Abstract
Botulinum neurotoxin (BoNT) binds peripheral neurons at the neuromuscular junction through a dual-receptor mechanism that includes interactions with ganglioside and protein receptors. The receptor identities vary depending on BoNT serotype (A-G). BoNT/B and BoNT/G bind the luminal domains of Synaptotagmin (Syt)-I and SytII, homologous synaptic vesicle proteins. We observe conditions in which BoNT/B binds both Syt isoforms, but BoNT/G only binds SytI. Both serotypes bind ganglioside GT1b. The BoNT/G receptor-binding domain crystal structure provides a context for examining these binding interactions and a platform to understand the physiological relevance of different Syt receptor isoforms in vivo.
Botulinum neurotoxin (BoNT) is the causative agent of botulism, a potentially lethal neuroparalytic condition in humans (1). The extreme potency of BoNT (LD50 value ∼ 0.1 ng kg-1) stems from the toxin's high affinity for neuronal receptors at the neuromuscular junction and enzymatic inhibition of neurotransmitter release (2). BoNT's are produced as single chain proteins in seven antigenically distinct forms (serotypes A-G). Most BoNT serotypes undergo post-translational cleavage to form a dichain molecule composed of a light- and heavy- chain linked by a disulfide bond. The light chain (LC) is a zinc metalloprotease that cleaves SNARE proteins to inhibit neurotransmitter vesicle fusion to the plasma membrane (3). The N-terminal half of the heavy chain (HCT) is involved in translocation of the LC across the endosomal membrane, and the C-terminal half of the heavy chain (HCR) is involved in binding receptors (4).
BoNT targets the neuromuscular junction through specific interactions with both ganglioside and protein receptors (5). BoNTs bind GD1a and gangliosides in the G1b series and show the highest affinity for the trisialoganglioside, GT1b (6). The protein receptor can vary with BoNT serotype. Synaptotagmin (Syt)-I and SytII mediate the internalization of BoNT/B and/G, but not BoNT/A or/E, into neuronal cells (7-9). SytI and SytII are homologous calcium sensors that couple neuronal calcium influx to the fast phase of neurotransmitter release (10). BoNT/B and/G bind to the luminal domains of SytI and SytII following the fusion of synaptic vesicles with the plasma membrane. The ability of a peptide, corresponding to 20 amino acids of the SytII luminal domain, in conjunction with gangliosides, to neutralize BoNT/B toxicity in mice is consistent with the SytII luminal domain being the neuronal receptor for BoNT/B (7).
BoNT/G is a recently discovered serotype whose HCR shares a high degree of primary amino acid conservation with BoNT/B (50% identical, 71% similar). Despite the homology between BoNT/B and BoNT/G and between SytI and SytII, several differences exist in their interactions with neurons. BoNT/B binds SytII independent of ganglioside but requires ganglioside to bind SytI (7). BoNT/G binds SytI and SytII in a ganglioside-independent manner (8). The ganglioside requirement is thought to reflect a situation where the BoNT-protein receptor affinity is too weak to promote a stable, productive interaction. This hypothesis is supported by isothermal titration calorimetry showing that in the absence of ganglioside the affinity of BoNT/B for SytII is 34 nM, about 100-fold greater than that for SytI. GST pull-down experiments showed that BoNT/G HCR bound both SytI and SytII. This study did not report a preference for either Syt isoform in BoNT/G HCR binding or BoNT/G neutralization (8). In this study, we analyze the crystal structure of BoNT/G HCR to better understand how BoNT/B and BoNT/G differ in their specificities for the two synaptotagmin isoforms and how they recognize their ganglioside co-receptor.
Experimental Procedures
Generation of the BoNT/G HCR expression vector
A previously described BoNT/G HCR pET28a expression vector (11) was modified to delete a 3xFLAG sequence from the N-terminus. This change was made to remove unstructured regions of the protein but also resulted in a significant decrease in protein solubility. In an effort to improve solubility, residues 1080-1084 were mutated from the naturally occurring SSLYW BoNT/G sequence to the sequence observed in BoNT/B, EERYK. Mutations were made with the QuickChange Protocol for site-directed mutagenesis and verified by sequencing.
