Abstract
Cells require ribonucleotide reductase (RNR) activity for DNA replication. In bacteria, electrons can flow from NADPH to RNR by either a thioredoxin-reductase- or a glutathione-reductase-dependent route. Yeast and plants artificially lacking thioredoxin reductases exhibit a slow-growth phenotype, suggesting glutathione-reductase-dependent routes are poor at supporting DNA replication in these organisms. We have studied proliferation of thioredoxin-reductase-1 (Txnrd1)-deficient hepatocytes in mice. During development and regeneration, normal mice and mice having Txnrd1-deficient hepatocytes exhibited similar liver growth rates. Proportions of hepatocytes that immunostained for PCNA, phosphohistone H3 or incorporated BrdU were also similar, indicating livers of either genotype had similar levels of proliferative, S and M phase hepatocytes, respectively. Replication was blocked by hydroxyurea, confirming that RNR activity was required by Txnrd1-deficient hepatocytes. Regenerative thymidine incorporation was similar in normal and Txnrd1-deficient livers, further indicating that DNA synthesis was unaffected. Using genetic chimeras in which a fluorescently marked subset of hepatocytes was Txnrd1-deficient while others were not, we found that the multigenerational contributions of both hepatocyte types to development and to liver regeneration were indistinguishable. We conclude that, in mouse hepatocytes, a Txnrd1-independent route for the supply of electrons to RNR can fully support DNA replication and normal proliferative growth.
Keywords: Ribonucleotide reductase, Liver regeneration, Partial hepatectomy, DNA precursors, Cell cycle, Replication, Proliferation
Introduction
Conversion of ribonucleotides into deoxyribonucleotides by ribonucleotide reductase (RNR) is essential for genomic DNA replication (Arner and Eriksson, 1995; Holmgren, 1981; Nicander and Reichard, 1983; Thelander and Reichard, 1979). Thioredoxin was first discovered as a source of electrons for RNR in E. coli (Laurent et al., 1964) and thioredoxin reductase was identified as the flavin-containing enzyme that used electrons from NADPH to restore the reduced state of oxidized thioredoxin (Moore et al., 1964). Subsequent studies showed that a glutathione-reductase-dependent source of electrons could also support RNR activity and DNA replication. In this pathway, glutathione reductase transfers electrons from NADPH to glutathione, glutathione reduces glutaredoxin, and glutaredoxin donates the electrons to RNR (Holmgren, 1976; Miranda-Vizuete et al., 1994; Miranda-Vizuete et al., 1996).
Similar to bacteria, yeast and plants can survive in the absence of a functional thioredoxin-thioredoxin reductase pathway by using the glutathione-reductase-dependent source of electrons to fuel RNR (Koc et al., 2006; Meyer et al., 2009; Reichheld et al., 2007). Biochemical studies showed that mammalian RNR can also obtain electrons from the glutathione-reductase-dependent pathway (Luthman et al., 1979; Luthman and Holmgren, 1982) and a recent kinetic study showed that either route supports similar RNR enzymatic efficiency in vitro (Avval and Holmgren, 2009). Thus, it appears that most life forms might be able to use either thioredoxin reductase- or glutathione-reductase-dependent pathways to drive RNR-dependent DNA precursor production. However, none of the mammalian biochemical systems used to date have been able to approach the rate of RNR activity that would be required to support S phase replication in vivo (Avval and Holmgren, 2009), and the effectiveness of either source is not necessarily equivalent in all species (Arner, 2009; Fernandes and Holmgren, 2004).
Although the electrons supplied to RNR by either the thioredoxin reductase- or the glutathione-reductase-dependent route can support DNA replication, eukaryotic systems studied to date tended to favor the thioredoxin-reductase-dependent pathway (Arner, 2009; Arner and Holmgren, 2006a; Berndt et al., 2007; Lu and Holmgren, 2009). For example, yeast lacking thioredoxins exhibit a reduced rate of proliferation, skewed deoxyribonucleotide pools, and a protracted S phase, consistent with the glutathione-reductase-dependent pathway being a poor provider of DNA precursors (Koc et al., 2006). Measurements of the in vivo redox state of RNR verified that thioredoxin is normally the major direct provider of electrons for RNR in yeast (Camier et al., 2007). Similarly, plants lacking thioredoxin reductases show a slow-growth phenotype that is consistent with compromised replication (Reichheld et al., 2007).
The biological world contains two known families of thioredoxin reductase enzymes (Arner, 2009). One, a smaller protein with high substrate fidelity for thioredoxin, is found in Archaea, eubacteria, plants, protists and fungi. The other is a larger enzyme that evolved independently from glutathione reductase, has higher substrate promiscuity, and has been found universally in metazoan animals (Aleksunes and Manautou, 2007; Arner, 2009; Lobanov et al., 2009; Lu and Holmgren, 2009; Meyer et al., 2009). It remains unclear why an ancestral pre-metazoan animal might have discarded its thioredoxin reductase genes (both cytoplasmic and mitochondrial forms of the enzyme) and generated a new lower fidelity family of enzymes from glutathione reductase; however, this event might underlie fundamental differences in the thioredoxin-reductase-pathway functions between animals and other life forms.
Previous studies suggested that, like yeast and plants, metazoans might also preferentially use their thioredoxin-reductase-dependent pathway to fuel RNR. Thus, although mice lacking glutathione reductase are viable (Rogers et al., 2004), mice lacking thioredoxin 1 (Txn1) (Matsui et al., 1996) or thioredoxin reductase 1 (Txnrd1) (Bondareva et al., 2007; Jakupoglu et al., 2005) are embryonic lethal. Lethality, however, does not necessarily implicate an RNR activity deficiency. Txnd1 also participates in reducing protein disulfides and supplying electrons to homeostatic and antioxidant systems, including peroxiredoxins, methionine sulfoxide reductases and others (Arner, 2009; Fomenko et al., 2009; Kim and Gladyshev, 2007; Rundlof and Arner, 2004). We recently showed that disruption of Txnrd1 in mouse hepatocytes or fibroblasts results in chronic induction of cytoprotective pathways, which are thought to compensate for the loss of Txnrd1 in homeostatic antioxidant functions (Suvorova et al., 2009). Embryonic lethality in Txnrd1-deficient mouse embryos occurs only after replication has generated several thousand (Bondareva et al., 2007) or more (Jakupoglu et al., 2005) Txnrd1-deficient cells, again suggesting that functions other than DNA precursor production underlie lethality. Similarly, disruption of the genes encoding either mitochondrial thioredoxin (Txn2) (Nonn et al., 2003) or mitochondrial thioredoxin reductase (Txnrd2) (Conrad et al., 2004) are also embryonic lethal despite these proteins being generally located in a compartment (mitochondria) separate from that of ribonucleotide reduction for genomic DNA replication (cytosol) (Arner, 2009).
Our recent analyses of mice having hepatocyte-specific disruption of the Txnrd1 gene showed strikingly normal liver development, including apparently normal levels of endoreplication and acytokinetic mitosis in mature Txnrd1-deficient hepatocytes (Suvorova et al., 2009). Also, although RNAi knockdown of Txnrd1 in mouse cancer cell cultures inhibits self-sufficient growth and other cancer-related characteristics of the cells, it does not block proliferation (Yoo et al., 2006; Yoo et al., 2007). These studies could not rule out the possibility that replication was supported by Txnrd1 enzyme remaining from maternal (Bondareva et al., 2007; Jakupoglu et al., 2005), pre-hepatocytic (Suvorova et al., 2009) or residual (Yoo et al., 2006; Yoo et al., 2007) sources; however, they provided the first in vivo evidence to suggest that Txnrd1-deficient mammalian cells might be able to undergo genome replication. To determine if this was, indeed, true, we evaluated the requirement of Txnrd1 for replication in hepatocytes.
In the current study, we tested whether hepatocytes in mouse liver were able to proliferate in the absence of Txnrd1. Experimental systems were designed in which cells had been Txnrd1-deficient for extended periods and, in some cases, for many cell cycles, to rule out any possibility that persisting pre-formed Txnrd1 supported replication. Results showed that Txnrd1−/− hepatocytes were able to proliferate and, unlike proliferation in Txnrd-deficient yeast or plants (Koc et al., 2006; Reichheld et al., 2007), hepatocyte proliferative indexes were not measurably affected by loss of Txnrd1. Our results indicate that the Txnrd1-independent route supplying electrons for RNR can support normal replication in a mammalian system. The implications of this finding should be considered in therapies using drugs that inhibit or otherwise affect Txnrd1 activity.
Several different abbreviations are currently used for thioredoxin reductases, including Txnrd, TxnR, TR and TRR. We have used the abbreviations for mouse enzymes recommended by the National Center for Biotechnology Information (NCBI) and the Mouse Genome Informatics database.
Genetic designations used in this paper are as follows. Gene loci are italicized and separated by a semicolon. Alleles are designated as superscript and separated by a slash. Transgenes are followed by a superscript to indicate whether they are homozygous present (2), hemizygous (1) or absent (0). For example, Txnrd1+/−;AlbCre0 is heterozygous for a null (−) allele of Txnrd1 and does not carry an AlbCre transgene.
