, dendrite, spine, synapse, synaptogenesis, Xlr
Introduction
Neurons of the nervous system establish complex and stereotyped patterns of connectivity and the number and strength of the synapses are precisely regulated. In this process, the development of specific dendritic structures determines the functions and specializations of neuronal subclasses. Dendritic branching specifies the connectivity with selected axonal inputs, while spine density and morphology determines the number, strength and stability of synaptic contacts, thereby shaping neuronal circuits and influencing cognition (Parrish et al., 2007; Tada and Sheng, 2006). The essential role of dendritic structures is reflected by the fact that dendrite and spine alterations are often the only morphological defects that can be detected in post-mortem studies of patients affected by non-syndromic forms of mental retardation (Dierssen and Ramakers, 2006).
The regulation of dendrite structures generates neuronal diversity and determines neuronal function, but how the specific dendritic morphologies of the distinct neuronal subclasses are specified is largely unknown. As with other subclass-specific neuronal features, dendritic architecture is thought to be instructed in part by the restricted expression of transcription factors (TFs). However, very few of such TFs are actually known to control dendrite development in vertebrates (Parrish et al., 2007). In addition, it is unclear whether subclass specific TFs can influence the establishment of dendritic spines and the maturation and strength of the synapses, or whether these aspects of neuronal function depend solely on the action of external signals (Tada and Sheng, 2006).
The vertebrate cortex is functionally organized into distinct layers. Pyramidal neurons in each cortical layer have distinct molecular identities and marked differences in dendritic morphology (Ballesteros-Yanez et al., 2006; DeFelipe, 1988). In recent years, several cortical layer-specific TFs have been described (Molyneaux et al., 2007), but only the expression of Fezf2/Zfp312 in layer V neurons has been shown to regulate dendrite formation (Chen et al., 2005). The regulation of upper layer neurons of the cerebral cortex is of particular interest. Layer II–III neurons develop elaborated dendritic trees and abundant dendritic spines, which enable them to integrate numerous intracortical inputs (DeFelipe, 1988). Upper cortical neurons are also the last to appear during development and evolution, likely contributing to the increased cognitive capacity of the mammalian brain. Besides, these neurons are particularly highly elaborated in higher primates, including humans (Marin-Padilla, 1992). In the mouse, upper cortical layers are identified by the expression of the TFs Cux1 and Cux2 (Nieto et al., 2004; Zimmer et al., 2004). While hCux2 also defines the upper layers of the human cerebral cortex (Arion et al., 2007), the expression patterns of hCux1 remain unknown. Cux1 and Cux2 encode the vertebrate homologues of the Drosophila homeobox transcription factor Cut (Quaggin et al., 1996; Sansregret and Nepveu, 2008), which controls the dendrite morphology of postmitotic populations in the peripheral nervous system (PNS) (Grueber et al., 2003; Jinushi-Nakao et al., 2007; Komiyama and Luo, 2007). In addition to the upper cortical layers, mammalian Cux genes are expressed in other neural populations in the central nervous system (CNS) and PNS (Iulianella et al., 2003). While Cux2 has been shown to participate in neural precursor proliferation (Cubelos et al., 2008a; Iulianella et al., 2008), to date there is no information regarding the role of Cux1 and Cux2 in postmitotic neurons.
In the cerebral cortex the highly overlapping patterns of Cux1 and Cux2 expression, and the high proportion of cells expressing either protein, indicate co-expression of both genes and suggest their possible redundant functions (Nieto et al., 2004). Indeed, the cortical and brain organization of single Cux1−/− and Cux2−/− knock-outs (ko) is overall normal and they show no changes in the expression of upper layer markers or in that of the reciprocal Cux homologue (Cubelos et al., 2008a), although there are more upper layer neurons in Cux2−/−, but not in Cux1−/−, due to increased proliferation of SVZ cells (Cubelos et al., 2008a). Cux1−/−; Cux2−/− double ko mice suffer highly penetrant early embryonic lethality, but the few double ko mice that survive until birth show no defects in neuronal migration or in the expression of layer specific proteins (Cubelos et al., 2008b). Thus, Cux genes do not appear to affect early specification programs but rather, may regulate later aspects of differentiation, including a possible conserved role in dendritogenesis along with Cut.
Here we show that the mouse Cux genes play a critical role in controlling dendritic branching and the formation of the dendritic spines and functional synapses in layer II–III neurons of the cortex. We also demonstrate that Cux genes intrinsically regulate the number and differentiation of the dendritic spines by binding and regulating the expression of Xlr4b and Xlr3b, two chromatin remodeling genes previously implicated in cognitive defects. Suggestive of functional consequences, the observed dendritic and synaptic alterations in Cux2−/− animals correlate with working memory deficiencies. Our results therefore reveal an important role of Cux genes in regulating neuronal function and cognition by controlling dendritic structures, and identify novel mechanisms involved in neuronal specification.
Results
Cux genes control dendrite branching and the number of dendritic spines in pyramidal neurons of the upper cortical layers
Previous studies suggested that Cux genes may regulate late aspects of neuronal differentiation (Cubelos et al., 2008a; Cubelos et al., 2008b). To investigate whether the homeobox Cux proteins play a role in dendritogenesis, we analyzed the dendritic morphology of individual layer II–III neurons in the somatosensory cortex of WT, single Cux1−/− and single Cux2−/− mice, using the Golgi-Cox impregnation method (Ramon Moliner, 1970). The total length of all the dendrite processes was assessed as a measure of dendritic complexity, and the numbers and length of the primary, secondary and tertiary branches was quantified in P60 animals. In WT animals, layer II–III neurons developed complex dendritic trees, with profuse apical and basal branching (Fig 1a, c, e, f). Strikingly, layer II–III neurons of the single Cux1−/− or Cux2−/− mice had much simpler morphologies, with a significant decrease in the dendritic length and the number of branches (Fig 1a, c, e, f). Furthermore, the density of the dendritic spines on layer II–III neurons of Cux1−/− and Cux2−/− mice was severely reduced by more than 50% when compared with upper layer neurons from WT mice (Fig 1a, d). By contrast, the upper layer neurons of Cux1+/− and Cux2+/−, and Cux1+/−; Cux2+/− compound heterozygote animals did not display defects in dendritic differentiation. Moreover, the defects in layer II–III neurons from Cux2−/−; Cux1+/− compound heterozygous were equivalent to those in the neurons from Cux2−/− (not shown). These observations suggest that Cux proteins are expressed normally in heterozygous animals. All these aspects of dendritic structures were affected to a similar extent in the upper layers of the Cux1−/− and Cux2−/− mice, indicating that the two genes fulfill necessary functions and that they contribute similarly to the regulation of dendrite development. These similarities also strongly support that dendritic defects do not relate to the increased number of upper layer neurons observed only in Cux2−/− mice, but not in Cux1−/− animals. Importantly, Cux deficiency did not affect dendrite branching and spine numbers in layers V (Fig 1b, c, d) and VI (not shown). Together, these results suggest that Cux TFs are specific determinants of dendritogenesis in the postmitotic neuronal populations where they are expressed.
Figure 1. Cux1 and Cux2 control the dendritic morphology and spine number of upper cortical pyramidal neurons.
a, b) Golgi-Cox stained individual neurons in WT, Cux2−/− and Cux1−/− animals. a) Pyramidal neurons in upper cortical layers II–III show fewer dendritic branches and spines in Cux2−/− and Cux1−/− mutants than in the WT animals, (upper panels). High optical magnification images of dendritic spines (lower panels). b) No differences were observed in the dendritic morphology of pyramidal neurons in cortical layer V (upper panels) or in their dendritic spines (lower panels). Bars represent 50 µm (upper panels) and 20 µm (lower panels). c) Total cumulative length of dendritic processes per neuron in cortical layers II–III and V of the somatosensory cortex of WT, Cux1−/− and Cux2−/− mice. d) Dendritic spine density in layers II–III and layer V. e) Total cumulative dendrite length of primary, secondary and tertiary branches per neuron in layers II–III. f) Total number of primary, secondary and tertiary dendrite branches per neuron in layers II–III. WT (n= 16), Cux1−/− (n=15) and Cux2−/− (n=15). * p<0.05 and ** p<0.01 between WT and mutant cortex.