Protein expression and purification
(BoNT/G HCR) E. coli BL21(DE3) harboring the BoNT/G HCR expression vector were grown in LB broth supplemented with 50 μg/mL kanamycin at 37 °C with shaking. At OD600 of ∼0.6, the temperature was reduced to 16 °C and 1 mM IPTG was added. Cells were harvested after 16-20 hours by centrifugation and lysed by French Press. The protein was bound to HIS-Select™ nickel affinity gel (Sigma-Aldrich) and washed with buffers containing 50 mM KPi pH 8.5, 10 mM imidazole, and 500-700 mM NaCl. BoNT/G HCR was eluted with a buffer containing 50 mM KPi pH 8.5, 50 mM NaCl, and 150 mM imidazole. The resulting eluate was applied to a GE Healthcare HiTrap™ Q HP ion exchange column. The flowthrough was incubated in 6 mM sodium iodoacetate for 30 minutes and immediately injected onto a GE Healthcare HiLoad™ 16/60 Superdex™ 200 size exclusion column using a buffer of 20 mM Tris pH 7.5, 100 mM KCl. (BoNT/A and BoNT/B HCR) The construction of plasmids encoding the BoNT/A and BoNT/B HCR domains (pET-HCR/A and pET-HCR/B) with an N-terminal 6xHis and 3xFLAG sequence has been previously described (11). E. coli BL21(DE3) harboring these plasmids were grown in LB broth supplemented with 50 μg of kanamycin / ml at 30 °C for 2 hours shaking. At OD600 of ∼0.6, the temperature was reduced to 16 °C and 0.1 mM IPTG was added. Cells were harvested after ∼16 hours by centrifugation and lysed with a French press. Proteins were purified using nickel affinity gel and eluted with buffer containing 0.25 M imidazole. Eluted proteins were subjected to S200HR gel filtration (Sephacryl, Sigma, 600 mL resin equilibrated in 10mM Tris-HCl pH 7.9, 0.2 M NaCl, 0.5mM EDTA and 0.1% Triton-X100). The peak fractions were pooled and concentrated using nickel affinity gel. Purified HCR/A and HCR/B were dialyzed into 10 mM Tris-HCl pH 7.9, 0.2 M NaCl and 40% glycerol for storage.
Synaptotagmin binding assay
Three peptides were synthesized with an N-terminal cysteine residue: SytI (cgegkedafsklkekfmnelhk), SytII (cgesqedmfaklkeklfneink), and a scrambled negative control (ckeankgdlkemseglkfehfk) (Biosyn). Peptides were diluted to 10 μg/ml in binding buffer (0.1 M sodium phosphate, 0.15 M sodium chloride, 10 mM EDTA; pH 7.2) and adsorbed to maleimide activated 96-well plates (Pierce) overnight. Unreacted maleimide groups were quenched with a cysteine solution and the wells were washed three times with wash buffer (0.1 M sodium phosphate, 0.15 M sodium chloride, 0.05% Tween®-20; pH 7.2). BoNT HCR proteins were normalized for protein concentration, diluted to 25 μg/ml in wash buffer and bound to wells for 2 hours. Wells were washed and then incubated with 0.5 μM His-Probe (an NTA-HRP conjugate sold by Pierce). The presence of HRP was detected using the 1-Step Ultra TMB-ELISA substrate (Pierce). Each well was assayed 5 times and averaged and the experiment has been repeated numerous times with multiple concentrations of peptide bound and protein incubated.
Ganglioside binding assay
One hundred μl of GT1b (0.1mg/ml, Matreya, Pleasant Gap, PA) in methanol was added to a 96-well, non-binding microtiter plate (Corning, Corning, NY) and allowed to dry overnight. The plate was washed 3× with PBS then blocked with 1 % BSA in 50 mM NaCO3 (pH 9.6) for 1 hr at 4°C. The plate was washed 3× with PBS then the indicated WT-HCR-His-probe conjugate (100ul/well) PBS + 1% BSA 1 hr at 4°C. After washing 3× with PBS, the plate was developed at RT with TMB substrate (100ul/well, Thermo Scientific) for 30 min, stopped with 1M H2SO4, and absorbance was read at 450nm. Data were measured in duplicate, averaged, and non-specific binding in a well without GT1b was subtracted. Data were analyzed using Excel (Microsoft, Seattle, WA) and ranges of HCR yielding dose-dependent GT1b binding were displayed using GraphPad Prism (GraphPad Software Inc, La Jolla, CA).