Results
Txnrd1-deficient hepatocyte populations expand and replicate DNA during liver development
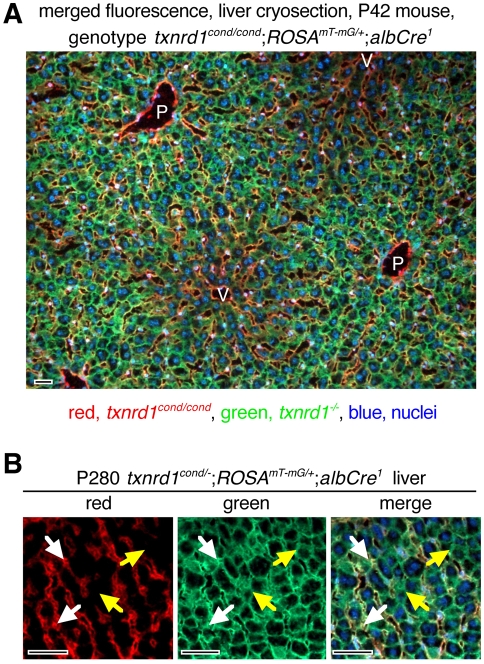
We previously established a mouse (Mus musculus, strain C57Bl/6J) system in which all differentiated hepatocytes lack Txnrd1 (Suvorova et al., 2009). Briefly, female animals that are homozygous for the functionally wild-type yet Cre-inactivatable (‘conditional’) Txnrd1cond allele (Bondareva et al., 2007) and for a two-color Cre-responsive reporter gene, ROSAmT-mG (Muzumdar et al., 2007) (genotype Txnrd1cond/cond;ROSAmT-mG/mT-mG) were mated with males that were heterozygous for the null allele of Txnrd1 (Bondareva et al., 2007) and carried two copies of an albumin promoter/enhancer-driven Cre transgene (Postic et al., 1999) (genotype Txnrd1+/−;AlbCre2). Offspring were either Txnrd1cond/−;ROSAmT-mG/+;AlbCre1 (hereafter referred to as experimental) or Txnrd1cond/+;ROSAmT-mG/+;AlbCre1 (control). The AlbCre transgene drives Cre expression in all differentiated hepatocytes (Postic and Magnuson, 2000; Postic et al., 1999) including those that arise in fetal stages (Weisend et al., 2009). The ROSAmT-mG allele, in its non-recombined state, drives strong membrane-targeted accumulation of the red fluorescent protein tdTomato in all cell types (Muzumdar et al., 2007). Exposure to Cre deletes the tdTomato cistron and uncovers a cistron that yields a membrane-associated modified green fluorescent protein (GFP). Thus, in experimental animals, all cells are Txnrd1cond/− and red fluorescent, except differentiated hepatocytes, which are Txnrd1−/− and green fluorescent (Suvorova et al., 2009). Previous studies showed that livers from these mice expressed almost no functional Txnrd1 mRNA or protein, suggesting all hepatocytes completely lacked Txnrd1 (Suvorova et al., 2009). In control animals all cells are red fluorescent and Txnrd1cond/+ except differentiated hepatocytes, which are green-fluorescent and Txnrd1+/− (Suvorova et al., 2009). In these studies as well as in previous studies, we have been unable to measure a difference in phenotype between Txnrd1+/−, Txnrd1cond/−, Txnrd1cond/+, Txnrd1cond/cond and Txnrd1+/+ systems (Bondareva et al., 2007; Suvorova et al., 2009) (data not shown), suggesting that hepatocytes hemizygous for an active Txnrd1 allele (i.e. either Txnrd1+/− or Txnrd1cond/−) represent a valid Txnrd1-containing control. Sections of liver from control (not shown) or experimental (Fig. 1A,B) adults showed that nearly all hepatocytes, which are identified as very large and often cuboidal cells with large heterogeneous nuclei, had green fluorescent membranes (Fig. 1B, center panel). Endothelial cells surrounding capillaries, which are identified as very small cells that form tubular networks between hepatocytes, and other non-hepatocyte cell types had red membranes (Fig. 1B, left panel) (Suvorova et al., 2009; Weisend et al., 2009). Hepatocytes in zones surrounding portal circulation exhibited stronger green fluorescence than those in zones surrounding venous circulation, whereas red fluorescence in endothelial cells appeared uniform (Fig. 1A and not shown). This gives the impression that venous zones are less green than portal zones, however, careful examination indicated that nearly all hepatocyte membranes were purely green (Fig. 1, and not shown).
Fig. 1.
Hepatocyte-specific allelic conversion in mouse liver by AlbCre. (A) Low magnification micrograph showing merged fluorescence and DAPI staining of a cryosection of liver from an adult (P42) animal of genotype Txnrd1cond/cond;ROSAmT-mG/+;AlbCre1. All hepatocytes in this image, identified as large and generally cuboidal cells with abundant cytoplasm with large heterogeneous weakly DAPI-stained nuclei, and frequently having two nuclei or oversized polyploid nuclei, have green fluorescent membranes, which indicates that AlbCre efficiently directed Cre-mediated allelic conversion in hepatocytes. All non-hepatocyte cells, in particular the endothelial cells, identified as very small irregularly shaped cells with small strongly DAPI-stained nuclei, which form tubular networks surrounding blood vessels and capillaries, exhibited red fluorescence. Note that the intensity of green fluorescence varies zonally as a function of the relative distance from portal (P; arterial) versus venous (V) circulation. Because the intensity of red fluorescence in non-hepatocyte cells did not vary similarly, the peri-venous zones appear darker and redder. We are uncertain of the causes of this hepatocyte-specific zonal fluorescence variation, although it might reflect zonal differences in metabolic activity of hepatocytes resulting from natural gradients in oxygen tension or nutrient availability. Careful examination revealed that nearly all hepatocytes in all zones had exclusively green membranes. (B) High-magnification fluorescent micrographs of a DAPI-stained liver cryosection from an aged adult (P280) animal of genotype Txnrd1cond/−;ROSAmT-mG/+;AlbCre1. The red channel (left) shows endothelial membranes surrounding capillaries. The green channel (middle) shows hepatocyte membranes. The merged channel (right) shows that hepatocyte-endothelial interfaces have both red and green membranes (white arrows), whereas hepatocyte-hepatocyte interfaces have only green membranes (yellow arrows). Scale bars: 50 μm.
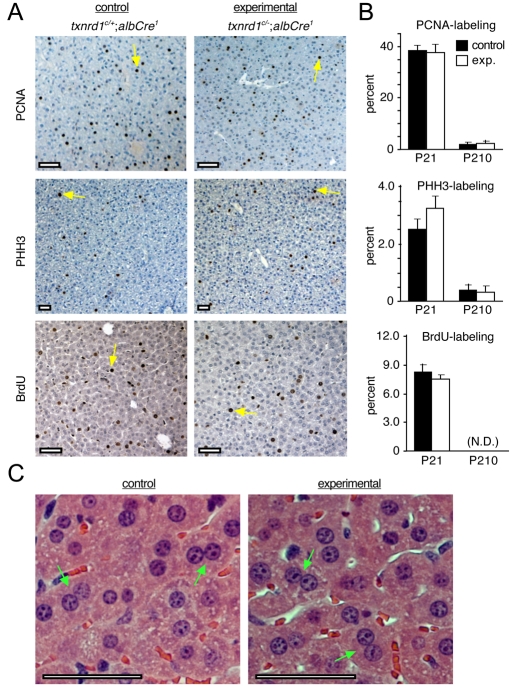
Adult experimental livers were subtly larger than control livers (see below). Juvenile experimental livers showed normal proportions of proliferative cells, as distinguished by the presence of mitotic figures (data not shown), immunostaining for proliferative cell nuclear antigen (PCNA, a marker of proliferative and recently proliferative cells), immunostaining for phosphohistone H3 (PHH3; a marker of M phase cells), and incorporation of bromodeoxyuridine (BrdU; a marker of S phase cells) in hepatocytes (Fig. 2A,B). This suggested that proliferative genesis of new hepatocytes was unaffected by disruption of Txnrd1. However, new hepatocytes can arise either by proliferation of existing differentiated hepatocytes or by differentiation from a self-renewing pre-hepatocytic progenitor or stem-cell population, sometimes referred to as ‘oval cells’ or ‘bi-potential cells’ (Michalopoulos, 2007; Snykers et al., 2009; Suzuki et al., 2008). Here we counted only cells having a differentiated hepatocyte morphology (Fig. 2A, yellow arrows, and see below). Also, histological comparisons of livers from adult experimental and control mice showed similar numbers of bi- and large-nucleate hepatocytes (Fig. 2C). These are polyploid cells that arise normally by acytokinetic mitosis or endoreplication, respectively, of differentiated hepatocytes (Evans, 1976; Melchiorri et al., 1993; Severin et al., 1984; Watanabe, 1970). Since only differentiated hepatocytes, not pre-hepatocytic cells, are known to undergo these atypical cell cycles, the presence of polyploid hepatocytes in experimental mice strongly suggested that genome replication occurred in the absence of Txnrd1 (see below).
Fig. 2.
Developmental proliferation of Txnrd1-deficient hepatocytes. (A) Mice of genotypes Txnrd1cond/+;AlbCre1 (left panels, controls) or Txnrd1cond/−;AlbCre1 (right panels, experimentals) were killed at P21 and liver sections were immunostained for PCNA (upper panels; marker of proliferative cells), for PHH3 (middle panels; M phase marker), or for BrdU inorporation (lower panels; S phase marker). Yellow arrows indicate representative hepatocytes that stained for PCNA (upper panels), PHH3 (middle panels), or BrdU (lower panels). (B) Proliferative indexes during liver development. The percentage of hepatocytes labeled for each proliferative marker in P21 livers were quantified; non-hepatocyte cells were excluded from the counts. Values are means and s.e.m. of at least three animals under each condition, as described in Materials and Methods. N.D., not determined. Statistical analysis indicated that none of the differences between control and experimental samples was significant (P>0.05). (C) Histology of control and experimental livers. Green arrows denote representative bi-nucleate hepatocytes. Scale bars: 100 μm in all panels.