Cux1 and Cux2 additive functions instruct early dendrite development
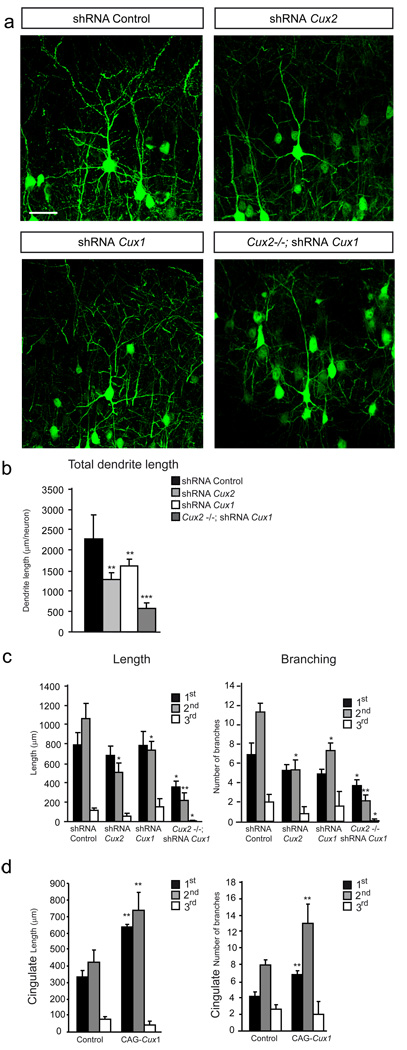
Although dendrite branching and spine density can be influenced by presynaptic axonal inputs (Cline and Haas, 2008; Parrish et al., 2007), the absence of detectable defects in the major axonal tracks of Cux1−/− and Cux2−/− brains, such as the corpus callosum or the anterior commissure (Cubelos et al., 2008a; Luong et al., 2002), suggests potential intrinsic roles in dendritogenesis. Nevertheless, to confirm a cell intrinsic function of Cux genes in otherwise intact brain and to rule out the possible contribution of subtle defects in the afferents targeting the upper layers, we knocked down Cux1 and Cux2 in discrete neuronal populations within layer II–III. shRNA lentiviral constructs were electroporated in utero in E15.5 WT embryos and co-electroporation with GFP allowed visualization of the morphology of the targeted neurons at P21. Effective down-regulation of the targeted proteins, as well as the correct migration and generation of electroporated neurons, was confirmed in the cortex of P4 and P21 animals (not shown and Fig S1A and Fig S1B). Neurons electroporated with control shRNA or with CAG-GFP alone displayed the highly branched morphology characteristic of upper layer neurons (Fig 2a). Remarkably, while most axonal inputs to the electroporated neurons should have remained unaffected, dendritic branching was visibly and quantitatively reduced by the knockdown of Cux1 or Cux2 (Fig 2a, b, c), closely matching the alterations observed in Cux1−/− and Cux2−/− mice (compare Fig 2 with Fig 1a, c, e, f). These changes were specific because dendritic morphology was not affected when Cux targeting shRNAs were electroporated with their respective mutated resistant form (Fig S1C, data not shown and S experimental procedures), excluding possible off-target effects. Moreover, examination of the effect of Cux2 knockdown on dendrite development in early differentiating neurons at P4 demonstrated a clear reduction in branch number and neurite length (Fig S1D). Hence, these data demonstrated an early intrinsic control of Cux2 on dendrite development, independent of synapse activity and irrespective of any possible effects on dendrite remodeling and pruning.
Figure 2. Cux1 and Cux2 proteins stimulate dendrite development via cell intrinsic and additive mechanisms.
a) Confocal micrographs showing GFP-expressing layer II–III neurons in the P21 cortex. Neuronal morphology was analyzed at P21 after in utero electroporation at E15.5. Knock-down of Cux1 or Cux2 with shRNA lentiviral constructs decreases the dendrite complexity of layer II–III neurons compared to control shRNA electroporated neurons. Knock-down of Cux1 in Cux2−/− layer II–III neurons induces still simpler dendrite morphologies. Bar represents 25 µm. b) Total cumulative lengths of dendritic processes per GFP-positive neuron in layers II–III. c) Cumulative dendrite length of primary, secondary and tertiary branches (left) and the average number of primary, secondary and tertiary dendrite branches (right) per neuron. Control shRNA (n= 19), shRNA Cux1 (n=15) and shRNA Cux2 (n=22), shRNA Cux1 in Cux2−/− (n=12). d) Overexpression of Cux1 in neurons of the cingulate cortex stimulates dendritic branching. Cumulative dendrite length of primary, secondary and tertiary branches (left) and the number of primary, secondary and tertiary dendrite branches (right) per GFP positive layer II–III neuron control (n= 15), CAG Cux1 (n=15) * p<0.05, ** p<0.01 and *** p<0.001 compared with controls.
The knockdown experiments indicated that Cux1 and Cux2 exerted cell autonomous control of dendrite development. On the other hand, the requirement for Cux1 and Cux2 during dendrite development suggested converging downstream mechanisms. Indeed, overexpression of Cux1 in the upper layer neurons of Cux2−/− animals reverted dendritic defects to normal morphologies, suggesting some equivalent functions (Fig S1F). However, staining in the somatosensory cortex indicated that a large proportion of neurons co-express both Cux1 and Cux2 proteins (Cubelos et al., 2008a; Ferrere et al., 2006) (Fig S1E), and we therefore next investigated the effect of loss of function of both Cux genes on dendrite development. Using the in utero electroporation system to knockdown Cux1 in neurons of the upper cortical layer of Cux2−/− embryos, we overcame the embryonic lethality of the double Cux1;Cux2 knockout and analyzed neuronal morphology. Knockdown of Cux1 in Cux2−/− upper layer neurons of the somatosensory cortex produced a dramatic reduction in branching and total dendrite length (Fig 2 a, b, c), demonstrating an additive effect of the two factors. In contrast to the somatosensory areas, late born neurons of the cingulate cortex have simple dendritic morphologies (Fig 2d and Fig S1G) and express Cux2, but only low levels of Cux1 (Ferrere et al., 2006; Nieto et al., 2004). Forced overexpression of Cux1 protein in cingulate neurons resulted in a significant increase in dendritic complexity (Fig 2d and Fig S1G), further indicating additive activities. Altogether, these experiments demonstrate the related and additive function of the two Cux genes and suggest that the final pattern of dendritic complexity in neurons of the upper layers depends on the combinatorial expression of both Cux1 and Cux2 proteins.
Synaptic defects in Cux2−/− upper layer cortical neurons
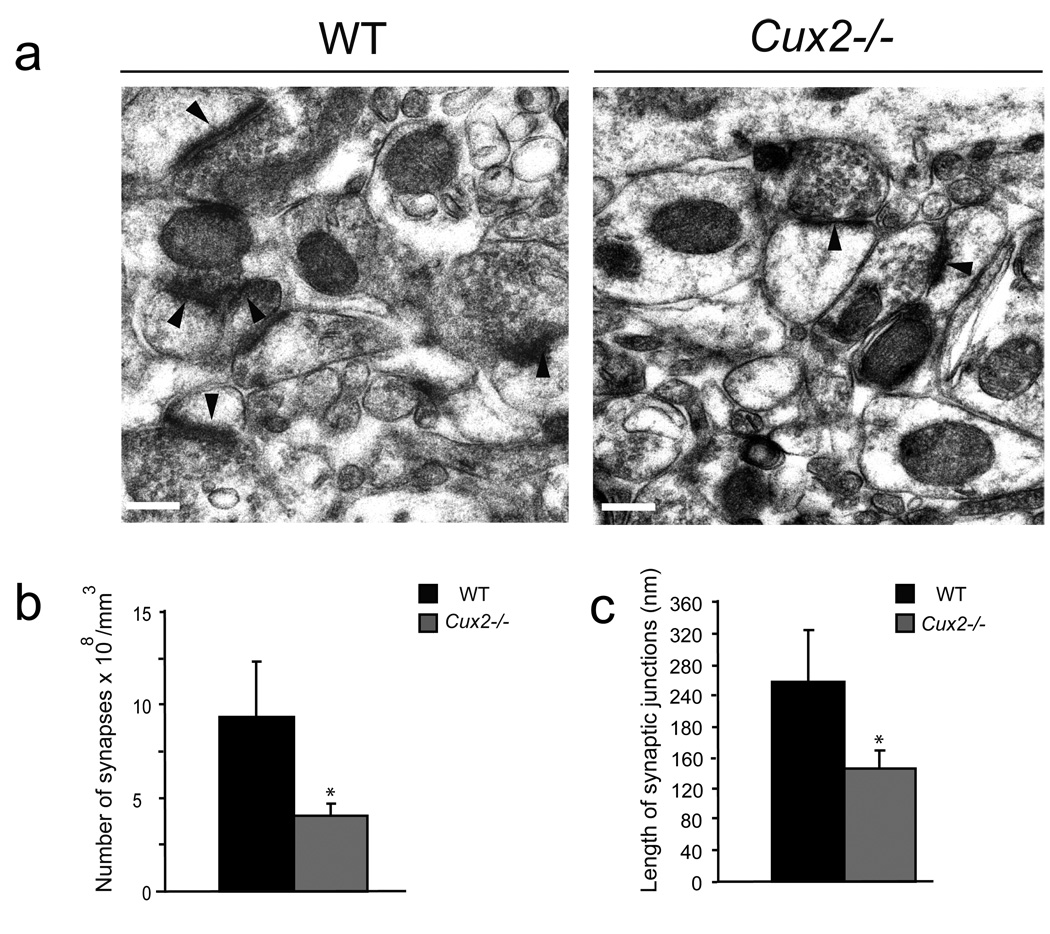
Dendritic spines are the site of synaptic contacts. Often, reductions in the density of dendritic spines, such those found in layer II–III neurons of Cux1−/− and Cux2−/− cortex, are a consequence of defects in the establishment and/or stabilization of the synapse. Thus, we studied the formation of synapses in layer II–III neurons by electron microscopy analysis. These and all subsequent analyses were confined to the study of WT and Cux2−/− animals because most Cux1−/− animals die perinatally due to defects unrelated to the nervous system (Luong et al., 2002). The very few Cux1−/− animals that survived past P21 were used for the Golgi analysis (Fig 1). Electron microscopy showed that the density of asymmetric synaptic contacts was approximately 2-fold lower in layer II–III neurons of Cux2−/− cortex when compared with WT animals (Fig 3a, b), and hence accompanied the reduction in the number of dendritic spines (Fig 1d). More importantly, we found a significant reduction in the average length of the synaptic junction apposition surface in synapses of Cux2−/− layer II–III neurons (Fig 3c). The synaptic apposition surface correlates with spine head size and characterizes the strength and stability of the synapse (Sabatini et al., 2001; Tada and Sheng, 2006). Therefore, these data suggested that Cux regulate mechanism of synaptogenesis.