Crystallization
Purified BoNT/G HCR was concentrated to 11.5 ± 2 mg/mL and mixed 1:1 with mother liquor for hanging drop vapor diffusion crystallization. Reproducible crystals formed with mother liquor containing 12%-15% (w/v) polyethylene glycol 3350, 20 mM Bis-Tris buffer pH 5.75-6.50 and 20-25 mM MgCl. Crystals were preserved in a cryo-protectant of 15% ethylene glycol before being plunged in liquid nitrogen.
Structure determination
X-ray data were obtained from a single crystal at 100K on beamline 21-ID-G of the Advanced Photon Source. Diffraction data were indexed, integrated, scaled and merged with HKL2000 (Table 1) (12). The crystal was space group P212121 and had unit cell dimensions of 57.63 Å, 90.16 Å, and 91.89 Å. Phases were determined by molecular replacement using the BoNT/B HCR structure (13) and the program MOLREP (14). The structure was subjected to iterative rounds of model building in Coot (15) and refinement in Phenix (16). Three TLS groups were assigned 1-213, 214-367, and 368-433 based on tlsmd (17, 18). The refined model (Rcryst=17.47% Rfree=22.08%) consists of amino acids 867-920, 931-1036, 1039-1087, 1090-1157, 1165-1249, 1259-1299 and 348 water molecules.
Table 1. X-ray data collection and refinement statistics.
| Data Collection | Native |
|---|---|
| Wavelength, Å | 0.9780 |
| Resolution (outer shell), Å | 50-2.0 (2.03-2.0) |
| Rmerge*, % | 10.4 (58.5) |
| Mean I/σI | 16.8 (3.4) |
| Completeness, % | 99.9 (100.0) |
| Redundancy | 4.2 (4.0) |
| Unique observations | 33,608 (1,631) |
| Refinement | |
| Rcryst/Rfree, % † | 17.47/22.08 |
| No. protein atoms | 3380 |
| No. ligand atoms | 0 |
| No. solvent waters | 348 |
| Bond length rmsd, Å | 0.007 |
| Bond angle rmsd, ° | 1.070 |
| Avg. protein B, Å2 | 43.5 |
| Ramachandran plot, %‡ | |
| Most favored | 319 (87.9%) |
| Allowed | 44 (12.1%) |
| Generously allowed | 0 (0.0%) |
| Disallowed | 0 (0.0%) |
Outer resolution bin statistics are given in parentheses.
Rmerge = Shkl(Si|Ihkl,i - <Ihkl>))/Shkl,i<Ihkli>, where Ihkl,i, is the intensity of an individual measurement of the reflection with Miller indices h, k and l, and <Ihkl> is the mean intensity of that reflection.
Rcryst = S‖Fobs, hkl| - |Fcalc, hkl‖/|Fobs, hkl|, where |Fobs, hkl| and |Fcalc, hkl| are the observed and calculated structure factor amplitudes. Rfree is equivalent to Rcryst but calculated with reflections (5%) omitted from the refinement process.
Results and Discussion
Initial studies developed a direct binding assay to measure interactions between BoNT/G HCR and either Syt or ganglioside. Synthetic peptides corresponding to the luminal domains of SytI and SytII (or a scrambled negative control) were tethered to a maleimide-activated solid support through an N-terminal cysteine residue. BoNT/G- or BoNT/B- HCR was added in excess and the relative amount of bound HCR was determined. Syt peptide interactions were measured in the absence of ganglioside. This analysis showed that while BoNT/B HCR bound both isoforms of Syt, BoNT/G HCR bound only the SytI peptide (Figure 1a).