Contributions of Txnrd1-deficient hepatocytes versus Txnrd1-containing progenitor cells to liver regeneration
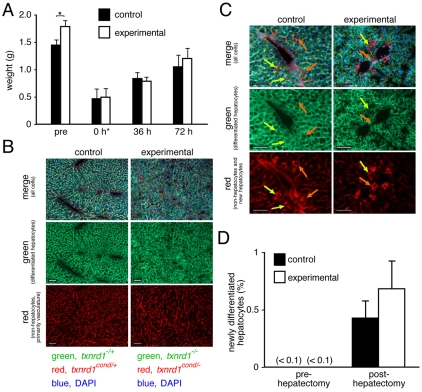
To induce global proliferation of differentiated hepatocytes, we performed partial hepatectomies on adult mice. Following surgical removal of two-thirds of normal rodent liver, most differentiated hepatocytes rapidly enter the cell cycle and proliferate to regenerate liver mass (Higgins and Anderson, 1931; Martins et al., 2008; Mitchell and Willenbring, 2008; Watanabe, 1970). DNA synthesis peaks around 36 hours post-hepatectomy as the first semi-synchronous wave of hepatocytes transits S phase, and proliferation remains elevated until regeneration is complete, after 7-10 days (Mitchell and Willenbring, 2008). At hepatectomy, experimental liver masses were ~24% greater than control liver masses; rates of liver mass accumulation following a two-thirds hepatectomy were similar in experimental and control mice (Fig. 3A), suggesting that Txnrd1-deficient livers were not compromised in regenerative tissue growth.
Fig. 3.
Contributions of Txnrd1−/− hepatocytes to liver regeneration. (A) Liver mass accumulation following two-thirds hepatectomy of adult mice of genotype Txnrd1cond/+;ROSAmT-mG/+;AlbCre1 (control) or Txnrd1cond/−;ROSAmT-mG/+;AlbCre1 (experimental). Bars at the left (‘pre’) show liver masses of mice not subjected to hepatectomy (n=41 controls and 31 experimentals). For post-hepatectomy samples, mice were two-third-hepatectomized and harvested after either 36 (±4) or 72 (±4) hours of recovery (n=3-5 mice for each condition). Traces of necrotic tissue distal to the hepatectomy ligations were carefully removed and livers were weighed. Freshly hepatectomized (0 h*) liver mass was not measured, but rather, was estimated by subtraction of the total mass of the resected lobes from the average mass of resting livers (‘pre’ values). Because small amounts of tissue distal to the points of ligation are always left behind, these values somewhat overestimate the mass of live liver remaining after hepatectomy. Pre-hepatectomy experimental livers were significantly larger than control livers (mean 1.46 g versus 1.81 g; P<0.05). None of the differences between post-hepatectomy control and experimental samples was significant. (B) Liver regeneration in adult mice having normal (controls, left panels) or Txnrd1-deficient (experimentals, right panels) hepatocytes. Adult (P42) mice were two-thirds hepatectomized. Livers were harvested 48 hours later and DAPI-stained cryosections were evaluated by fluorescence microscopy. Based on our independently calibrated 10-day chronometer of red-to-green conversion in hepatocytes (see text), all newly differentiating hepatocytes during a 48-hour regeneration period would have predominantly red membranes. Results showed that nearly all hepatocytes in both experimental and control mice were green, indicating that they were of cell lineages that had differentiated into hepatocytes prior to the hepatectomy. At the bottom the genotypes that can be inferred from the fluorescence staining of the membranes is indicated. (C,D) Contributions of newly differentiated hepatocytes to liver regeneration. Isolated ‘pockets’ of newly differentiated hepatocytes could be found in scattered periportal regions of regenerating livers of either genotype. Orange and light-green arrows indicate apparent hepatocytes with membranes that were predominantly or partially red fluorescent, respectively. These cells were quantified in arbitrary fields of view (C). Statistical analysis indicated that the difference between control and experimental post-hepatectomy samples was not significant (P>0.05). Scale bars: 100 μm in all panels; all graphical data show mean and s.e.m. for at least three animals.
Liver growth can occur by either proliferation of differentiated hepatocytes or proliferation of pre-hepatocytic progenitor cells followed by differentiation into hepatocytes (Oertel and Shafritz, 2008). Because allelic conversion was driven by AlbCre, which is induced coincident with hepatocyte differentiation (Postic et al., 1999; Weisend et al., 2009), proliferating progenitor cells would contain Txnrd1. To distinguish contributions of Txnrd1-containing pre-hepatocytic progenitor cells from proliferation of Txnrd1-deficient differentiated hepatocytes, we developed a novel ‘ten-day chronometer’ of hepatocyte lineage age based on conversion of the ROSAmT-mG allele by AlbCre (S. V. Iverson, K.M.C., C.M.W., J.A.K. and E.E.S., unpublished data). Briefly, activation of the AlbCre transgene during hepatocyte differentiation (Postic et al., 1999; Weisend et al., 2009) will simultaneously convert both the Txnrd1cond allele and the ROSAmT-mG allele (Suvorova et al., 2009; Weisend et al., 2009). Cre-mediated recombination of ROSAmT-mG initiates a progressive conversion in membrane fluorescence from red to green as GFP accumulates and tdTomato protein decays. Fluorescent marker conversion in hepatocyte membranes required roughly 10 days to complete [(Muzumdar et al., 2007) and data not shown]. Therefore, 48 hours after partial hepatectomy, hepatocytes arising from pre-hepatocytic progenitor cells (0- to 48-hours post-onset of Cre expression) will exhibit low ratios of green/red fluorescence and could be readily distinguished from the purely green hepatocytes arising from proliferation of preexisting hepatocytes. At this time point, nearly all hepatocyte membranes exhibited purely green fluorescence (Fig. 3B). At low incidences, fields of view in some peri-portal regions could be found with newly differentiated hepatocytes [i.e. retaining some red membrane fluorescence; Fig. 3C, orange arrows denote younger (redder) hepatocytes than the light-green arrows], verifying that contributions of progenitor cells to liver regeneration could be detected using this system. Importantly, frequencies of newly differentiated hepatocytes were low during regeneration (~1% of total hepatocytes) and did not differ significantly between experimental and control livers (Fig. 3C,D). Most hepatocytes 2 days after partial hepatectomy arose from cell lineages that had differentiated into hepatocytes prior to hepatectomy; pre-hepatocytic progenitor cells were only minor contributors.
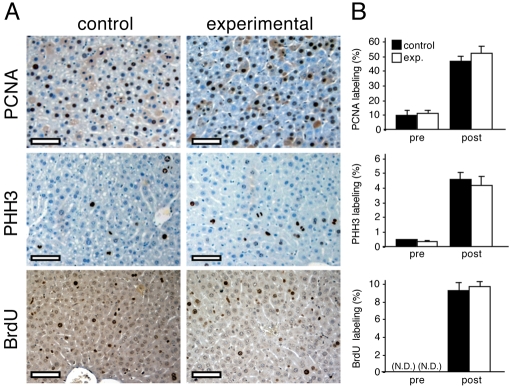
Thioredoxin-deficient yeast are able to proliferate; however they exhibit a protracted S phase and a prolonged cell cycle (Koc et al., 2006). Static frequencies of S phase hepatocytes cannot, alone, indicate either differences in cell cycle duration or in S phase duration; however, by measuring multiple static proliferative indexes representing different cell cycle phases, one can more accurately determine whether the cell cycle is perturbed (supplementary material Fig. S1). To test whether disruption of Txnrd1 in hepatocytes affected cell cycle progression, we measured the proliferative index by PCNA staining, the S phase index by BrdU labeling, and the M phase index by staining for PHH3 (Fig. 4A). Results showed no measurable differences in any of these proliferative indexes between hepatocytes within regenerating control and experimental livers (Fig. 4B).
Fig. 4.
Proliferation of Txnrd1-deficient hepatocytes during liver regeneration. (A) Proliferative marker staining in post-hepatectomy livers. Adult (P42) mice of genotypes Txnrd1cond/+;AlbCre1 (left panels, controls) or Txnrd1cond/−;AlbCre1 (right panels, experimentals) were two-thirds hepatectomized. For PCNA (upper panels) and PHH3 (middle panels) staining, livers were harvested 48 hours post-hepatectomy when the first ‘wave’ of proliferative hepatocytes would be around G2-M in the cell cycle. The abundant hepatocytes with diffuse cytoplasmic PCNA staining (upper panel) are presumably in M or early G1 phase, after nuclear membrane dissolution allows PCNA protein to redistribute into the cytoplasm. Hepatocytes with nuclear or cytoplasmic staining were scored as proliferative, as both types were either within or just exiting from a proliferative cell cycle. PCNA-positive cell counts in liver 36 hours post-hepatectomy were similar (not shown) albeit with less cytoplasmic staining. For BrdU staining (lower panels), BrdU was administered intraperitoneally at 36 hours post-hepatectomy when the first wave of regenerating hepatocytes were in S phase, and mice were killed 1 hour later. Scale bars: 100 μm in all panels. (B) Proliferative indexes for resting (pre-hepatectomy; pre) or regenerating (post-hepatectomy; post) liver. N.D., not determined. Values are means and s.e.m. for counts on three animals. Statistical analysis indicated that none of the differences between control and experimental samples was significant.
Based on previous studies in lower eukaryotes (Koc et al., 2006; Reichheld et al., 2007), it was surprising that mouse hepatocyte proliferation was not measurably affected by disruption of Txnrd1. As a control, we confirmed that the RNR inhibitor hydroxyurea entirely prevented regenerative replication of both normal and Txnrd1-deficient hepatocytes (Fig. 5A). Also, in vivo post-hepatectomy metabolic labeling showed that thymidine incorporation during liver regeneration did not differ between normal and Txnrd1-deficient livers (Fig. 5B). In combination, the data above show that proliferative liver growth was driven primarily by hepatocyte proliferation, not by proliferation of Txnrd1-containing progenitor cells, and they indicate that the ability of RNR to support hepatocyte DNA replication was not measurably compromised by disruption of Txnrd1.
Fig. 5.