Figure 3. Altered synapse formation in the upper layers of Cux2−/− mice.
a)Electron micrographs showing the synapses (arrowheads) in sections of cortical layers II–III of the somatosensory cortex of WT and Cux2−/− animals. Bar represents 0.25 µm. b) Quantification of synapse density in layers II–III of WT and Cux2−/− animals. c) Average length of the synaptic junction apposition surface in layers II–III of WT and Cux2−/− animals. * p<0.001 compared with WT.
Cux1 and Cux2 regulate the morphology of dendritic spines
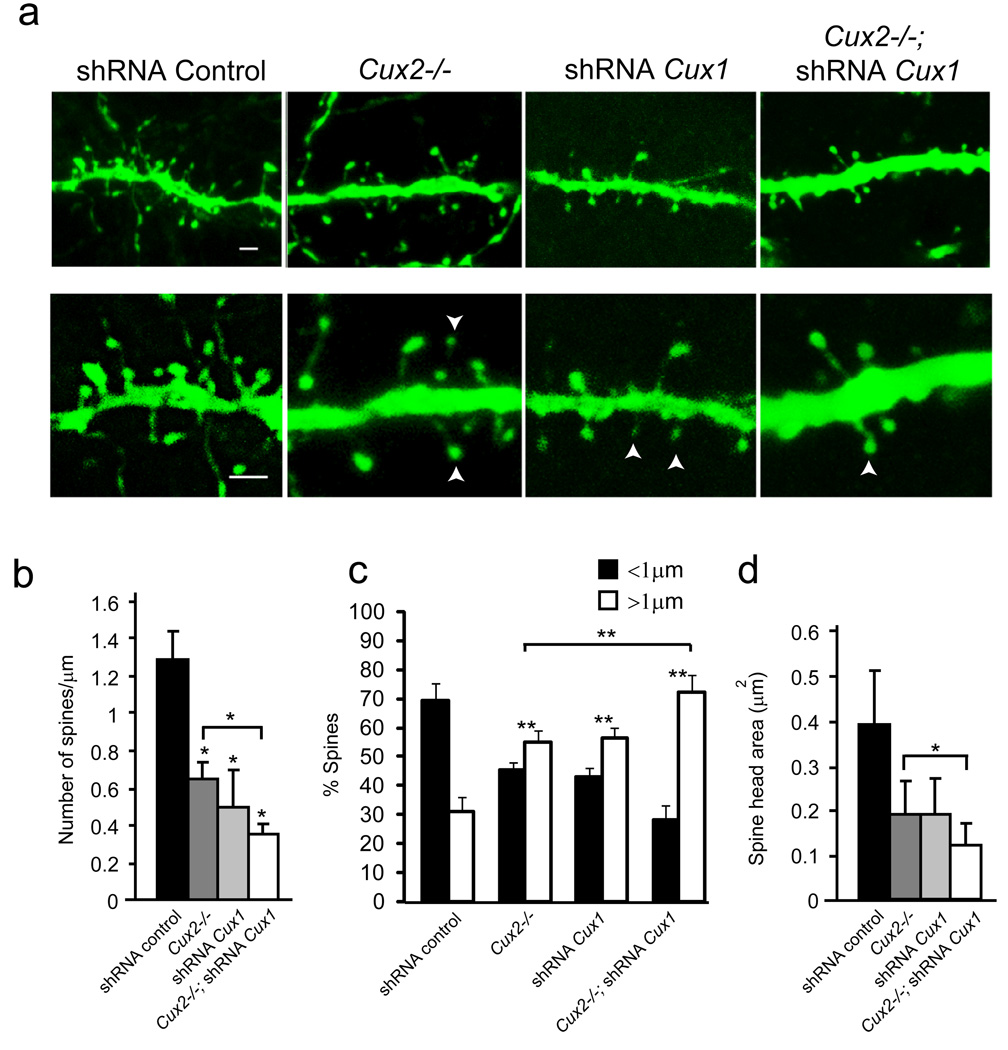
Mechanisms of synaptogenesis are intimately linked to the regulation of spine morphology. The dendritic spine can function as a structural regulator of the synapse, and in turn, can also reflect its activity (Bourne and Harris, 2007; Sabatini et al., 2001; Tada and Sheng, 2006; Yuste et al., 2000). Hence, we investigated whether abnormal synapses in Cux2−/− layer II–III neurons correlated with changes in spine morphology. Spine density, the surface of the head, and the length of the spine were estimated in GFP electroporated neurons. Dendritic spines were classified as short (<1 µm) and long (>1 µm) (Ballesteros-Yanez et al., 2006). Upper layer neurons of WT mice electroporated with control shRNA or GFP alone showed a profusion of spines with the typical range of thin, stubby and mushroom morphologies (Fig 4a and video S1). Comparative analysis of the dendritic spines (morphology and density) of WT upper layer neurons electroporated with GFP or filled intracellular with lucifer yellow (LY) gave equivalent results (Fig S2), showing a majority of short spines (69%) (Fig 4c), as previously described (Ballesteros-Yanez et al., 2006). This confirmed the reliability of our analysis. Analysis of Cux2−/− layer II–III neurons electroporated with GFP confirmed the decreased spine density observed in Golgi studies (Fig 4a, b). Remarkably, this decreased spine density was associated with aberrant morphologies, with the majority of the spines (55%) developing long necks with small heads (Fig 4 a, c, d and video S2). This type of morphology characterizes immature spines and weak synapses. Importantly, nearly identical changes in spine density and morphology were observed in WT neurons after in utero knockdown of Cux1 (Fig 4a, b, c, d and video S3) or Cux2 (not shown). Dendritic spine morphology and numbers were not affected when shRNAs targeting Cux were electroporated with their respective mutated resistant forms (Fig S1C, not shown and Supplementary Experimental Procedures). Knockdown of Cux1 in the Cux2−/− cortex caused a sharp reduction in spine density, and a further increase in the proportion of long spines (72%) associated with an even greater reduction in spine head size (Fig 4a, b, c, d and video S4). Thus, these data show that Cux genes control not only the number of dendritic spines, but also their morphological characteristics, a key aspect in synapse regulation.
Figure 4. Cux1 and Cux2 regulate dendritic spine number and spine morphology.
a) Confocal images, showing dendritic spines of GFP-positive layer II–III neurons expressing control, Cux1, or Cux2 shRNAs and of WT or Cux2−/− P21 cortex. Bar represents 1 µm. Arrowheads point to small spine heads. b, c, d) Quantitative analysis of dendritic spine defects. n≥ 15 dendrite segments and n≥ 500 spines for each sample. * p<0.01 and ** p<0.001 compared to WT or Cux2−/− (brackets).
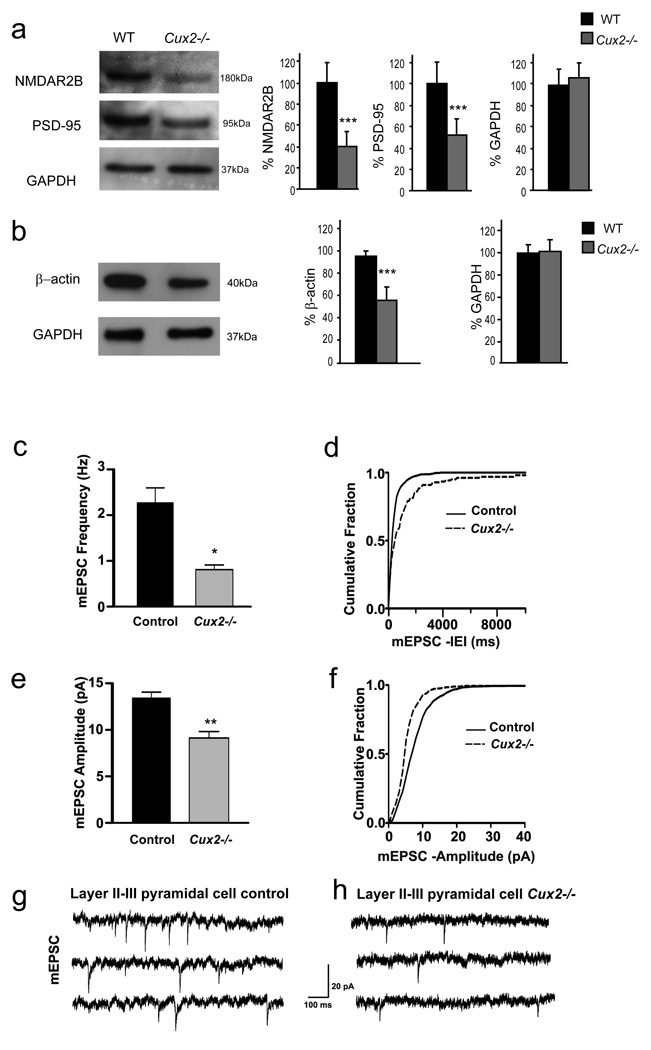
The effects of Cux genes in dendritic spine development prompted us to analyzed the expression of proteins known to modulate the number and morphology of the spine, such as PSD95 and NMDA receptor (NMDAR) (El-Husseini et al., 2000; Tada and Sheng, 2006; Ultanir et al., 2007). Western blot demonstrated a pronounced reduction of both PSD95 and the 2B subunit of NMDAR (NMDAR2B), normally abundant in the upper layers (Rudolf et al., 1996), in total lysates from adult Cux2−/− cortex (Fig 5a). By contrast, the expression of other receptors such as Glutamate receptors 1 and 2 (GluR1 and GluR2) and NMDAR1 (Fig S3A) was unaltered. Furthermore, the expression of β-actin, which is also crucial for both dendrite branching and the formation and stabilization of spines and synapses (Ammer and Weed, 2008; Cingolani and Goda, 2008), was also 30% lower in the Cux2−/− cortex (Fig 5b). In contrast, the expression of other cytoskeletal components and regulators implicated in synapse formation, such as focal adhesion kinase (FAK) (Cingolani and Goda, 2008) or N-Wasp (Wegner et al., 2008) was normal (data not shown). These results indicated that Cux genes may modulate directly or indirectly the expression of synaptic proteins in layer II–III neurons.