Figure 1. Direct assessment of BoNT/G binding to Syt and GT1b.
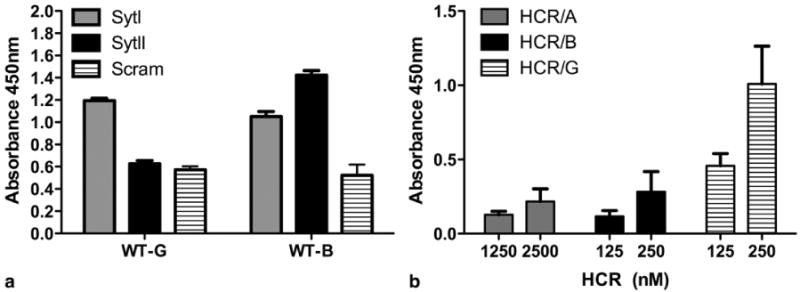
a) Peptides from the luminal domains of SytI or SytII (or a scrambled negative control) were covalently linked to individual wells of maleimide-activated 96-well plates through N-terminal cysteine residues. Unbound peptide was washed away, unreacted maleimide groups were quenched, and wells were incubated with excess BoNT/G- and BoNT/B- HCR (25 μg/ml) for 2 hours at room temperature. Wells were washed 5 times and incubated with His-Probe (an NTA-horse radish peroxidase fusion). Addition of a TMB-ELISA substrate allowed colorimetric detection of bound protein. The average (with standard deviation) for five replicates is shown. b) Non-binding ELISA plates were coated with 10 μg of GT1b / well in methanol and dried overnight. Plates were blocked in carbonate buffer, washed 3× with PBS and incubated with the indicated HCR-Hisprobe conjugate for 1 hr at 4°C. The plate was washed and developed with TMB substrate for 30 min, stopped with 1M H2SO4, and absorbance read at 450nm. Duplicate wells were averaged and subtracted from absorbance detected in an uncoated well. Data are the average of two independent experiments. In both experiments, the mutated form of BoNT/G containing the EERYK solubility sequence was used. The 6xHis sequences are located at the N-terminus of the HCR domains and are not expected to interfere with Syt or ganglioside binding.
That BoNT/B showed a greater interaction with SytII over SytI is consistent with previous reports (7-9). The fact that gangliosides were not required in either interaction could reflect an enhanced sensitivity for the ELISA-based method. We were surprised, however, to see that in our assay, BoNT/G only interacted with SytI. Previous binding interaction studies have been assessed with a GST-pulldown in which GST-Syt fusion proteins are displayed on a glutathione sepharose bead support. Bound proteins are eluted from the beads and subjected to SDS-PAGE (7-9). We assume that if the ELISA-based method were in fact more sensitive, that we would have detected BoNT/G binding to both isoforms of Syt. Since the conditions for binding, washing, and detection differ significantly between the two assays, we are left with the conclusion that the mode in which BoNT/B and BoNT/G recognize SytI and SytII likely differs.
The capacity for BoNT/G HCR to interact with gangliosides (GT1b) was also determined (Figure 1b). BoNT/B and BoNT/G HCRs showed a similar, measureable association, which was greater than the interaction of BoNT/A HCR with GT1b. This was somewhat unexpected, since the BoNT/A HCR is the only serotype that has been co-crystallized in the presence of a GT1b head group (19). This suggested that HCR/B and HCR/G bound GT1b with a higher affinity than HCR/A and motivated our investigation into the structural basis for how BoNT/G interacts with ganglioside.
Thus, the next goal was to obtain a structural context for understanding the Syt and ganglioside binding properties of BoNT/G HCR. However, initial structural studies of BoNT/G HCR were hampered by protein insolubility. In an effort to increase solubility, residues 1080-1084 of BoNT/G HCR were mutated from SSLYW to EERYK. Residues 1080-1084 are located in the α-helix connecting the N-terminal β-barrel domain and C-terminal β-trefoil domain and therefore were not expected to affect the receptor binding sites that are located at the C-terminal region of the HCR. This substitution enhanced the solubility of BoNT/G HCR and allowed conditions to be established to produce a crystal that diffracted to 2.0 Å (Table 1, Figure 2). The BoNT/G HCR structure aligns to the structure of the BoNT/B HCR with an r.m.s.d. of 0.64 Å. The principle differences are located in surface loops of the β-trefoil domain and are highlighted in Figure 2a. Coincident with the efforts in our laboratory, another group determined a structure of the BoNT/G HCR using a different crystal form (20). The structures align with an r.m.s.d. of 0.44 Å and differ in the positions of some surface loops (Figure 2b).