Sensitivity to hydroxyurea and [3H]thymidine incorporation during regeneration of control and Txnrd1-deficient livers. (A) RNR requirement for replication. Adult (P42) mice of genotypes Txnrd1cond/+;AlbCre1 (left panels, controls) or Txnrd1cond/−;AlbCre1 (right panels, experimentals) were two-thirds hepatectomized. After 35 hours of recovery, mice received a single IP inoculation of either HU in saline (lower panels; + HU) or saline alone (upper panels; – HU). This was followed 30 minutes later with an injection of BrdU + HU (lower panels) or BrdU alone (upper panels). After a 1-hour labeling period, livers were harvested. Regardless of genotype, no BrdU-positive cells were detected in HU-treated animals. Scale bars: 100 μm. (B) Quantitative thymidine incorporation. Mice of the same genotypes and ages as in A were two-thirds heptectomized and, 33 hours later, received 12.5 μCi of [3H]thymidine IP. Livers were harvested 6 hours later and the specific activity of nuclear acid-precipitated DNA was determined using 0, 0.1, 0.3 and 1.0 mg nucleic-acid equivalents of nuclei. Values for all animals were plotted and best-fit lines for each genotype were assigned by least squares regression.
Contributions of Txnrd1-containing versus Txnrd1-deficient cell lineages to development and regeneration of genetically chimeric livers
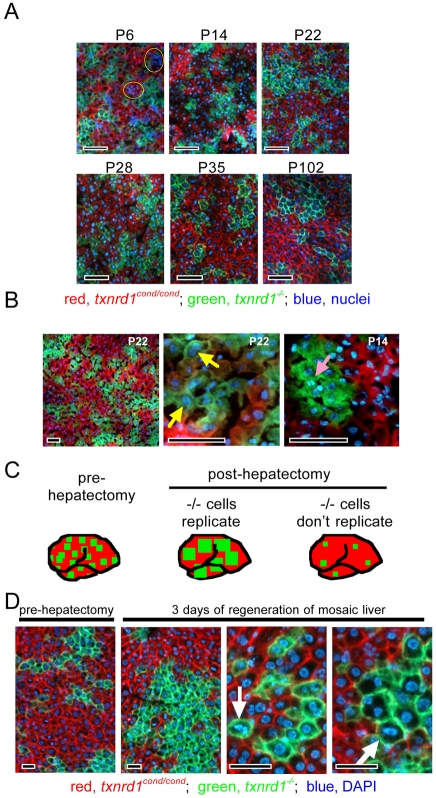
For a potentially more sensitive comparison of relative proliferative potentials, we generated fetally induced genetically chimeric mice in which individual livers contained both red-fluorescent-tagged normal and green-fluorescent-tagged Txnrd1-deficient hepatocytes. Using these mice, we evaluated the contributions of both hepatocyte types to developmental and regenerative liver growth. By this approach, the size of the adult liver can be seen as a target niche that the two types of hepatocytes will compete to populate. This results in a ‘multi-generational relay-race’ in which cells with a subtle genetic growth advantage will pass this advantage to their daughter cells, thus amplifying the proliferative difference each generation. To produce the mosaics, we used the Tamoxifen-inducible ROSACreER allele (Badea et al., 2003). Dams of genotype Txnrd1cond/cond;ROSAmT-mG/mT-mG were mated to sires of genotype Txnrd1cond/cond;ROSACreER/CreER. In this mating, the dams themselves lack the ROSACreER allele and do not exhibit Cre activity in response to Tamoxifen; however, all progeny are genotype Txnrd1cond/cond;ROSAmT-mG/CreER so that fetal cells exposed to Tamoxifen can convert to Txnrd1−/− and from red to green fluorescence. A time and dose study established that intraperitoneal (IP) administration of 0.5 mg of 4-hydroxytamoxifen (4OHT) to pregnant dams at embryonic day 14.5 (E14.5) resulted in 100% survival of pups to term (C.M.W., J.A.K. and E.E.S., unpublished data). By this dosage protocol, pups uniformly exhibited 30-50% green (Txnrd1−/−) hepatocytes as neonates (Fig. 6A and not shown), whereas frequencies of green cells in other organs were much lower (ranging from a low of <0.1% in brain to a high of ~10% in pancreas, with most organs ~1%; C.M.W. and E.E.S., unpublished data).
Fig. 6.
Analysis of the relative contributions of control and Txrnd1-deficient cells to development and regeneration of genetically chimeric livers. E14.5 fetuses of genotype Txnrd1cond/cond;ROSAmT-mG/CreER were exposed in utero to 4OHT by a single IP inoculation of the pregnant dam at a dose that caused ~30-50% of the hepatocytes to convert to Txnrd1−/− (green membrane fluorescence; see Materials and Methods). This provided a fetal ‘time stamp’ on liver cell lineages; all green cells at times thereafter would either be, or be descendents of, cells that had converted to Txnrd1−/− during the brief period of 4OHT exposure. (A) Developmental contributions. Livers were harvested at the indicated postnatal ages and the relative contributions of red (Txnrd1-containing) and green (Txnrd1-deficient) hepatocytes to the hepatocyte populations at each stage were determined. Yellow ovals circumscribe clusters of hematopoietic cells that persist in livers of very young mice (Weisend et al., 2009). (B) Active proliferation in juvenile Txnrd1-deficient hepatocytes. Fluorescence micrographs show mosaic livers that were generated as in A and harvested at P14 (right panel) or P22 (left and middle panels). The lower magnification panel on left shows relative contributions of red (normal) and green (Txnrd1-deficient) ‘clones’ of hepatocytes to P22 liver in mosaics. The middle panel shows that green clones have abundant binucleate cells, a characteristic of some differentiated hepatocytes (yellow arrows). The right panel shows that some cells within green clones exhibit mitotic figures (magenta arrow). (C) Schematic of possible outcomes for regeneration in mosaic livers. The left image depicts a resting adult mouse liver with the typical distribution of red (normal) and green (Txnrd1-deficient) hepatocyte ‘clones’, as in A. The middle and right images depict the expected results following liver regeneration if Txnrd1-deficent hepatocytes can (middle image) or cannot (right image) contribute to regeneration. (D) Regeneration in mosaic livers. Adult mice (P42) of genotype Txnrd1cond/cond;ROSAmT-mG/CreER that had been exposed in utero (E14.5) to 4OHT as in A were two-thirds hepatectomized and allowed to recover for 72 hours. DAPI-stained cryosections were examined by fluorescence microscopy. Left panel shows a field of a resected lobe (‘pre-hepatectomy’). Middle and right panels show two representative fields of the post-hepatectomy liver. Arrows indicate mitotic figures in green cells. Scale bars in all panels: 100 μm.
Following maternal administration of 4OHT at E14.5, cryosections of livers harvested from neonatal [postnatal day (P) 6], juvenile (P14-P28), young adult (P35) and older adult (P102) offspring showed similar mosaic distributions of ~50-70% red (normal) and ~30-50% green (Txnrd1-deficient) hepatocytes (Fig. 6A), indicating that Txnrd1-containing hepatocytes did not have a growth advantage over Txnrd1-deficient hepatocytes despite having 5 days of pre-natal and 102 days of post-natal development to manifest a competitive advantage. At P14 and P22, when liver is predominantly composed of hepatocytes (Weisend et al., 2009) yet still undergoing active proliferative growth, bi-nucleate cells (Fig. 6B, center panel, yellow arrows) and mitotic figures (Fig. 6B, right panel, pink arrow) were found in Txnrd1-deficient (green) hepatocytes, verifying that Txnrd1-deficient hepatocytes had replicated their genomes.
To more accurately estimate how many consecutive cell cycles the hepatocytes had completed, we quantified the cellular dynamics of liver development. Fresh whole livers harvested from E14.5 fetal mice from our colony weighed 0.0315±0.0032 g (mean ± s.e.m., n=6). Previously, we showed that E14.5 liver is composed of <10% differentiated hepatocytes by mass (Weisend et al., 2009). In adults, the liver weighs ~1.6 g (Fig. 3A) and is composed of ~95% differentiated hepatocytes by mass (Schmidt and Schibler, 1995; Weisend et al., 2009). Thus, there is a roughly 500-fold increase in mass of total liver hepatocytes during development. Although a portion of this is accounted for by the increase in average liver cell size that occurs during rodent development [~fivefold during post-natal development in rats (Schmidt and Schibler, 1995)], we estimate that there is also a 30- to 100-fold developmental increase in the number of hepatocyte genomes in mice, which accounts for the majority of this growth. This increase in hepatocyte genomes, in the absence of any cell death, would require five to seven consecutive cell doublings (25=32; 27=128). Since relative representation of both the E14.5-converted Txnrd1−/− (green) and the non-converted Txnrd1cond/cond (red) hepatocytes was similar in mosaic juvenile and adult livers (Fig. 6A,B), we concluded that, by adulthood the cell lineages leading to both hepatocyte types had undergone a minimum of five to seven consecutive replicative cycles since their Cre-mediated genetic divergence without showing a measurable difference in proliferative potential.
Next, we tested whether adult Txnrd1-deficient hepatocytes that arose from lineages that had been converted at E14.5 retained the same capacity as normal hepatocytes to contribute to the rapid proliferative burst that follows a two-thirds hepatectomy (i.e. 1– 1.5 more replicative cycles). As modeled in Fig. 6C, we predicted that, if the Txnrd1-deficient hepatocytes could contribute to regeneration, then the ratio of red to green hepatocytes would be similar before and after hepatectomy; if not, we would observe an effective dilution of green cells. Results showed that the relative representation of red and green cells was similar before and after regeneration (Fig. 6D). Moreover, at 72 hours post-hepatectomy, mitotic figures were found in Txnrd1-deficient (green) cells, verifying these cells were engaged in the cell cycle (Fig. 6D, white arrows). We conclude that normal and Txnrd1-deficient mouse hepatocytes have equivalent developmental and regenerative proliferative potentials.