Figure 5. Reduced expression of synaptic proteins and changes in layer II–III mEPSC in amplitude and frequency in Cux2−/−.
a, b) Reduced expression of synaptic proteins in Cux2−/−.Western blot analysis of the expression of NMDAR2B, PSD-95 (a) and β-actin (b) in total cortical lysates from WT (n=4) and Cux2−/− (n=4). Graphs show the mean and SD signal quantification of the relative amount of protein in WT and Cux2−/− cortices. *** p<0.001. c) Average frequency of mEPSC of layer II–III pyramidal cells from control (WT and Cux2+/−) and Cux2−/− mice. (* p< 0.0005, Student’s, unpaired t test, n=13 and 14 cells, respectively), d) Cumulative fraction curves of interevent intervals (IEIs) for mEPSC of layer II–III pyramidal cells showing longer IEIs in Cux2−/− compared with control (p < 0.0005, K–S test). e) Average amplitude of mEPSC in layer II–III pyramidal cells from Cux2−/− (** p < 0.0005, Student’s, unpaired t test, n=13 and 14 cells, respectively). f) Cumulative fraction curves of amplitude of layer II–III pyramidal cells showing smaller amplitude in Cux2−/− animals compared with control (p < 0.0005, K-S test). g, h) Representative traces of mEPSC from layer II–III pyramidal cells of control and Cux2−/− mice, respectively. Data in bar graphs depict mean + SEM; control: black bars; Cux2−/−: gray bars. IEI: Interevent interval. mEPSC: miniature excitatory postsynaptic current.
Changes in mEPSC amplitude and frequency in pyramidal neurons of the upper layers of Cux2−/− mice
To directly test whether the morphological changes observed in Cux deficient upper layer neurons correlate with reduced synaptic function we next obtained patch-clamp recordings from pyramidal cells of the upper layer of WT and Cux2−/− mice. Miniature excitatory postsynaptic currents (mEPSC) recorded from the pyramidal cells of P20 animals showed that cells from Cux2−/− mice had smaller amplitude and lower frequency mEPSC than those of control animals (Fig 5c–h). In contrast, mEPSC recordings from layer V neurons were undistinguishable between control and Cux2−/− animals (Fig S3B–G). These data support the correlation between the decreases in the number of spines, the appearance of structural immature morphologies, and reduced synaptic function. Thus, Cux proteins appear to modulate the formation of functional synapses likely by cell autonomous mechanisms.
Cux1 and Cux2 bind and regulate the expression of Xlr3b and Xlr4b
The results we had obtained indicated that Cux genes control dendritogenesis and target mechanisms of spine and synapse formation in layer II–III neurons. Thus, we next compared gene expression between the cortex of Cux2−/− and control Cux2+/− mice in RNA arrays to identify genes that may be potentially involved in these functions (www.ncbi.nlm.nih.gov/projects/geo; accession numbers: GSE14971). In accordance with the observed decrease in the expression of β-actin protein (Fig 5b), β-actin RNA transcripts levels were reduced in Cux2−/− cortex (Table SI, Table SII). This was the only gene among those differentially expressed, that had been previously implicated in neurite elongation and synapse formation (Ammer and Weed, 2008; Cingolani and Goda, 2008) (S Table I, S Table II).
Among up-regulated genes, X-linked lymphocyte regulated (Xlr) 3b and Xlr4b (Table SI) caught our attention. These genes belong to a family of closely and rapidly evolving homologues that encode highly similar proteins of uncertain function, but possibly involved in chromatin modification as suggested by their co-localization with SATB1 (Escalier et al., 1999). Xlr3b and 4b are expressed and paternally imprinted in the cortex and other brain regions (Davies et al., 2005; Raefski and O'Neill, 2005). Up-regulated expression of Xlr3b in the brain correlates with behavioral defects in a mouse model of Turner syndrome (Davies et al., 2005). No mechanism has been proposed to explain this association, but we reasoned that Xlr genes may be involved in the formation of dendrites and synapses (Chechlacz and Gleeson, 2003; Tada and Sheng, 2006).
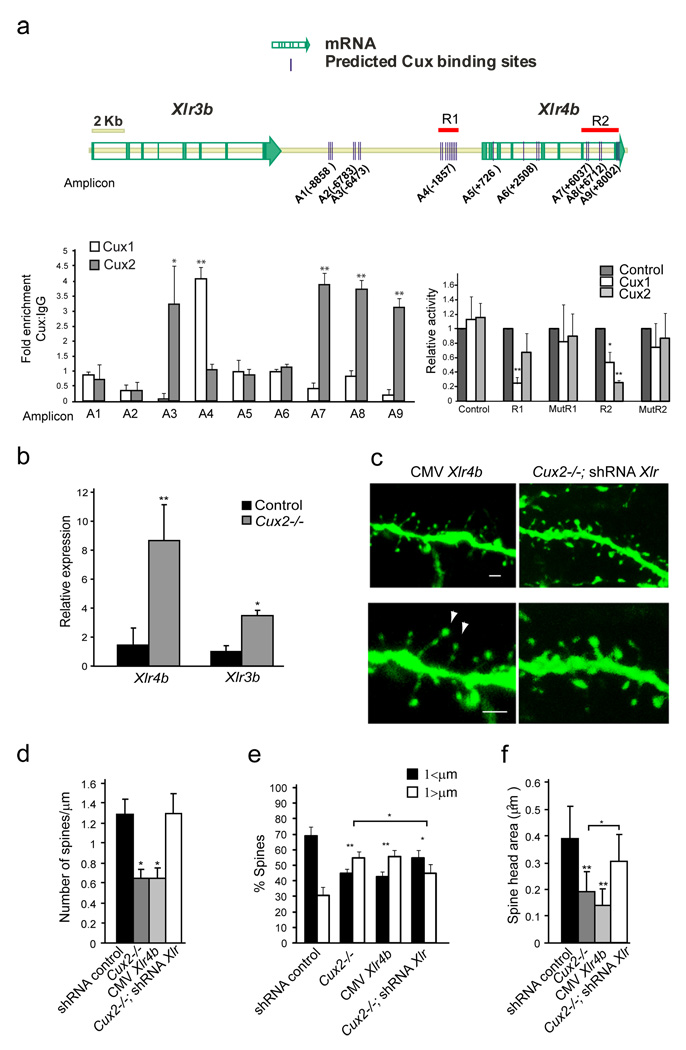
In order to analyze the potential functional relationship between Cux and Xlr3b and Xlr4b genes, we used in silico analysis with Genomatix MatInspector (www.genomatix.de) to identify consensus Cux binding sites. The 3´downstream and 5´upstream, and intronic regions of both Xlr3b and Xlr4b genes contained several consensus Cux binding sequences and some of these in close proximity to each other (Fig 6a). Although Cux proteins can also bind to matrix attachment regions (MARs) (Gingras et al., 2005; Sansregret and Nepveu, 2008), MARs were not identified using the SMARtest (www.genomatix.de). However, potential sites of stress-induced duplex destabilization (SIDD) required in MARs (http://www.genomecenter.ucdavis.edu/) were identified within these sequences, indicating the possibility of this type of transcriptional regulation. Chromatin immunoprecipitation (ChIP) assays with adult cortex demonstrate that both Cux1 and Cux2 proteins bind to regions that contained several consensus Cux binding sites in the Xlr4b locus in vivo (Fig 6a). Similar results were obtained with P7 brain extracts (not shown). Luciferase report assays performed in embryonic primary cortical cells demonstrate that Cux1 specifically repress transcription of a reporter construct containing 1Kb of the Xlr4b genomic locus. This region (R1) corresponds to that identified by ChIP as bound to Cux1 and it is rich in Cux consensus sites. Cux2 protein, and less efficiently Cux1, was able to repress a reporter containing 2.3 Kb (R2) spanning the genomic sequences that includes the three adjacent regions bound to Cux2 by ChIP. Cux1 and Cux2 failed to repress the transcription of mutated forms of these reporters in which Cux binding sites were abolished (mutR1 and mutR2) (Fig 6a). Thus, Cux1 and Cux2 can directly and differentially repress the function of regulatory regions in the Xlr4b locus. In WT cortex, Xlr4b and Xlr3b are expressed at very low levels in all layers (Fig S4A and Allen brain atlas, www.brain-map.org). However, the cortex of Cux2−/− showed an 8-and 1.8-fold increase in the respective expression of Xlr4b and Xlr3b as demonstrated by quantitative real time RT-PCR (Q-PCR) (Fig 6b). There were no significant differences in the levels of Xlr3a expression (not shown), which belongs to the same locus. A smaller increase in Xlr4b was observed in E18 Cux2−/− embryonic cortex, but not Cux1−/−, while Xlr3b expression was augmented in both single Cux1−/− and Cux2−/− E18 cortex (Fig S4C). Altogether, these results strongly suggest that Cux1 and Cux2 negatively and differentially regulate in a stage dependent manner the expression of Xlr3b and Xlr4b genes by direct DNA binding.