Figure 2. Comparison of BoNT/B and BoNT/G HCR Structures.
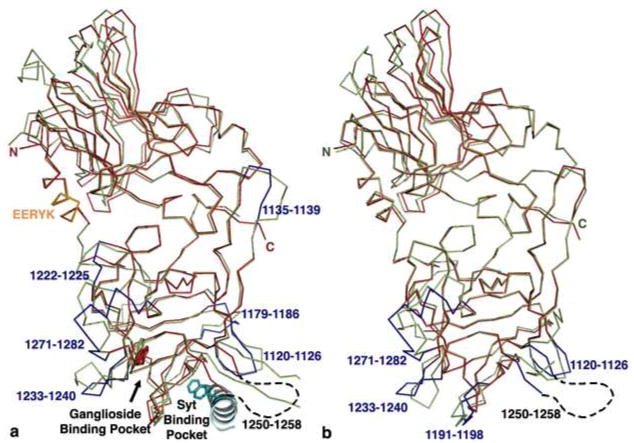
a) The structure of BoNT/G HCR (red) was aligned to the BoNT/B HCR (1EPW; green) and significant loop differences in BoNT/G were colored blue. The helix linking the N- and C- terminal sub-domains (orange) was mutated to EERYK to improve solubility. The position of the SytII helix (teal) was identified in the BoNT/B HCR -SytII crystal structure (2NM1). Two SytII phenylalanine residues (F47 and F54; shown in sticks) dominate the interaction with BoNT/B. A conserved tryptophan residue (shown in sticks) is known to be important for ganglioside binding. b) The structure of BoNT/G HCR (red and blue, this study) was aligned to that from a different crystal form (green, 2VXR). Significant loop differences in the C-terminal sub-domain are colored blue.
Alignments were made between the Syt binding sites of BoNT/B and BoNT/G. Previous structures of BoNT/B bound to a SytII peptide have revealed a two pocket-binding site on BoNT/B (21, 22). The pockets are defined by their interactions with one of two phenylalanine residues, F47 or F54, which are present in many SytI and SytII sequences. In BoNT/B, the pockets are lined by 12 amino acids, five of which are conserved in BoNT/G. In the BoNT/G structure, the five conserved amino acids (Y1189, F1202, A1204, P1205, and F1212) align closely with the position of the corresponding BoNT/B residues and line the F47 pocket (Figure 3). BoNT/B W1178, which is known to contact M46 of SytII, is changed to Y1186 in BoNT/G, which could bear relevance on Syt specificity since in SytI, position 46 is an Ala residue. This change to a smaller residue might better accommodate the charge of BoNT/G E1184 as well. For either isoform, binding will require rearrangement of BoNT/G residue N1207, which is currently poised to clash with F47.
Figure 3. Structural conservation in the Syt binding site.
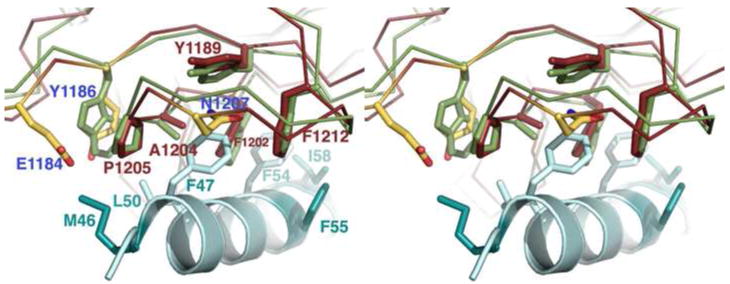
A cross-eye stereo close-up of the F47 binding pocket indicates that five of the 12 residues making contact between BoNT/B and SytII are conserved in both identity and position in the structure of BoNT/G (shown as red or green sticks to represent BoNT/G or BoNT/B, respectively). A sixth contact residue (BoNT/B W1178) is replaced by Y1186 in the BoNT/G structure. BoNT/G residues E1184 and N1207 (colored by atom) do not align with BoNT/B contact residues but are positioned so that they could impact Syt binding. The coordinates for BoNT/B were taken from the 1EPW structure while the SytII peptide (teal) is taken from the aligned 2NM1 structure. Residues that differ between SytI and SytII are shown in dark teal while conserved BoNT/B contact residues are shown in light teal.