Discussion
Txnrd1 disruption, selenium assimilation, and the hepatocyte selenoproteome
Mammalian Txnrds are selenoproteins, meaning they contain the atypical amino acid selenocysteine (Sec) (Gladyshev and Hatfield, 1999; Lobanov et al., 2009; Lu and Holmgren, 2009). Only 25 mammalian selenoproteins are known or predicted, all of which are oxidoreductases and all of which have Sec participating in active site electron transfer (Fomenko et al., 2007; Lobanov et al., 2007; Lobanov et al., 2009; Lu and Holmgren, 2009). For production of Sec, selenide is either acquired from organic dietary sources or as inorganic selenate and selenite. Dietary selenite is reduced to selenide by either the glutathione reductase (Gsr)-reduced glutathione-glutaredoxin or the Txnrd-Txn systems, whereas the mechanisms of selenate reduction remain uncertain (Lu et al., 2009; Lu and Holmgren, 2009). We have not tested whether disruption of Txnrd1 has global effects on the selenoproteome within individual cells, although the apparent redundancy of sources and reductase systems for mobilization of dietary selenium suggests Sec availability would not become limiting in Txnrd1-deficient cells.
Alternative pathways of supplying electrons to RNR
In all life forms studied to date, RNR activity is essential for DNA replication. RNR activity, in turn, is dependent on a source of electrons. Genetic malleability of bacterial, yeast and plant systems allowed identification of both thioredoxins and glutaredoxins as the electron donors for RNR in these systems; thioredoxins and glutaredoxins ultimately obtain the electrons from NADPH via thioredoxin reductase or via glutathione reductase and glutathione, respectively (Avval and Holmgren, 2009; Meyer et al., 2009). Previously, it had not been possible to test the relative contributions of these two pathways in vivo in mammals. It has long been known that mammalian glutaredoxin can provide electrons to RNR in vitro (Luthman et al., 1979; Luthman and Holmgren, 1982) and a recent study showed that both Txn1 and glutaredoxin are similarly efficient at supplying electrons to RNR in vitro (Avval and Holmgren, 2009). None of these biochemical assays could approach rates of deoxyribonucleotide production required during S phase, however, and no previous study definitively established that mammalian cells could replicate DNA and proliferate in the absence of Txnrd1. The survival and relatively normal physiology of mice lacking Gsr indicates that the Gsr-dependent pathway is not necessary in mammals (Rogers et al., 2004). Conversely, embryonic lethality of mice lacking either Txn1 (Matsui et al., 1996) or Txnrd1 (Bondareva et al., 2007; Jakupoglu et al., 2005) indicates that some aspect of the Txnrd1-dependent pathway is essential in mammals.
Although mice having a disrupted Txnrd1-Txn1 system are not viable, several lines of evidence suggested Txnrd1 is not essential for cell proliferation. First, cells in Txnrd1-deficient mouse embryos underwent at least twelve consecutive doublings prior to embryonic arrest (Bondareva et al., 2007; Jakupoglu et al., 2005). Second, knockdown of Txnrd1 in cell cultures did not affect cell proliferation (Yoo et al., 2006; Yoo et al., 2007). Third, mice lacking Txnrd1 in all differentiated hepatocytes have normal or slightly larger than normal sized livers (Fig. 3A) (Suvorova et al., 2009). It was possible that proliferation in these systems used cryptic sources of Txnrd1 (i.e. maternally derived in embryos, progenitor cell-derived in hepatocytes or residual following knockdown). Alternatively, a Txnrd1-independent pathway might have been supporting replication.
The most obvious alternative pathway for supporting RNR activity in Txnrd1-deficient hepatocytes would be the Gsr-dependent system that has been described in other eukaryotes and demonstrated in vitro. Alternatively, however, the mitochondrial Txnrd2 enzyme might be compensating for loss of Txnrd1. Although ribonucleotide reduction for genome replication occurs in the cytosol and there is no evidence of a mitochondrial source of electrons for this reaction (Arner, 2009; Arner and Holmgren, 2006a; Meyer et al., 2009), the Txnrd2 gene contains two alternative promoters and first exons capable of issuing mRNAs that would each encode cytosolic isoforms of Txnrd2 (Turanov et al., 2006). Accumulation of endogenous cytosolic Txnrd2 has not, to our knowledge, been shown. However, naturally occurring mRNAs encoding these isoforms have been described, and overexpression of minigenes encoding these isoforms fused to GFP in cultured cells results in cytosolic accumulation of the fluorescent Txnrd2-GFP fusion proteins (Turanov et al., 2006). Moreover, in vitro, recombinant Txnrd2 can reduce Txn1 (Turanov et al., 2006) and diminution of Txnrd1 activity in cell cultures by RNAi or inhibitory drugs does not lead to Txn1 oxidation (Watson et al., 2008).
The conditional hepatocyte-specific Txnrd1-deficient system provided a model in which prolonged temporal separation of gene disruption and proliferation could be achieved in vivo, thus eliminating the possibility that persistent pre-formed Txnrd1 was supporting replication. In the current study, we used this system to show that disruption of Txnrd1 had no measurable effect on the proliferative potential of hepatocytes in mice. Our results, combined with the normal physiology of Gsr-null mice (Rogers et al., 2004), lead us to conclude that, similar other life forms, animal cells can probably proliferate using either Txnrd- or Gsr-derived electrons to fuel RNR. Moreover, unlike yeast (Camier et al., 2007; Koc et al., 2006) and perhaps plants (Reichheld et al., 2007), we detected no evidence that the Txnrd1-dependent system was a preferential source of electrons. It is possible that this difference between species relates to the distinct evolutionary origins of metazoan compared with other eukaryotic thioredoxin reductases. Further studies will be required to distinguish the possible respective roles of Gsr versus cytosolic forms of Txnrd2 in supporting replication; however, intriguingly, the transcriptomes of both Txnrd1-deficient embryos (Bondareva et al., 2007) and livers (Suvorova et al., 2009) do not show any compensatory upregulation of mRNAs encoding either Gsr or Txnrd2. Thus, whatever pathway is supporting RNR activity during hepatocyte replication it might be sufficiently abundant and robust to fully support genomic DNA replication without further induction, and thus might be a major, as opposed to an alternative or compensatory, source of reducing potential for RNR. Although there might be some cell types or circumstances with different requirements, we find that either the Txnrd1-dependent or Txnrd1-independent route is probably equivalent in terms of supporting physiological RNR activity in hepatocytes.
Other roles and functions of the Txnrd1-Txn1 system
In addition to providing electrons to RNR, the Txnrd1-Txn1 system is a major protein thiol reductase; it provides reducing potential to enzymes including peroxiredoxins and methionine sulfoxide reductase, and it is generally considered to be a major participant in cellular antioxidant defense and redox regulation (Arner, 2009; Arner and Holmgren, 2006a; Meyer et al., 2009). We previously showed that disruption of Txnrd1 in embryos or hepatocytes resulted in a strong cytoprotective response, including induction of drug metabolism genes and upregulation the Nrf2 stress-response pathway (Bondareva et al., 2007; Suvorova et al., 2009). It is unclear how these responses might compensate for loss of Txnrd1 in cells. However, in the mutant livers where biochemical analyses could be performed, we observed no evidence of oxidative stress, suggesting this response was, indeed, compensatory (Suvorova et al., 2009). We are aware of no evidence that any of the cytoprotective pathways induced in Txnrd1-deficient hepatocytes could directly supply electrons to RNR.
Implications for Txnrd- and Txn-targeted pharmaceuticals
Considerable attention has been given to possible roles for Txnrd1, both favorable and deleterious, in cancer initiation and progression (Arner, 2009). Whereas Txnrd1 activity is thought to protect against the initiation of cancers (Arner, 2009; Arner and Holmgren, 2006b; Ganther, 1999), many aggressive cancers exhibit increased expression of Txnrd1 (Berggren et al., 1996; Nakamura et al., 2000; Rundlof and Arner, 2004), suggesting Txnrd1 activity supports cancer growth and progression (Arner, 2009; Arner and Holmgren, 2006a). Txnrd1 has been extolled as a ‘drugable target’ for cancer therapies (Arner, 2009; Arner and Holmgren, 2006a; Bindoli et al., 2009; Gandin et al., 2009). Numerous cancer drugs or drug candidates affect Txnrd1 activity, either as a primary target or, as in the case of cisplatin and other platinum-containing compounds (Sasada et al., 1999; Witte et al., 2005), as a collateral target. Some of these include arsenic trioxide (Lu et al., 2007), various organo-gold compounds (Bindoli et al., 2009; Casini et al., 2009; Coronnello et al., 2005; Gandin et al., 2009; Marzano et al., 2007; Rigobello et al., 2009), some mercury-containing compounds (Carvalho et al., 2008), and certain quinols (Chew et al., 2008), flavinoids (Du et al., 2009; Lu et al., 2006) and polyphenols (Du et al., 2009; Fang et al., 2005). Many other drugs will interact with the Txnrd1 pathway and, if used in combination therapies, might have their pharmacokinetics affected by compounds that inhibit Txnrd1. For example, selenium-containing compounds can be metabolically activated by Txnrd and/or Txn (Lu et al., 2009), which might be compromised if Txnrd1 is inhibited. Conversely, disruption of Txnrd1 causes increased expression of some drug metabolism enzymes (Suvorova et al., 2009), which might affect metabolic activation, stability, excretion, or toxicity of co-administered drugs in unexpected ways.