Figure 6. Cux1 and Cux2 regulate dendritic spine number and spine morphology through mechanisms that involve the repression of Xlr genes.
a) Cux putative binding sites identified (MatInspector (Genomatrix)) in the genomic region containing the Xlr gene cluster (see graphic). Left diagram, in vivo chromatin immunoprecipitation. 400bp average chromatin fragments were obtained from adult cortex and immunoprecipitation with Cux1 and Cux2 antibodies was performed. Binding to nine regions was tested by Q-PCR. Relative positions of the amplicons (A) to the Xlr4b ATG (+1) are indicated. Real Time PCR reactions were carried out in duplicates in three independent preparations of immunoprecipitated material from three cortexes. The fold enrichment for each tested region was normalized to control IgG.*p<0.01 and **p<0.001 compared to control IgG or region 1. Right graph, luciferase experiments performed in neuronal cells obtained from E12 cortex. Cux1 and Cux2 repress transcriptional activity of luciferase construct reporters containing regions R1 and R2 but not of these reporters when Cux putative sites are mutated (mutR1 and mutR2). *p<0.01 and **p<0.001 b) Up-regulation of Xlr4b and Xlr3b in the adult Cux2−/− cortex. Relative expression of Xlr4b and Xlr3b mRNA is shown in relation to one control sample normalized as 1. Expression of Xlr genes is shown as the ratio of the amounts of Xlr and GADPH transcripts measured by Q-PCR in total RNA obtained from the cortex of adult male Cux2+/− (n=4) and Cux2−/− (n=4) animals. * p<0.2 and ** p<0.05. c) Reduced number and aberrant morphologies of dendritic spines in GFP-positive layer II–III neurons over-expressing Xlr4b in WT animals (left panels). Reverted dendritic spine phenotypes in layer II–III neurons of Cux2−/− electroporated with shRNAs targeting Xlr genes (right panels). Bar represents 1 µm. Arrowheads point to small spine heads. d, e, f) Quantitative analysis of dendritic spine defects in GFP-positive layer II–III neurons with the indicated shRNAs. n≥ 15 dendrite segments and n≥ 500 spines for each sample.* p<0.01 and ** p<0.001 compared to WT or Cux2−/− (brackets).
Xlr genes are downstream effectors of Cux1 and Cux2 in controlling dendritic spine development
To determine whether Xlr4b and Xlr3b are indeed involved in dendrite and spine development downstream of Cux proteins, we asked whether Xlr4b could affect dendrite differentiation and revert the dendritic phenotypes of upper layer neurons of Cux2−/− mice. Xlr4b overexpression severely affected spine number and morphology (Fig 6c, d, e, f and video S5) while it had no effect on the number and length of dendrite branches (Fig S4D). The reduction in spine density upon Xlr4b overexpression was equal to that observed in Cux2-/- neurons or upon in utero knockdown of Cux1 (Fig 6c, d and 4a, b). The proportion of immature spines with long necks and smaller heads also increased after Xlr4b overexpression, beyond that induced by the suppression of Cux2 (Fig 6c, e, f). In contrast, efficient knockdown of Xlr genes in WT cortex with shRNA constructs targeting several of the highly conserved Xlr genes, including Xlr3b and Xlr4b, increased the spine head surface without affecting dendrite branching or dendritic spine density (Fig S4D, E, F), indicating that Xlr genes modulate dendritic spines and suggesting that they might positively regulate the strength and stability of the synapse.
Knockdown of the Xlr genes in layer II–III neurons of Cux2−/− mice and in neurons co-electroporated with shRNA targeting Cux1, rescued the effects of Cux1 or Cux2 suppression, reverting spine density to normal levels and significantly reducing the proportion of immature spines with long spines and small heads (Fig 6c, d, e, f Fig S4G and video S6). Dendritic spine phenotypes were not reverted in Cux2−/− upper layer neurons when Xlr targeting shRNAs were co-electroporated with a mutated resistant form of Xlr4b (Fig S4H and Supplementary Experimental Procedures) excluding possible off-target effects. These results therefore demonstrate that Cux1 and Cux2 control spine and synapse formation partly through the direct transcriptional regulation of Xlr genes, targeting a potentially important mechanism underlying cognition.
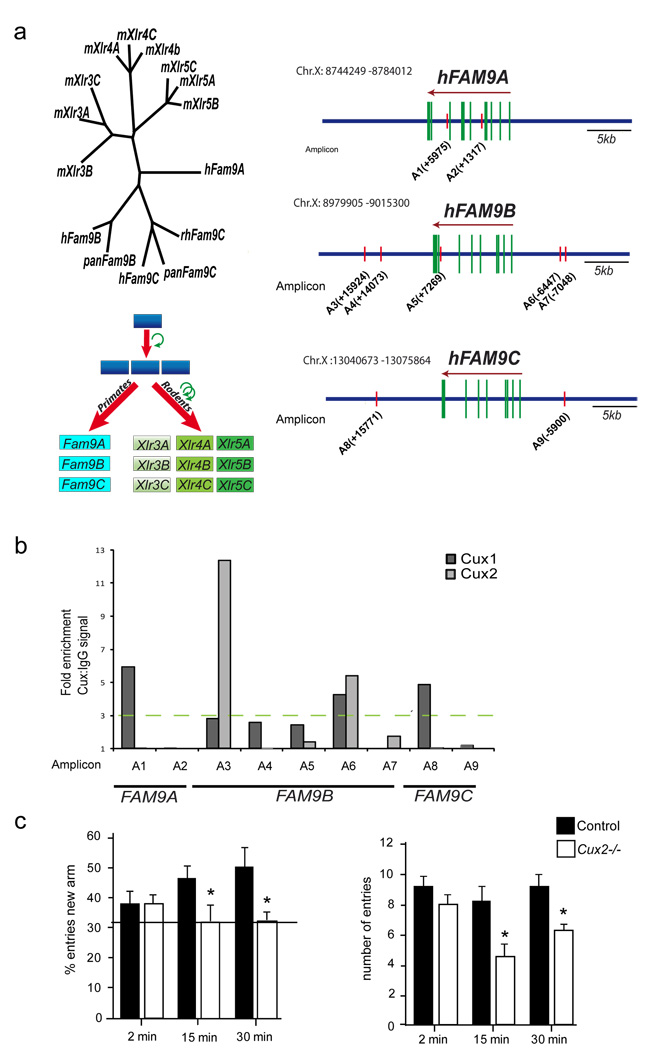
Xlr genes belong to the Cor1 superfamily of proteins (Dobson et al., 1994). Our phylogenetic analysis (Fig 7a) identify the FAM9 family (Martinez-Garay et al., 2002) as the closest orthologs of Xlr genes in humans and primates, as previously proposed (Davies et al., 2006), and indicates that Xlr genes and FAM9 genes may have arisen from common ancestor genes that later duplicated and rapidly evolved in rodents (Fig 7a). We searched for Cux binding sites in FAM9 gene loci and found that their regulatory regions contain potential Cux binding sites conserved between primates and humans (Fig 7a and Fig S5A). In vitro ChIP experiments in human neuroblastoma cell lines demonstrated binding of Cux1 and Cux2 proteins to these regions (Fig 7b). Since hCux2 expression defines the upper layer of the human cortex (Arion et al., 2007), it is possible that similar Cux mediated synaptic mechanisms act in humans.
Figure 7. Human FAM9 genes and cognitive defects.
a) Left diagram shows the phylogenetic relationship between Xlr and FAM9 superfamily members. Below, the possible duplication of an ancestral gene that gave rise to the Xlr and FAM9 orthologous genes. The upper right panel schematizes the location of putative Cux binding sites in FAM9A, B and C genes. b) Immunoprecipitation of the putative binding sites with anti-Cux1 and anti-Cux2 was tested in BE(2)-M17 human neuroblastoma cells transfected with Cux1 or Cux2 and by semi-quantitative PCR (representative experiment of three independent experiments). Relative positions of the amplicons (A) to each ATG (+1) are indicated. c) Cux2−/− mice have defects in working memory. Working memory was assessed in control and Cux2−/− mice with a two–trial memory task based on free-choice exploration of a Y-maze. ITI: inter-trial intervals (see Experimental Procedures). Histograms show the percentage of visits (left panel) and number of total visits to the new arm (right panel). Control and Cux-2−/− animals showed no differences in exploratory behavior (ITI=2 min), but working memory was impaired in Cux-2−/− mice (ITIs of 15 and 30 min).