Unlike the F47 pocket, where several surface residues are conserved, more differences were observed between the BoNT/B and BoNT/G residues lining the F54 pocket. In this pocket, none of the 6 BoNT/B residues that interacted with SytII were conserved in BoNT/G (Figure 4). Three of these residues (M1126, L1191, and Q1200) have already been tested in a detailed mutagenesis study (9). The individual mutation of each residue (M1126D, L1191R, Q1200E, Q1200K, and Q1200Y) resulted in a significant loss of binding to SytI and SytII in a GST pull-down assay and a detectable loss of toxicity in a mouse phrenic nerve assay. The most significant toxicity defect was observed for Q1200K, a mutation to the corresponding residue in BoNT/B. This led the authors to conclude that Q1200 is central to the specificity differences between BoNT/B and BoNT/G (9).
Figure 4. Structural divergence in the Syt binding site.
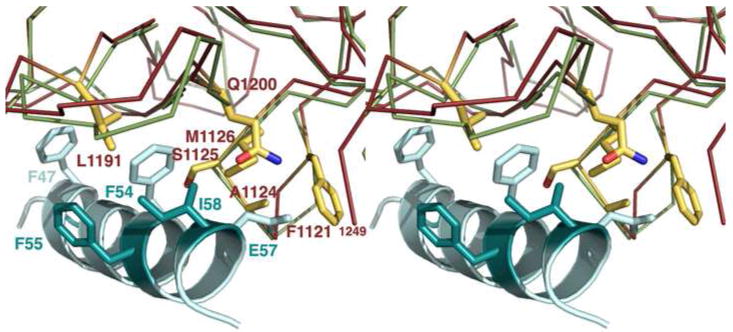
A cross-eye stereo close-up of the F54 binding pocket indicates that the remaining six BoNT/B contact residues differ in BoNT/G. (Only the BoNT/G sidechains are shown for clarity.) Four of the residues that differ (F1121, A1124, S1125, and M1126) are located on a loop whose backbone is positioned differently than the corresponding loop of BoNT/B. The structure from this study is colored red while BoNT/B (1EPW) is colored green. The SytII peptide (teal) is taken from the aligned 2NM1 structure. Residues that differ between SytI and SytII are shown in dark teal while conserved BoNT/B contact residues are shown in light teal.
The three contact residues that were not mutated in the previous study (F1121, A1124, and S1125) are located along with M1126 in the 1120-1126 (YFSKASM) sequence of BoNT/G (Figure 4). We propose that differences in both the side chain and backbone positions of this loop could impact the specificity and affinity for Syt receptors. Another loop that might locate in the vicinity of the F54 pocket was 1250-1258, however this loop was not observed in our electron density maps of BoNT/G HCR (Figure 2a) and was only partially visible and oriented differently in the other crystal form (Figure 2b) (20). In the BoNT/B-SytII structure, contacts were not observed between SytII and the corresponding1244-1253 loop of BoNT/B. Finally, we note that the side chain positions of many of the residues in the F54 pocket (F1121, M1126, L1191, Q1200, and F1202) differ between the two BoNT/G HCR structures. This is the likely result of differences in crystal contacts, present here in both crystal forms, and illustrates that this region is capable of reorganization with a potential for multiple binding modes depending on the nature of the available ligand.