One class of anti-cancer drug whose activity might have been expected to be downstream of the Txnrd1-Txn1 pathway is the RNR inhibitors, such as hydroxyurea (HU). Prior to the current study, it seemed likely that Txnrd1 would be a crucial supplier of electrons for RNR in mammalian systems, and therefore, that inhibition of Txnrd1 might also affect RNR activity. We show that genetic elimination of Txnrd1 has no measurable effect on RNR-dependent hepatocyte proliferation. This suggests that the anticancer effects of Txnrd1 inhibitors are not the result of preventing Txnrd1 from supplying electrons to RNR. Instead, these effects are probably the result of either inhibiting other activities of Txnrd1 or of interactions of these electrophilic compounds with cellular targets other than Txnrd1. Thus, in strategic design of combination therapies, these Txnrd1 inhibitors would not be redundant with RNR inhibitors and might, in some cases, be synergistic with them.
Materials and Methods
Mice and animal care
All animal procedures were approved by the Montana State University Institutional Animal Care and Use Committee (MSU-IACUC). All renewable resources in this study are available on request for unrestricted non-profit research unless regulated by another party. All mice were housed with specialized care conditions including sterilized feed, water and bedding in HEPA-filtered forced-air cages (Tecniplast) under a 14 hour:10 hour light:dark cycle. Feed (PicoLab mouse diet 5058, which contains selenium derived as a natural component of the grains and supplemented to 0.33 p.p.m. with sodium selenite) and water were unrestricted. All mutant alleles were followed by PCR analyses of genomic DNA samples using the primers and conditions described previously (Bondareva et al., 2007; Suvorova et al., 2009; Weisend et al., 2009). Mouse lines Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J (here abbreviated ‘ROSAmT-mG’) (Muzumdar et al., 2007), B6;129-Gt(ROSA)26Sortm1(cre/Esr1)Nat/J (‘ROSACreER’) (Badea et al., 2003), B6.Cg-Tg(Alb-Cre)21Mgn/J (‘AlbCre’) (Postic et al., 1999), and C57Bl/6J were obtained from Jackson Labs (stock numbers 007576, 004847, 003574 and 000664, respectively) and were maintained in our colony by back-crossing to C57Bl/6J. Mouse lines bearing the Txnrd1− and Txnrd1cond alleles have been described previously (Bondareva et al., 2007), as have all hybrids between these lines that were used in the current study (Suvorova et al., 2009). Timed pregnancies were monitored by checking for vaginal semen plugs each morning; noon on the day that a plug was found was defined as embryonic day 0.5 (E0.5). 4OHT (Sigma) was prepared as described previously (Badea et al., 2003) and was administered as an IP injection at the times and doses indicated in figures and legends.
Partial hepatectomies were performed as described previously (Mitchell and Willenbring, 2008) to remove the median and left-lateral lobes, which constitute approximately two-thirds of the mass of the liver. BrdU, when used, was administered as an IP inoculation of 50 mg/kg in sterile saline at the times indicated in figures and legends. To test the requirements of RNR for replication, HU was administered by IP inoculation at 0.0 (mock) or 1.3 mmol/kg in sterile saline followed 30 minutes later by an IP inoculation containing a mixture of 50 mg/kg BrdU with either 0.0 (mock) or 1.3 mmol/kg HU in sterile saline, as indicated in figures and legends. [3H]thymidine (MP Biomedicals #24059) was administered as a single IP dose of 12.5 μCi in 0.5 ml sterile saline as described in figures and legends.
Histology, microscopy, and image adjustments
Samples for cryosections were harvested into OCT compound (Tissue-Tek), frozen, and stored at −80°C; samples for paraffin embedding were harvested fresh into 10% neutral-buffered formalin. Processing and sectioning were performed by standard procedures as described previously (Bondareva et al., 2007; Suvorova et al., 2009; Weisend et al., 2009). Primary antibodies used for immunostaining were mouse-anti-PCNA monoclonal antibody (AbCam, no. A1029), rabbit-anti-PHH3 (Cell Signaling Technologies, no. 9701), and mouse-anti-BrdU-substituted DNA monoclonal antibody (Sigma, no. B2531). Secondary antibodies were all horseradish peroxidase conjugates, and were goat-anti-mouse (Pierce, no. 31430) or goat-anti-rabbit (GIBCO, no. 13859.012). All immunostaining was performed as per the suppliers' recommendations and our previously reported protocols (Suvorova et al., 2009). Labeling indexes were assessed by counting total hepatocytes and labeled hepatocytes in arbitrary fields of view. In general, counts were continued until at least 30 labeled hepatocytes were tallied (e.g. 30,000 cells counted for a 0.1% labeling index, 100 cells counted for a 30% labeling index). Morphologically non-hepatocytic cells were not included in counts. Counting newly differentiated hepatocytes (Fig. 3C) was complicated by the heterogeneous, clustered and intermittent distribution of these cells in liver (Fig. 3B) and by their low overall abundance. Arbitrary fields of view were counted until at least 30 red hepatocytes were tabulated and a minimum of 10 fields had been evaluated, or until a maximum of 30 fields had been evaluated. By these criteria, newly differentiated hepatocytes in resting adult (pre-hepatectomy samples) were determined to represent <0.1% of hepatocytes in these conditions (Fig. 3C).
All cryo and paraffin sections were cut 5.0 μm thick. Following immunostaining, paraffin sections for bright-field microscopy were counter-stained with Hematoxylin. Cryosections for fluorescence microscopy were counterstained using Fluoromount-G containing DAPI (Southern Biotech). All images were captured digitally on either a Nikon Eclipse E800 or an Olympus BX60 microscope. For all photomicrographs, the whole area shown was uniformly adjusted using the ‘Autolevels’ function in Photoshop CS3 software. Some micrographs were electronically enlarged or reduced in Photoshop CS3 software. Scale bars in figures were set by photographing a micrometer scale under each magnification used and subjecting these images to the same electronic enlargement or reduction as the biological images shown. For non-merged fluoromicrographs, the whole area shown was adjusted using the ‘Autocontrast’ function in Photoshop CS3. For merged fluoromicrographs, images were captured separately for each color channel and, prior to making any adjustments, were electronically layered using Photoshop CS3 software. For merging three-channel images (blue, red and green), the blue channel formed the background at 100% opacity, red was the next layer at 40% opacity, and green was the top layer at 50% opacity. Following layering, all visible layers were merged and were uniformly adjusted using the ‘Autolevels’ function in Photoshop CS3. With the exception of the images in Fig. 3C, all micrographs were representative of the whole organ. In the case of Fig. 3C, newly differentiated hepatocytes were clustered around widely scattered peri-portal regions. Images were taken to show these clusters, as arbitrarily chosen fields generally contained no red hepatocytes (e.g. Fig. 1A and Fig. 3B). All adjustments and manipulations made to images were performed uniformly to the entire image. None of these adjustments qualitatively or quantitatively affect the interpretations or conclusions arising from the data, but instead serve useful aesthetic functions and ease visualization and enumeration of important biological characteristics of each sample.
Nuclei isolation and DNA extraction for thymidine labeling studies
Following thymidine labeling, animals were killed by CO2 asphyxiation and livers were harvested into ice-cold PBS. Nuclei were isolated using protocols described previously (Schmidt and Schibler, 1995). Briefly, liver was blotted, weighed, minced and homogenized under final conditions of 10% (w/v) liver, 1.8 M sucrose, 9% (v/v) glycerol, 10 mM HEPES, pH 7.8, 15 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA, in a motor-driven Teflon and glass homogenizer. Lysate was layered on 0.5 ml cushions of 2.0 M sucrose, 10% glycerol, 10 mM HEPES, pH 7.8, 15 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA and were centrifuged at 116,000 gmax (26,000 r.p.m. in an SW-41 Ti rotor), 4°C, for 1 hour in an SW-41 rotor. Nuclei were resuspended in nucleus resuspension buffer (10% glycerol, 10 mM HEPES, pH 7.8, 15 mM KCl, 0.15 mM spermine, 0.5 mM spermidine, 1 mM EDTA), total nucleic acid content was determined, and concentrations were adjusted to 10 mg/ml nucleic acid with nuclear resuspension buffer. Samples equivalent to 0.0 (no input nuclei, background controls), 0.1, 0.3 and 1.0 mg of nucleic acid were adjusted to 200 μl with nucleus resuspension buffer, boiled for 3 minutes, and cooled to room temperature. RNases A and T1 were added to final concentrations of 10 μg/ml and 350 U/ml, respectively, and samples were incubated at 42°C for 30 minutes. Twenty percent SDS (10 μl) and 0.5 μl of 20 mg/ml proteinase K (Sigma) were added to each sample and they were then incubated at 50°C for 15 minutes. Samples were diluted with 800 μl of water, and high molecular mass DNA was precipitated by adding 120 μl of 100% (w/v) trichloroacetic acid, and the samples were mixed, and incubated on ice 30 minutes. Precipitated DNA was collected by vacuum filtration onto 0.5 cm glass-fiber filter discs. Discs were washed sequentially with a large volume of 10% trichloroacetic acid followed by 70% ethanol, and were assayed by liquid scintillation counting.
Statistical analyses
Statistical analyses used at least three biological replicates (i.e. from different animals) for each condition. Data in graphs are presented as means with error bars representing the standard error of the mean (s.e.m.). For proliferative indexes, significance was determined using the Student's t-test with a minimum P-value of 0.05. For [3H]thymidine incorporation (Fig. 6B), each sample (three input nuclei concentrations from three different animals from each genotype) was counted for four 1-minute periods by liquid scintillation counting. Counts per minute values for each sample were averaged and the average background (scintillation fluid only, no input nuclei) was subtracted. Data were plotted and a best-fit line was assigned by least-squares regression; line fit was evaluated by the coefficient of determination, r2. Slope represents thymidine incorporation per input nucleus.