Abnormal cortical dendrite differentiation in Cux2−/− mice correlates with cognitive defects
Neuronal function and synaptic remodeling in the prefrontal and entorhinal cortex, as well as in the hippocampus, are required for working memory and novelty recognition (Bourne and Harris, 2007; Compte et al., 2000). Cux2 is not expressed in the hippocampus, which appears histologically normal in Cux2−/− mice, and that also shows normal distribution of interneuronal subpopulations (Cubelos et al., 2008a; Nieto et al., 2004) (and unpublished results). Although other subtle and yet undetected developmental defects may exist, we evaluated possible behavioral consequences of the dendritic and spine defects observed in Cux2 cortical deficient neuronal populations, including those of the prefrontal and enthorinal cortex (Fig S5B). Working memory and exploration were evaluated in a Y maze two-trial assay (Dellu et al., 2000) in control and Cux2−/− animals. In the first trial, animals were allowed to explore only two arms of the maze. The ability of animals to recognize a new arm was then evaluated after different inter-trial intervals (ITIs). Exploration capability, assessed after an ITI of 2 min, was similar in control and Cux2−/− animals. However, after an ITI of 15 or 30 min, whereas control animals more often visited the new arm, Cux2−/− animals failed to distinguish the new arm and they entered each arm at random (33% of visits) (Fig 7c). These data demonstrate that working memory was severely impaired in the Cux2−/− mice and indicates that Cux2 influences circuits involved in cognition with potential implications for Cux and Xlr/FAM9 genes in human disorders.
Discussion
We demonstrate that Cux1 and Cux2 regulate fundamental aspects of late neuronal differentiation and control intrinsic mechanisms of dendrite development, spine formation and synaptic function in layers II–III of the cortex. Cux genes control dendrite branching and synaptogenesis by partly independent down-stream mechanisms (Fig 8). This is indicated by the early inhibition of neurite outgrowth induced by Cux down-regulation in P4 neurons, and the fact that the Xlr genes, Cux downstream targets, regulate spine number and morphologies but not branching. The combination of these mechanisms specifies upper layer neuron connectivity and is likely involved in the establishment of cognitive circuits. Our work adds Cux genes to the few TFs known to regulate dendrite branching patterns in vertebrate neuronal subclasses (Chen et al., 2005; Hand et al., 2005; Vrieseling and Arber, 2006). It also highlights novel and specific regulatory mechanisms of dendritic spine formation and synaptic function in restricted neuronal subpopulations.
Figure 8. Cux1 and Cux2 promote dendritic branching and spine differentiation.
Cux1 and Cux2 induce cell autonomous development of dendritic branches and promote dendritic spine development and stabilization in early differentiating neurons by at least partly independent mechanisms. Regulation of Xlr3b and Xlr4b gene expression by Cux proteins contributes to trigger dendritic spine differentiation.
Much of what we know about the development of the specific dendritic architecture of neuronal subclasses comes from studies in Drosophila (Corty et al., 2009; Parrish et al., 2007), but less is known about the specification of the more elaborate dendritic trees of vertebrate neurons (Chen et al., 2005; Hand et al., 2005; Parrish et al., 2007; Vrieseling and Arber, 2006). In Drosophila, increasing levels of Cut expression correlate with increased dendrite branching and number of dendritic spikes, while Cut null mutations have the opposite effect (Grueber et al., 2003). We demonstrate that Cux1 and Cux2 have complementary and additive functions instructing the final complexity of the dendritic arbor, as well as the number of spines. These additive functions and the combinatorial expression of both Cux genes may account for the differences in size of the dendritic arbor and spine densities of upper layer neurons in the specialized areas of the cortex (Benavides-Piccione et al., 2006), as we show for neurons of the cingulate cortex. It remains to be determined if a fine modulation of Cux levels further refines dendritic complexity, equivalent to the mechanisms of action of Drosophila Cut. Nevertheless, our results demonstrate novel interesting evolutionarily conserved role of Drosophila Cut and vertebrate Cux genes in the control of dendrite development of distinct neuronal subclasses (Parrish et al., 2007). It also suggests that the functions of Drosophila Cut specifying simpler neuronal types may have been co-opted to generate the more complex upper layers of the mammalian cortex.
Synaptic modulation and plasticity is considered essential to the formation of specialized circuits and for the regulation of cognitive processes. However, the regulators of these processes are poorly understood (Cingolani and Goda, 2008; Penzes and Jones, 2008). A few other TFs, such as MEF2, have been implicated in activity dependent spine formation and synaptogenesis (Flavell et al., 2006; Shalizi et al., 2006; Tada and Sheng, 2006), but to our knowledge the existence of intrinsic mechanisms functioning specifically in neuronal subclasses have not been proposed or explored. We demonstrate that Cux TFs exert an additive control of the number and morphology of the spine. Importantly, we confirmed that these synapses have the expected decreased in amplitude and frequency in mEPSC predicted by their immature morphology (Fig 5c–h). Thus, the homeobox Cux genes may provide the first examples of neuronal TFs regulating synaptogenesis and the strength of the synapse in a selected subclass of neurons. This suggests that intrinsic neuronal determinants exert an influence in synaptic activity over and above that expected.
Our results demonstrate that Cux genes promote synaptic stability and maturation by mechanisms involving indirect down-regulation of the expression of NMDAR2B, PSD95 proteins (El-Husseini et al., 2000; Ultanir et al., 2007), and more importantly, by direct transcriptional control of Xlr4b and Xlr3b. In vivo and in vitro binding and transcriptional repression of Cux proteins to the regulatory regions of this gene cluster indicates direct mechanisms of gene repression, either by active transcriptional regulation or by the chromatin remodeling action of Cux proteins through binding to MARs, as previously described (Liu et al., 1999; Sansregret and Nepveu, 2008).
The Xlr3, 4 and 5 are a family of highly homologous genes that encode nuclear proteins thought to regulate chromatin remodeling (Escalier et al., 1999; Garchon and Davis, 1989). The imprinted status of the Xlr3b and Xlr4b genes was shown to be temporally dynamic and to regulate their developmental expression in different brain regions (Davies et al., 2005; Raefski and O'Neill, 2005). Interestingly, our results implicate the potential chromatin remodeling functions of Xlr genes in dendritic spine development and synaptogenesis, which may explain the greater behavioral inflexibility associated with the up-regulated expression of Xlr3b genes in a model of Turner syndrome (Davies et al., 2005). Upper layer neurons integrate neuronal circuits that likely contributed to the expansion of mammalian cortical circuits (Hill and Walsh, 2005) and thus, the fine control of their dendritic and synaptic structures seems to have critical consequences. We show that FAM9 genes are the human orthologues of murine Xlr genes. The functions of FAM9 genes are unknown but it is worth mentioning that microdeletions encompassing FAM9B have been noted in cases of autism (Thomas et al., 1999) and schizophrenia (Milunsky et al., 1999). In human cortex, Cux2 expression is restricted to the upper layers (Arion et al., 2007) and we demonstrated that Cux proteins can bind to the conserved Cux binding sequences of human FAM9 genes in neuroblastoma cell lines. These data suggest that similarly to the mouse upper layers,Cux2 might regulate mechanisms of synaptogenesis in human neuronal subpopulations. Finally, although we cannot exclude the contribution of other developmental defects in the circuitry, the cognitive deficiencies of Cux2−/− mice likely reflect both the abnormal branching and synaptic regulation. Our results therefore converge in the idea that Cux genes target developmental mechanisms of dendritogenesis and synaptogenesis relevant for cognition. These developmental mechanisms in turn, specify the functions of the upper layer neurons.
Experimental procedures
Animals
All animal procedures were approved by the Centro Nacional de Biotecnología Animal Care and Use Committee, in compliance with National and European Legislation. Cux2−/− mice (C57BL6 background) have been described previously (Cubelos et al., 2008a). Cux1+/− mice were obtained from A.J. van Wijnen (University of Massachussetts Medical School, MA, USA) (Luong et al., 2002). Morning of the day of the appearance of the vaginal plug was defined as embryonic day (E) 0.5.
Golgi staining, electron microscopy and confocal microscopy
Brains of P60 animals were processed and stained using the FD rapid Golgi Stain kit (FD Neurotechnologies, Inc, MD), and stained sections were matched. Electron microscopy studies weres as described (Cubelos et al., 2005). Quantification of synaptic density and the average length of synaptic junctions was performed as described (DeFelipe et al., 1999). Confocal microscopy was performed with a TCS-SP5 (Leica) Laser Scanning System on a Zeiss Axiovert 200 microscope and 50 µm sections were analyzed by taking 0.2 µm serial optical sections with the Lasaf v1.8 software (Leica).
Morphological analysis
Dendritic processes, spine number, the length and spine head surface of the spines of individual neurons of the somatosensory cortex were measured with LaserPix software (Bio-Rad) in Golgi photographs or confocal reconstructions. Except mentioned, measurements were performed on the primary sensory cortex (Interaural 3.10-2.46, Bregma −0.82∓1.34, according to the mouse atlas of Paxinos and Franklin, 1997). For branching, measurements were only made on neurons with the main apical process parallel to the plane of section, contacting layer I and with at least three basal processes. The cumulative dendritic length of total branches, and the number and cumulative length of primary, secondary and tertiary branches was also measured.