In summary, while BoNT/B is capable of binding both SytI and SytII, our assay of SytI/SytII binding indicates that the BoNT/G HCR specifically binds SytI. Analysis of the BoNT/B and BoNT/G Syt binding sites suggest that specificity differences are likely to stem from differences in the ‘F54 subsite.’ In addition to a number of amino acid differences that have been noted in point mutagenesis studies, we note a significant difference in the position of the BoNT/G 1120-1126 loop relative to the position of the corresponding loop in BoNT/B. We also note that a crystal structure of BoNT/G from another crystal form has a number of structural differences due to crystal contacts. This observation suggests that the region is malleable and can change its structure in response to the properties of its ligand. In light of these data, we suggest that future efforts in deciphering the molecular basis of specificity differences consider the impact of entire loop substitutions in addition to that of single point mutants.
Our assay of ganglioside binding indicates that the interaction of GT1b with BoNT/A is measureable, but of lower affinity than seen with BoNT/B and BoNT/G. There are a number of structural differences in the GT1b binding site that could account for this difference in affinity. A crystal structure of BoNT/A bound to GT1b revealed 8 amino acids involved in direct contacts with the carbohydrate head group (Figure 5a). Only three of these residues are conserved in BoNT/G. Interestingly, two of these residues (W1268 and R1281) in BoNT/G are in different conformations from what has been observed in other structures (Figure 5b). This could be the result of a crystallization contact that likely impacts the position of the R1271-R1281 loop. The altered position of the 1271-1282 loop causes R1281 to be located within the ganglioside binding pocket. This position is different from the other BoNT/G HCR crystal form but is notable in its aligned proximity to BoNT/A R1276, a key residue in the interaction of BoNT/A with ganglioside (Figure 5a). In the other crystal form of BoNT/G HCR, this position is occupied by R1272, suggesting the importance of arginine in supporting the key tryptophan of the ganglioside binding site. We propose that rearrangements in the 1271-1282 loop of BoNT/G could allow the toxin to recognize different ganglioside head groups. The structures of BoNT/B and BoNT/G have more space at the top of the ganglioside-binding site due to the substitution of BoNT/A residues Y1117 and F1252 with glycine and isoleucine, respectively. Finally, we note that BoNT/G has a Gly1246 residue located at the base of the carbohydrate binding pocket, while most BoNT serotypes encode a histidine residue at this position (Figure 5b).
Figure 5. The ganglioside binding site.
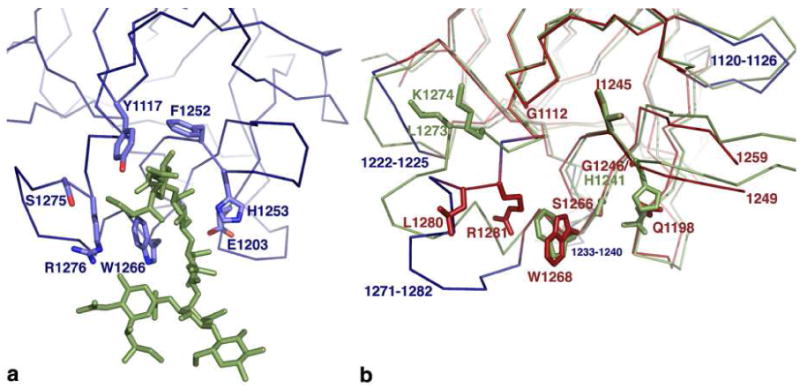
a) The structure of BoNT/A bound to GT1b revealed 8 residues (depicted as sticks) that had contact with the GT1b carbohydrate head group (green). b) The corresponding residues in BoNT/G and BoNT/B are colored red and green, respectively. Most notably, BoNT/G has a glycine (G1246) in a position where BoNT/A, BoNT/B, and TeNT have a Histidine residue. BoNT/G loops that differ significantly from the corresponding BoNT/B loops are colored in blue. The altered position of loop 1271-1282 (blue) more closely resembles that observed in the BoNT/A-GT1b structure and allows Arg1281 to buttress the tryptophan of the ganglioside-binding site.