Supplementary Material
Acknowledgments
The authors thank S. Iverson, J. Prigge and G. Callis for their technical contributions or scientific criticisms of this work; we thank C. O'Rourke for training us to perform partial hepatectomies. M.F.R. was supported in part by a research scholarship from University Scholars Program at Montana State University; D.M.v.d.H. was supported by the Howard Hughes Medical Institute's Undergraduate Research Program; K.M.C. was supported by the National Science Foundation's Research Experiences for Undergraduates program; E.E.S. and C.M.W. were supported in part by the Montana Agricultural Experiment Station; infrastructure support came from NIH COBRE grant P20 RR020185 to VMB; other support was provided by Montana State University and by grants from the NIH and NSF to E.E.S. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/14/2402/DC1
References
- Aleksunes L. M., Manautou J. E. (2007). Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 35, 459-473 [DOI] [PubMed] [Google Scholar]
- Arner E. S. (2009). Focus on mammalian thioredoxin reductases – important selenoproteins with versatile functions. Biochim. Biophys. Acta 1790, 495-526 [DOI] [PubMed] [Google Scholar]
- Arner E. S., Eriksson S. (1995). Mammalian deoxyribonucleoside kinases. Pharmacol Ther 67, 155-186 [DOI] [PubMed] [Google Scholar]
- Arner E. S., Holmgren A. (2006a). The thioredoxin system in cancer. Semin. Cancer Biol. 16, 420-426 [DOI] [PubMed] [Google Scholar]
- Arner E. S., Holmgren A. (2006b). The thioredoxin system in cancer – introduction to a thematic volume of Seminars in Cancer Biology. Semin. Cancer Biol. 16, 419 [DOI] [PubMed] [Google Scholar]
- Avval F. Z., Holmgren A. (2009). Molecular mechanisms of thioredoxin and glutaredoxin as hydrogen donors for mammalian S phase ribonucleotide reductase. J. Biol. Chem. 284, 8233-8240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea T. C., Wang Y., Nathans J. (2003). A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J. Neurosci. 23, 2314-2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren M., Gallegos A., Gasdaska J. R., Gasdaska P. Y., Warneke J., Powis G. (1996). Thioredoxin and thioredoxin reductase gene expression in human tumors and cell lines, and the effects of serum stimulation and hypoxia. Anticancer Res. 16, 3459-3466 [PubMed] [Google Scholar]
- Berndt C., Lillig C. H., Holmgren A. (2007). Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 292, H1227-H1236 [DOI] [PubMed] [Google Scholar]
- Bindoli A., Rigobello M. P., Scutari G., Gabbiani C., Casini A., Messori L. (2009). Thioredoxin reductase: A taret for gold compounds acting as potential anticancer drugs. Coord. Chem. Rev. 253, 1692-1707 [Google Scholar]
- Bondareva A. A., Capecchi M. R., Iverson S. V., Li Y., Lopez N. I., Lucas O., Merrill G. F., Prigge J. R., Siders A. M., Wakamiya M., et al. (2007). Effects of thioredoxin reductase-1 deletion on embryogenesis and transcriptome. Free Radic. Biol. Med. 43, 911-923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camier S., Ma E., Leroy C., Pruvost A., Toledano M., Marsolier-Kergoat M. C. (2007). Visualization of ribonucleotide reductase catalytic oxidation establishes thioredoxins as its major reductants in yeast. Free Radic. Biol. Med. 42, 1008-1016 [DOI] [PubMed] [Google Scholar]
- Carvalho C. M., Chew E. H., Hashemy S. I., Lu J., Holmgren A. (2008). Inhibition of the human thioredoxin system. A molecular mechanism of mercury toxicity. J. Biol. Chem. 283, 11913-11923 [DOI] [PubMed] [Google Scholar]
- Casini A., Kelter G., Gabbiani C., Cinellu M. A., Minghetti G., Fregona D., Fiebig H. H., Messori L. (2009). Chemistry, antiproliferative properties, tumor selectivity, and molecular mechanisms of novel gold(III) compounds for cancer treatment: a systematic study. J. Biol. Inorg. Chem. 14, 1139-1149 [DOI] [PubMed] [Google Scholar]
- Chew E. H., Lu J., Bradshaw T. D., Holmgren A. (2008). Thioredoxin reductase inhibition by antitumor quinols: a quinol pharmacophore effect correlating to antiproliferative activity. FASEB J. 22, 2072-2083 [DOI] [PubMed] [Google Scholar]
- Conrad M., Jakupoglu C., Moreno S. G., Lippl S., Banjac A., Schneider M., Beck H., Hatzopoulos A. K., Just U., Sinowatz F., et al. (2004). Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 24, 9414-9423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronnello M., Mini E., Caciagli B., Cinellu M. A., Bindoli A., Gabbiani C., Messori L. (2005). Mechanisms of cytotoxicity of selected organogold(III) compounds. J. Med. Chem. 48, 6761-6765 [DOI] [PubMed] [Google Scholar]
- Du Y., Wu Y., Cao X., Cui W., Zhang H., Tian W., Ji M., Holmgren A., Zhong L. (2009). Inhibition of mammalian thioredoxin reductase by black tea and its constituents: implications for anticancer actions. Biochimie 91, 434-444 [DOI] [PubMed] [Google Scholar]
- Evans I. H. (1976). Polyploidization in rat liver: the role of binucleate cells. Cytobios 16, 115-124 [PubMed] [Google Scholar]
- Fang J., Lu J., Holmgren A. (2005). Thioredoxin reductase is irreversibly modified by curcumin: a novel molecular mechanism for its anticancer activity. J. Biol. Chem. 280, 25284-25290 [DOI] [PubMed] [Google Scholar]
- Fernandes A. P., Holmgren A. (2004). Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox Signal. 6, 63-74 [DOI] [PubMed] [Google Scholar]
- Fomenko D. E., Xing W., Adair B. M., Thomas D. J., Gladyshev V. N. (2007). High-throughput identification of catalytic redox-active cysteine residues. Science 315, 387-389 [DOI] [PubMed] [Google Scholar]
- Fomenko D. E., Novoselov S. V., Natarajan S. K., Lee B. C., Koc A., Carlson B. A., Lee T. H., Kim H. Y., Hatfield D. L., Gladyshev V. N. (2009). MsrB1 (methionine-R-sulfoxide reductase 1) knock-out mice: roles of MsrB1 in redox regulation and identification of a novel selenoprotein form. J. Biol. Chem. 284, 5986-5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V., Fernandes A. P., Rigobello M. P., Dani B., Sorrentino F., Tisato F., Bjornstedt M., Bindoli A., Sturaro A., Rella R., et al. (2009). Cancer cell death induced by phosphine gold(I) compounds targeting thioredoxin reductase. Biochem. Pharmacol. 79, 90-101 [DOI] [PubMed] [Google Scholar]
- Ganther H. E. (1999). Selenium metabolism, selenoproteins and mechanisms of cancer prevention: complexities with thioredoxin reductase. Carcinogenesis 20, 1657-1666 [DOI] [PubMed] [Google Scholar]
- Gladyshev V. N., Hatfield D. L. (1999). Selenocysteine-containing proteins in mammals. J. Biomed. Sci. 6, 151-160 [DOI] [PubMed] [Google Scholar]
- Higgins G. M., Anderson R. M. (1931). Experimental pathology of the liver: restoration of the liver of the white rat following partial surgical removal. Arch. Pathol. 12, 186-202 [Google Scholar]
- Holmgren A. (1976). Hydrogen donor system for Escherichia coli ribonucleoside-diphosphate reductase dependent upon glutathione. Proc. Natl. Acad. Sci. USA 73, 2275-2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. (1981). Regulation of ribonucleotide reductase. Curr. Top. Cell Regul. 19, 47-76 [DOI] [PubMed] [Google Scholar]
- Jakupoglu C., Przemeck G. K., Schneider M., Moreno S. G., Mayr N., Hatzopoulos A. K., de Angelis M. H., Wurst W., Bornkamm G. W., Brielmeier M., et al. (2005). Cytoplasmic thioredoxin reductase is essential for embryogenesis but dispensable for cardiac development. Mol. Cell. Biol. 25, 1980-1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. Y., Gladyshev V. N. (2007). Methionine sulfoxide reductases: selenoprotein forms and roles in antioxidant protein repair in mammals. Biochem. J. 407, 321-329 [DOI] [PubMed] [Google Scholar]
- Koc A., Mathews C. K., Wheeler L. J., Gross M. K., Merrill G. F. (2006). Thioredoxin is required for deoxyribonucleotide pool maintenance during S phase. J. Biol. Chem. 281, 15058-15063 [DOI] [PubMed] [Google Scholar]
- Laurent T. C., Moore E. C., Reichard P. (1964). Enzymatic synthesis of deoxyribonucleotides. IV. Isolation and characterization of thioredoxin, the hydrogen donor from Escherichia coli B. J. Biol. Chem. 239, 3436-3444 [PubMed] [Google Scholar]
- Lobanov A. V., Fomenko D. E., Zhang Y., Sengupta A., Hatfield D. L., Gladyshev V. N. (2007). Evolutionary dynamics of eukaryotic selenoproteomes: large selenoproteomes may associate with aquatic life and small with terrestrial life. Genome Biol. 8, R198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobanov A. V., Hatfield D. L., Gladyshev V. N. (2009). Eukaryotic selenoproteins and selenoproteomes. Biochim. Biophys. Acta 1790, 1424-1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Holmgren A. (2009). Selenoproteins. J. Biol. Chem. 284, 723-727 [DOI] [PubMed] [Google Scholar]
- Lu J., Papp L. V., Fang J., Rodriguez-Nieto S., Zhivotovsky B., Holmgren A. (2006). Inhibition of Mammalian thioredoxin reductase by some flavonoids: implications for myricetin and quercetin anticancer activity. Cancer Res. 66, 4410-4418 [DOI] [PubMed] [Google Scholar]