Immunohistochemistry and Western blotting
Perfused brains were processed and sections were stained as described (Cubelos et al., 2008a). Anti-Cux2 was a gift from A. Nepveu (Gingras et al., 2005). SDS–PAGE and Western blotting was performed as described (Cubelos et al., 2005). Anti-NMDR2B (BD transduccion laboratories), rabbit polyclonal anti-GluR1 (Abcam); anti-GluR2 mouse monoclonal (L21/32, NeuroMab, CA); anti-NMDAR1 mouse monoclonal (Upstate), anti-PSD95, anti-GADPH (clone sc-32233, Santa Cruz Biotechnology, Inc, CA), and anti-β-actin (Sigma, St Louis). Bands were visualized by ECL and quantified by densitometry (Molecular Dynamics Image Quant vs. 3.0)
In utero electroporation
In utero electroporation was as described previously (Tabata and Nakajima, 2001). shRNA plasmids (1 µg/µl) were mixed with pCAG-GFP (1 µg/µl). Xlr4b cDNA (GenBank accession BC025576) was from the IMAGE Consortium. Lentiviral shRNA constructs were obtained from Sigma-Aldrich and Open Biosystems (Inc). Mutated resistant forms for Cux1, Cux2 and Xlr4b are described in supplemental experimental procedures. A non-targeting shRNA containing 5 base pair mismatches to any known mouse gene (Sigma-Aldrich) was used as a negative control.
Gene array and Real-time quantitative RT-PCR (Q-PCR)
The microarray data are available on the Gene Expression Omnibus (GEO) website: http://www.ncbi.nlm.nih.gov/projects/geo (accession numbers: GSE14971). 1 µg of total RNA from the cerebral cortex of 3 months old male (Invitrogen) was reverse transcribed with random primers and the superscript reverse transcriptase (Life Technologies). PCR reaction mixtures containing DNA Master Sybr green I mix (Applied Biosystems) were incubated at 95°C for 5 min followed by 40 PCR cycles (5s at 95°C, 45s at 60°C, 90s at 68°C) in an Abi-prism 7000 detector (Applied Biosystems). Specific primers for Xlr4b, Xlr3a, and Xlr3b have been previously described (Davies et al., 2005; Raefski and O'Neill, 2005). The results were normalized as indicated by the parallel amplification of GADPH (5'-TGACGTGCCGCCTGGAGAAA-3', 5'-AGTGTAGCCCAAGATGCCCTTCAG-3’).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed with a commercial kit (Catalog # 17–611, Millipore). The cortex from wild-type mice were minced and cross-linked in 1% formaldehyde (F8775, Sigma) for 15 minutes and stopped adding glycine (0.125 M). Nuclei were precipitated, lysated and sonicated on ice 10 times for 10 seconds (duty cycle 40%, microtip limit 4) (Vibra-Cell V 50, Sonics Materials) (average fragment size of 400bp). 1% of supernatant was saved as input. The immunoprecipitating antibodies were a polyclonal anti-Cux1 (CDP, C-20; sc-6327, Santa Cruz Biotechnology) (see supplemental experimental procedures) and an unrelated goat IgG. The serum of a rabbit immunized against Cux2 (see supplemental experimental procedures) and the serum of the non immunized rabbit. Immunoprecipitates were mixed with protein G magnetic beads and incubated overnight at 4°C, washed and protein/DNA complexes were eluted and cross-links reversed by incubating in ChIP elution buffer plus proteinase K 2 hours at 62°C. DNA was purified using spin columns and analyzed in duplicate by Q-PCR using specific amplicons of 100bp. Primer sequences for amplicons are described in supplemental experimental procedures. Fold enrichment is expressed as the ratio of Cux1 or Cux2 signal to IgG signal 2−(ΔΔCt), where ΔΔCt=CtCux−CtIgG. Results show data obtained from male adult brains and equivalent data was confirmed using adult female brains. Binding of Cux1 and Cux2 protein to human sequences was assessed in human neuroblastoma cells BE(2)-M17. Specific primers on FAMB genes are described in supplemental experimental procedures.
Luciferase reporter assays
Sequence containing Xlr4b regulatory regions (see below) corresponding to those identified in the Chip assays were cloned into the pGL4.23 luciferase vector (Promega). Luciferase activity experiments were performed on neuronal cultures of E12.5 primary cortical cells as described in supplemental experimental procedures.
Electrophysiology
Electrophysiology was performed as described in supplementary experimental procedures from male and female control (WT and Cux2+/−) and Cux2−/− mice (P20) (n=15). Whole cell voltage-clamp recordings were obtained from layer II–III pyramidal cell neurons visually identified using an IR-DIC video microscopy system (Nikon). Cells were filled with Lucifer yellow and analyzed post hoc to confirm morphology and location inlayer II–III. During the recordings each slice was pursued with normal artificial CSF (nACSF) containing 10 mM bicuculline and 1 mm Tetrodotoxine (TTX) to isolate the miniature EPSC (mEPSC) and recorded as described in supplemental experimental procedures. Results are presented as the mean ± SEM. To compare results between cells from different animals, we used unpaired student t-test, and cumulative probability curves with Kolmogorov-Smirnov (K.S.) statistical test with significance level of p < 0.05.
Y-maze protocol
A two-trial memory task, based on free-choice exploration in a Y-maze, was used to study recognition processes and working memory in male individuals as described previously (Dellu et al., 2000). During the first trial (acquisition), the animal is allowed to visit two arms of a Y-maze, the third being blocked with a door. During the second trial (retrieval), the door is opened, and the animal has access to all arms. Discrimination of novelty versus familiarity can then be studied by comparing exploration of the novel arm versus the known arms. Memory can be tested by evaluating the influence on recognition of varying intertrial interval (ITI) between acquisition and retrieval. Exploration was measured after a short (2 min) ITI, while memory was examined at longer ITIs (15 min, 30 min).
Statistical analysis
All results are expressed as the mean ± SD. Experimental groups were compared with Student's two-sample t test and the P values are indicated in figure legends. For analysis of gene expression, raw data were quantile normalized and expression values (log2 transformed) were obtained for each probe. Next, differential expression was assessed using the linear modelling features of the limma package, a package of Bioconductor: http://www.bioconductor.org/.
Supplementary Material
Acknowledgments
We thank B. Alarcón, S. Bartlett, M. Gómez Vicentefranqueira, L. Menendez de la Prida and H. M. van Santen for critical reading of the manuscript and for their experimental advice, A. Nepveu for anti-cux2 antibody and A.J. van Wijnen for the Cux1 mutant mice. We are grateful to S. Gutierrez-Erlandsson M.T. Rejas, M. Guerra F. Ocaña, R. Gutierrez and A. Morales for excellent technical assistance.