Our data suggest that BoNT/G binds GT1b with a higher affinity than what has been observed for BoNT/A and that SytI and SytII be considered, not as homologs, but as distinct modes of entry for the BoNT/B and BoNT/G serotypes. A molecular understanding of the host-toxin interaction for botulism will require a detailed knowledge of which receptors are selected preferentially for entry in vivo. Efforts to dissect these in vivo entry mechanisms based on structure-guided mutagenesis are underway.
Acknowledgments
Funding: JTB wishes to acknowledge membership within and support from Region V ‘Great Lakes RCE’ (NIH award 2-U54-AI-057153). DBL was supported by NIH NIAID AI075259.
Abbreviations
- BoNT
botulinum neurotoxin
- Syt
synaptotagmin
- LC
light chain
- HCT
heavy chain translocation domain
- HCR
heavy chain receptor-binding domain
Footnotes
Coordinates and data have been deposited in the Protein Data Bank, entry 3MPP.
References
- 1.Simpson LL. The origin, structure, and pharmacological activity of botulinum toxin. Pharmacol Rev. 1981;33:155–188. [PubMed] [Google Scholar]
- 2.Gill DM. Bacterial toxins: a table of lethal amounts. Microbiol Rev. 1982;46:86–94. doi: 10.1128/mr.46.1.86-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger AT, Jin R, Breidenbach MA. Highly specific interactions between botulinum neurotoxins and synaptic vesicle proteins. Cell Mol Life Sci. 2008;65:2296–2306. doi: 10.1007/s00018-008-8088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montecucco C, Papini E, Schiavo G. Bacterial protein toxins penetrate cells via a four-step mechanism. FEBS Lett. 1994;346:92–98. doi: 10.1016/0014-5793(94)00449-8. [DOI] [PubMed] [Google Scholar]
- 5.Montecucco C. How do tetanus and botulinum neurotoxins bind to neuronal membranes? Trends in Biochemical Sciences. 1986;11:314–317. [Google Scholar]
- 6.Kitamura M, Iwamori M, Nagai Y. Interaction between Clostridium botulinum neurotoxin and gangliosides. Biochim Biophys Acta. 1980;628:328–335. doi: 10.1016/0304-4165(80)90382-7. [DOI] [PubMed] [Google Scholar]
- 7.Dong M, Richards DA, Goodnough MC, Tepp WH, Johnson EA, Chapman ER. Synaptotagmins I and II mediate entry of botulinum neurotoxin B into cells. J Cell Biol. 2003;162:1293–1303. doi: 10.1083/jcb.200305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rummel A, Karnath T, Henke T, Bigalke H, Binz T. Synaptotagmins I and II act as nerve cell receptors for botulinum neurotoxin G. J Biol Chem. 2004;279:30865–30870. doi: 10.1074/jbc.M403945200. [DOI] [PubMed] [Google Scholar]
- 9.Rummel A, Eichner T, Weil T, Karnath T, Gutcaits A, Mahrhold S, Sandhoff K, Proia RL, Acharya KR, Bigalke H, Binz T. Proceedings of the National Academy of Sciences of the United States of America. Vol. 104. 2007. Identification of the protein receptor binding site of botulinum neurotoxins B and G proves the double-receptor concept; pp. 359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 11.Baldwin MR, Tepp WH, Przedpelski A, Pier CL, Bradshaw M, Johnson EA, Barbieri JT. Subunit vaccine against the seven serotypes of botulism. Infect Immun. 2008;76:1314–1318. doi: 10.1128/IAI.01025-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 13.Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nature structural biology. 2000;7:693–699. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- 14.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 15.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 17.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 18.Painter J, Merritt EA. A molecular viewer for the analysis of TLS rigid-body motion in macromolecules. Acta Crystallogr D Biol Crystallogr. 2005;61:465–471. doi: 10.1107/S0907444905001897. [DOI] [PubMed] [Google Scholar]
- 19.Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenmark P, Dong M, Dupuy J, Chapman ER, Stevens RC. Crystal structure of the botulinum neurotoxin type G binding domain: insight into cell surface binding. J Mol Biol. 397:1287–1297. doi: 10.1016/j.jmb.2010.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- 22.Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]