- Lu J., Chew E. H., Holmgren A. (2007). Targeting thioredoxin reductase is a basis for cancer therapy by arsenic trioxide. Proc. Natl. Acad. Sci. USA 104, 12288-12293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Berndt C., Holmgren A. (2009). Metabolism of selenium compounds catalyzed by the mammalian selenoprotein thioredoxin reductase. Biochim. Biophys. Acta 1790, 1513-1519 [DOI] [PubMed] [Google Scholar]
- Luthman M., Holmgren A. (1982). Glutaredoxin from calf thymus. Purification to homogeneity. J. Biol. Chem. 257, 6686-6690 [PubMed] [Google Scholar]
- Luthman M., Eriksson S., Holmgren A., Thelander L. (1979). Glutathione-dependent hydrogen donor system for calf thymus ribonucleoside-diphosphate reductase. Proc. Natl. Acad. Sci. USA 76, 2158-2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins P. N., Theruvath T. P., Neuhaus P. (2008). Rodent models of partial hepatectomies. Liver Int. 28, 3-11 [DOI] [PubMed] [Google Scholar]
- Marzano C., Gandin V., Folda A., Scutari G., Bindoli A., Rigobello M. P. (2007). Inhibition of thioredoxin reductase by auranofin induces apoptosis in cisplatin-resistant human ovarian cancer cells. Free Radic. Biol. Med. 42, 872-881 [DOI] [PubMed] [Google Scholar]
- Matsui M., Oshima M., Oshima H., Takaku K., Maruyama T., Yodoi J., Taketo M. M. (1996). Early embryonic lethality caused by targeted disruption of the mouse thioredoxin gene. Dev. Biol. 178, 179-185 [DOI] [PubMed] [Google Scholar]
- Melchiorri C., Chieco P., Zedda A. I., Coni P., Ledda-Columbano G. M., Columbano A. (1993). Ploidy and nuclearity of rat hepatocytes after compensatory regeneration or mitogen-induced liver growth. Carcinogenesis 14, 1825-1830 [DOI] [PubMed] [Google Scholar]
- Meyer Y., Buchanan B. B., Vignols F., Reichheld J. P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335-367 [DOI] [PubMed] [Google Scholar]
- Michalopoulos G. K. (2007). Liver regeneration. J. Cell Physiol. 213, 286-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Vizuete A., Martinez-Galisteo E., Aslund F., Lopez-Barea J., Pueyo C., Holmgren A. (1994). Null thioredoxin and glutaredoxin Escherichia coli K-12 mutants have no enhanced sensitivity to mutagens due to a new GSH-dependent hydrogen donor and high increases in ribonucleotide reductase activity. J. Biol. Chem. 269, 16631-16637 [PubMed] [Google Scholar]
- Miranda-Vizuete A., Rodriguez-Ariza A., Toribio F., Holmgren A., Lopez-Barea J., Pueyo C. (1996). The levels of ribonucleotide reductase, thioredoxin, glutaredoxin 1, and GSH are balanced in Escherichia coli K12. J. Biol. Chem. 271, 19099-19103 [DOI] [PubMed] [Google Scholar]
- Mitchell C., Willenbring H. (2008). A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3, 1167-1170 [DOI] [PubMed] [Google Scholar]
- Moore E. C., Reichard P., Thelander L. (1964). Enzymatic synthesis of deoxyribonucleotides.V. purification and properties of thioredoxin reductase from Escherichia coli B. J. Biol. Chem. 239, 3445-3452 [PubMed] [Google Scholar]
- Muzumdar M. D., Tasic B., Miyamichi K., Li L., Luo L. (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593-605 [DOI] [PubMed] [Google Scholar]
- Nakamura H., Bai J., Nishinaka Y., Ueda S., Sasada T., Ohshio G., Imamura M., Takabayashi A., Yamaoka Y., Yodoi J. (2000). Expression of thioredoxin and glutaredoxin, redox-regulating proteins, in pancreatic cancer. Cancer Detect. Prev. 24, 53-60 [PubMed] [Google Scholar]
- Nicander B., Reichard P. (1983). Dynamics of pyrimidine deoxynucleoside triphosphate pools in relationship to DNA synthesis in 3T6 mouse fibroblasts. Proc. Natl. Acad. Sci. USA 80, 1347-1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonn L., Williams R. R., Erickson R. P., Powis G. (2003). The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol. Cell. Biol. 23, 916-922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertel M., Shafritz D. A. (2008). Stem cells, cell transplantation and liver repopulation. Biochim. Biophys. Acta 1782, 61-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postic C., Magnuson M. A. (2000). DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis 26, 149-150 [DOI] [PubMed] [Google Scholar]
- Postic C., Shiota M., Niswender K. D., Jetton T. L., Chen Y., Moates J. M., Shelton K. D., Lindner J., Cherrington A. D., Magnuson M. A. (1999). Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305-315 [DOI] [PubMed] [Google Scholar]
- Reichheld J. P., Khafif M., Riondet C., Droux M., Bonnard G., Meyer Y. (2007). Inactivation of thioredoxin reductases reveals a complex interplay between thioredoxin and glutathione pathways in Arabidopsis development. Plant Cell 19, 1851-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigobello M. P., Gandin V., Folda A., Rundlof A. K., Fernandes A. P., Bindoli A., Marzano C., Bjornstedt M. (2009). Treatment of human cancer cells with selenite or tellurite in combination with auranofin enhances cell death due to redox shift. Free Radic. Biol. Med. 47, 710-721 [DOI] [PubMed] [Google Scholar]
- Rogers L. K., Tamura T., Rogers B. J., Welty S. E., Hansen T. N., Smith C. V. (2004). Analyses of glutathione reductase hypomorphic mice indicate a genetic knockout. Toxicol. Sci. 82, 367-373 [DOI] [PubMed] [Google Scholar]
- Rundlof A. K., Arner E. S. (2004). Regulation of the mammalian selenoprotein thioredoxin reductase 1 in relation to cellular phenotype, growth, and signaling events. Antioxid. Redox. Signal. 6, 41-52 [DOI] [PubMed] [Google Scholar]
- Sasada T., Nakamura H., Ueda S., Sato N., Kitaoka Y., Gon Y., Takabayashi A., Spyrou G., Holmgren A., Yodoi J. (1999). Possible involvement of thioredoxin reductase as well as thioredoxin in cellular sensitivity to cis-diamminedichloroplatinum (II). Free Radic. Biol. Med. 27, 504-514 [DOI] [PubMed] [Google Scholar]
- Schmidt E. E., Schibler U. (1995). Cell size regulation, a mechanism that controls cellular RNA accumulation: consequences on regulation of the ubiquitous transcription factors Oct1 and NF-Y and the liver-enriched transcription factor DBP. J. Cell Biol. 128, 467-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin E., Willers R., Bettecken T. (1984). Flow cytometric analysis of mouse hepatocyte ploidy. II. The development of polyploidy pattern in four mice strains with different life spans. Cell Tissue Res. 238, 649-652 [DOI] [PubMed] [Google Scholar]
- Snykers S., De Kock J., Rogiers V., Vanhaecke T. (2009). In vitro differentiation of embryonic and adult stem cells into hepatocytes: state of the art. Stem Cells 27, 577-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova E. S., Lucas O., Weisend C. M., Rollins M. F., Merrill G. F., Capecchi M. R., Schmidt E. E. (2009). Cytoprotective Nrf2 pathway is induced in chronically txnrd 1-deficient hepatocytes. PLoS ONE 4, e6158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Sekiya S., Onishi M., Oshima N., Kiyonari H., Nakauchi H., Taniguchi H. (2008). Flow cytometric isolation and clonal identification of self-renewing bipotent hepatic progenitor cells in adult mouse liver. Hepatology 48, 1964-1978 [DOI] [PubMed] [Google Scholar]
- Thelander L., Reichard P. (1979). Reduction of ribonucleotides. Annu. Rev. Biochem. 48, 133-158 [DOI] [PubMed] [Google Scholar]
- Turanov A. A., Su D., Gladyshev V. N. (2006). Characterization of alternative cytosolic forms and cellular targets of mouse mitochondrial thioredoxin reductase. J. Biol. Chem. 281, 22953-22963 [DOI] [PubMed] [Google Scholar]
- Watanabe M. (1970). DNA synthesis in polyploid and binucleate hepatic cells in the regenerating rat liver of different ages. Nagoya J. Med. Sci. 33, 1-11 [PubMed] [Google Scholar]
- Watson W. H., Heilman J. M., Hughes L. L., Spielberger J. C. (2008). Thioredoxin reductase-1 knock down does not result in thioredoxin-1 oxidation. Biochem. Biophys. Res. Commun. 368, 832-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisend C. M., Kundert J. A., Suvorova E. S., Prigge J. R., Schmidt E. E. (2009). Cre activity in fetal albCre mouse hepatocytes: Utility for developmental studies. Genesis 47, 789-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte A. B., Anestal K., Jerremalm E., Ehrsson H., Arner E. S. (2005). Inhibition of thioredoxin reductase but not of glutathione reductase by the major classes of alkylating and platinum-containing anticancer compounds. Free Radic. Biol. Med. 39, 696-703 [DOI] [PubMed] [Google Scholar]
- Yoo M. H., Xu X. M., Carlson B. A., Gladyshev V. N., Hatfield D. L. (2006). Thioredoxin reductase 1 deficiency reverses tumor phenotype and tumorigenicity of lung carcinoma cells. J. Biol. Chem. 281, 13005-13008 [DOI] [PubMed] [Google Scholar]
- Yoo M. H., Xu X. M., Carlson B. A., Patterson A. D., Gladyshev V. N., Hatfield D. L. (2007). Targeting thioredoxin reductase 1 reduction in cancer cells inhibits self-sufficient growth and DNA replication. PLoS ONE 2, e1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.