This work was supported by the MICINN grants (SAF2005-0094; SAF2008-00211; PIE-200820I166; BFU2007-61774), a grant from Mutua Madrileña Automovilística (0328-2005), and a grant from the Spanish Comunidad de Madrid CCG08-CSIC/SAL-3464. B. Cubelos holds a fellowship from the CSIC (JAEDoc2008-020) and A. Sebastian-Serrano from the MICINN (BES-2006-13901). C.A. W was supported by 2RO1 NS032457 from the NINDS and JM. Redondo by grant SAF2006-08348. C.A.W. is an Investigator of the Howard Hughes Medical Institute. The Centro Nacional de Investigaciones Cardiovasculares is supported by the MICINN and the Pro-CNIC Foundation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ammer AG, Weed SA. Cortactin branches out: roles in regulating protrusive actin dynamics. Cell motility and the cytoskeleton. 2008;65:687–707. doi: 10.1002/cm.20296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arion D, Unger T, Lewis DA, Mirnics K. Molecular markers distinguishing supragranular and infragranular layers in the human prefrontal cortex. The European journal of neuroscience. 2007;25:1843–1854. doi: 10.1111/j.1460-9568.2007.05396.x. [DOI] [PubMed] [Google Scholar]
- Ballesteros-Yanez I, Benavides-Piccione R, Elston GN, Yuste R, DeFelipe J. Density and morphology of dendritic spines in mouse neocortex. Neuroscience. 2006;138:403–409. doi: 10.1016/j.neuroscience.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Benavides-Piccione R, Hamzei-Sichani F, Ballesteros-Yanez I, DeFelipe J, Yuste R. Dendritic size of pyramidal neurons differs among mouse cortical regions. Cereb Cortex. 2006;16:990–1001. doi: 10.1093/cercor/bhj041. [DOI] [PubMed] [Google Scholar]
- Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Current opinion in neurobiology. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nature reviews. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Cline H, Haas K. The regulation of dendritic arbor development and plasticity by glutamatergic synaptic input: a review of the synaptotrophic hypothesis. The Journal of physiology. 2008;586:1509–1517. doi: 10.1113/jphysiol.2007.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compte A, Brunel N, Goldman-Rakic PS, Wang XJ. Synaptic mechanisms and network dynamics underlying spatial working memory in a cortical network model. Cereb Cortex. 2000;10:910–923. doi: 10.1093/cercor/10.9.910. [DOI] [PubMed] [Google Scholar]
- Corty MM, Matthews BJ, Grueber WB. Molecules and mechanisms of dendrite development in Drosophila. Development (Cambridge, England) 2009;136:1049–1061. doi: 10.1242/dev.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Gimenez C, Zafra F. Localization of the GLYT1 glycine transporter at glutamatergic synapses in the rat brain. Cereb Cortex. 2005;15:448–459. doi: 10.1093/cercor/bhh147. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Kim S, Moreno-Ortiz C, Redondo JM, Walsh CA, Nieto M. Cux-2 controls the proliferation of neuronal intermediate precursors of the cortical subventricular zone. Cereb Cortex. 2008a;18:1758–1770. doi: 10.1093/cercor/bhm199. [DOI] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Kim S, Redondo JM, Walsh C, Nieto M. Cux-1 and Cux-2 control the development of Reelin expressing cortical interneurons. Developmental neurobiology. 2008b;68:917–925. doi: 10.1002/dneu.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechlacz M, Gleeson JG. Is mental retardation a defect of synapse structure and function? Pediatric neurology. 2003;29:11–17. doi: 10.1016/s0887-8994(03)00152-8. [DOI] [PubMed] [Google Scholar]
- Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17792–17797. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nature genetics. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- Davies W, Isles AR, Burgoyne PS, Wilkinson LS. X-linked imprinting: effects on brain and behaviour. Bioessays. 2006;28:35–44. doi: 10.1002/bies.20341. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Marco P, Busturia I, Merchan-Perez A. Estimation of the number of synapses in the cerebral cortex: methodological considerations. Cereb Cortex. 1999;9:722–732. doi: 10.1093/cercor/9.7.722. [DOI] [PubMed] [Google Scholar]
- DeFelipe JaJ, G E. Cajal on the Cerebral Cortex. New York Oxford: Oxford university Press; 1988. [Google Scholar]
- Dellu F, Contarino A, Simon H, Koob GF, Gold LH. Genetic differences in response to novelty and spatial memory using a two-trial recognition task in mice. Neurobiology of learning and memory. 2000;73:31–48. doi: 10.1006/nlme.1999.3919. [DOI] [PubMed] [Google Scholar]
- Dierssen M, Ramakers GJ. Dendritic pathology in mental retardation: from molecular genetics to neurobiology. Genes, brain, and behavior. 2006;2(5 Suppl):48–60. doi: 10.1111/j.1601-183X.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. Journal of cell science. 1994;107(Pt 10):2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science (New York, N.Y. 2000;290:1364–1368. [PubMed] [Google Scholar]
- Escalier D, Allenet B, Badrichani A, Garchon HJ. High level expression of the Xlr nuclear protein in immature thymocytes and colocalization with the matrix-associated region-binding SATB1 protein. J Immunol. 1999;162:292–298. [PubMed] [Google Scholar]
- Ferrere A, Vitalis T, Gingras H, Gaspar P, Cases O. Expression of Cux-1 and Cux-2 in the developing somatosensory cortex of normal and barrel-defective mice. The anatomical record. 2006;288:158–165. doi: 10.1002/ar.a.20284. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science (New York, N.Y. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Garchon HJ, Davis MM. The XLR gene product defines a novel set of proteins stabilized in the nucleus by zinc ions. The Journal of cell biology. 1989;108:779–787. doi: 10.1083/jcb.108.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras H, Cases O, Krasilnikova M, Berube G, Nepveu A. Biochemical characterization of the mammalian Cux2 protein. Gene. 2005;344:273–285. doi: 10.1016/j.gene.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Hand R, Bortone D, Mattar P, Nguyen L, Heng JI, Guerrier S, Boutt E, Peters E, Barnes AP, Parras C, et al. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron. 2005;48:45–62. doi: 10.1016/j.neuron.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Hill RS, Walsh CA. Molecular insights into human brain evolution. Nature. 2005;437:64–67. doi: 10.1038/nature04103. [DOI] [PubMed] [Google Scholar]
- Iulianella A, Sharma M, Durnin M, Vanden Heuvel GB, Trainor PA. Cux2 (Cutl2) integrates neural progenitor development with cell-cycle progression during spinal cord neurogenesis. Development (Cambridge, England) 2008;135:729–741. doi: 10.1242/dev.013276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iulianella A, Vanden Heuvel G, Trainor P. Dynamic expression of murine Cux2 in craniofacial, limb, urogenital and neuronal primordia. Gene Expr Patterns. 2003;3:571–577. doi: 10.1016/s1567-133x(03)00123-6. [DOI] [PubMed] [Google Scholar]
- Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW. Knot/Collier and cut control different aspects of dendrite cytoskeleton and synergize to define final arbor shape. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- Komiyama T, Luo L. Intrinsic control of precise dendritic targeting by an ensemble of transcription factors. Curr Biol. 2007;17:278–285. doi: 10.1016/j.cub.2006.11.067. [DOI] [PubMed] [Google Scholar]
- Liu J, Barnett A, Neufeld EJ, Dudley JP. Homeoproteins CDP and SATB1 interact: potential for tissue-specific regulation. Molecular and cellular biology. 1999;19:4918–4926. doi: 10.1128/mcb.19.7.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luong MX, van der Meijden CM, Xing D, Hesselton R, Monuki ES, Jones SN, Lian JB, Stein JL, Stein GS, Neufeld EJ, van Wijnen AJ. Genetic ablation of the CDP/Cux protein C terminus results in hair cycle defects and reduced male fertility. Molecular and cellular biology. 2002;22:1424–1437. doi: 10.1128/mcb.22.5.1424-1437.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Padilla M. Ontogenesis of the pyramidal cell of the mammalian neocortex and developmental cytoarchitectonics: a unifying theory. The Journal of comparative neurology. 1992;321:223–240. doi: 10.1002/cne.903210205. [DOI] [PubMed] [Google Scholar]
- Martinez-Garay I, Jablonka S, Sutajova M, Steuernagel P, Gal A, Kutsche K. A new gene family (FAM9) of low-copy repeats in Xp22.3 expressed exclusively in testis: implications for recombinations in this region. Genomics. 2002;80:259–267. doi: 10.1006/geno.2002.6834. [DOI] [PubMed] [Google Scholar]
- Milunsky J, Huang XL, Wyandt HE, Milunsky A. Schizophrenia susceptibility gene locus at Xp22.3. Clinical genetics. 1999;55:455–460. doi: 10.1034/j.1399-0004.1999.550610.x. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nature reviews. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Nieto M, Monuki ES, Tang H, Imitola J, Haubst N, Khoury SJ, Cunningham J, Gotz M, Walsh CA. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II-IV of the cerebral cortex. The Journal of comparative neurology. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- Parrish JZ, Emoto K, Kim MD, Jan YN. Mechanisms that regulate establishment, maintenance, and remodeling of dendritic fields. Annual review of neuroscience. 2007;30:399–423. doi: 10.1146/annurev.neuro.29.051605.112907. [DOI] [PubMed] [Google Scholar]
- Penzes P, Jones KA. Dendritic spine dynamics--a key role for kalirin-7. Trends in neurosciences. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaggin SE, Heuvel GB, Golden K, Bodmer R, Igarashi P. Primary structure, neural-specific expression, and chromosomal localization of Cux-2, a second murine homeobox gene related to Drosophila cut. The Journal of biological chemistry. 1996;271:22624–22634. doi: 10.1074/jbc.271.37.22624. [DOI] [PubMed] [Google Scholar]
- Raefski AS, O'Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nature genetics. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- Ramon Moliner E. Comparative Methods in Neuroanatomy. New york: Springer; 1970. [Google Scholar]
- Rudolf GD, Cronin CA, Landwehrmeyer GB, Standaert DG, Penney JB, Jr, Young AB. Expression of N-methyl-D-aspartate glutamate receptor subunits in the prefrontal cortex of the rat. Neuroscience. 1996;73:417–427. doi: 10.1016/0306-4522(96)00048-6. [DOI] [PubMed] [Google Scholar]
- Sabatini BL, Maravall M, Svoboda K. Ca(2+) signaling in dendritic spines. Current opinion in neurobiology. 2001;11:349–356. doi: 10.1016/s0959-4388(00)00218-x. [DOI] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science (New York, N.Y. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Current opinion in neurobiology. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Thomas NS, Sharp AJ, Browne CE, Skuse D, Hardie C, Dennis NR. Xp deletions associated with autism in three females. Human genetics. 1999;104:43–48. doi: 10.1007/s004390050908. [DOI] [PubMed] [Google Scholar]
- Ultanir SK, Kim JE, Hall BJ, Deerinck T, Ellisman M, Ghosh A. Regulation of spine morphology and spine density by NMDA receptor signaling in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:19553–19558. doi: 10.1073/pnas.0704031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieseling E, Arber S. Target-induced transcriptional control of dendritic patterning and connectivity in motor neurons by the ETS gene Pea3. Cell. 2006;127:1439–1452. doi: 10.1016/j.cell.2006.10.042. [DOI] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ. N-wasp and the arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. The Journal of biological chemistry. 2008;283:15912–15920. doi: 10.1074/jbc.M801555200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Majewska A, Holthoff K. From form to function: calcium compartmentalization in dendritic spines. Nature neuroscience. 2000;3:653–659. doi: 10.1038/76609. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron MC, Bodmer R, Cremer H. Dynamics of Cux2 expression suggests that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.