Abstract
The cell cycle is tightly orchestrated during normal development. Embryonic stem (ES) cells have a unique cell cycle structure, where the G1/S restriction is largely absent enabling cells to rapidly move through G1 and enter S phase. This hastened cell cycle allows the early embryo to rapidly grow. Recent experiments suggest that small non-coding RNAs, the microRNAs, play a central role in achieving this unique cell cycle structure. The responsible microRNAs function by suppressing multiple inhibitors of the G1/S transition. Expression of these miRNAs drops dramatically as the ES cells differentiate and the G1 phase extends. Some of the same miRNAs are overexpressed in cancers where they can promote tumor growth suggesting common mechanisms of miRNA regulated cell cycle control in ES cells and cancers. This review discusses these recent findings in the context of broader knowledge of cell cycle control in normal and abnormal development.
G1/S transition in somatic cells
The G1 phase is a gap period between cytokinesis and DNA replication. During the G1 phase a cell senses its environment for the presence of growth factors and nutrients as well as evaluates the integrity of its genome. These roles are accomplished through a restriction or check point at the G1/S transition (1). Following the restriction point, a cell can pass through S phase and mitosis independent of mitogens. The G1 restriction point requires the sequential activation of the Cdk4/6 and the Cdk2 kinases, which are expressed throughout the cell cycle but only activated upon binding of their specific cyclins. During the early G1 phase, the mitogenic factors stimulate the expression of the D-type cyclins. The Cdk4/6–Cyclin D complex then phosphorylates proteins of the retinoblastoma (pRB) family. This event leads to a partial inhibition of RB and release of the E2F transcription factors, increasing the transcription of the E2F targets. Among the E2F targets there are the E-type cyclins, which activate Cdk2 further phosphorylating RB. This feed-forward loop fully releases E2F, leading to the transcription of genes required for progression through S phase. In addition, the Cdk2–Cyclin E also phosphorylates several other targets important in the progression through S phase (2, 3). Upstream inhibitors including members of the INK (p15, p16 and p18) and CIP families (p21, p27 and p57) modulate the activity of the Cdk–Cyclin complexes. Some of these inhibitors are induced upon stresses such as nucleotide depletion and DNA damage. For example, the DNA damage checkpoint pathway upregulates the expression of p21 through the post-translational modification of p53, which arrests cells in the G1 phase until feedback from the DNA repair machinery promotes transition into the S phase (4). Differential expression of the cell cycle regulatory factors including E2F, RB, Cdk, Cyclins and Cdk inhibitors shapes the G1/S transition kinetics in different cell types. Aberrations in the expression of these regulatory factors can lead to uncontrolled proliferation, the hallmark of cancer (5, 6).
miRNA biogenesis and function
miRNAs are a class of regulatory small RNAs important in a variety of developmental and physiological processes (7). These small RNAs (18-24 nucleotides in length) are broadly present in eukaryotic organisms and repress gene expression by destabilizing target mRNAs as well as inhibiting their translation. Mature miRNAs are generated through two sequential cleavages by RNase III enzymes (8). They are usually transcribed as a part of a long RNA transcript (pri-miRNA) by pol II. The first cleavage is conducted in the nucleus by the microprocessor complex (9, 10) consisting of the RNaseIII enzyme Drosha and its RNA binding partner DGCR8. The cleavage generates a short hairpin (pre-miRNA) around 60-75 nucleotides. The pre-miRNA is then exported into the cytoplasm by Exportin 5 in a Ran-GTP dependent manner. Another RNase III enzyme Dicer along with its partner TRBP conducts the second cleavage on the pre-miRNA to generate the mature miRNA duplex. The duplex enters a third protein complex called the RNA induced silencing complex (RISC), which produces and directs the mature miRNA to its targets. Mature miRNAs bind to the 3′UTR and coding regions of their target mRNAs by an imperfect Watson-Crick base pairing. In particular, miRNA targets are largely determined through base pairing between a small sequence of 7 nucleotides (the seed sequence) at the 5′ end of the miRNA and a matching sequence in the mRNA. This small degree of required complementarity enables a great deal of flexibility. Accordingly, miRNAs are expected to regulate a third of all protein-coding genes in human cells (11). Therefore it is not surprising that there exists a significant crosstalk between the miRNAs and the cell cycle regulatory factors, and that cancer cells often modify the miRNA-mediated regulation for their own proliferative advantage (12).
The link between miRNAs and cell cycle regulation in ES cells
ES cells have a very short G1 phase and lack a functional restriction or check point at the G1/S transition. In this respect they are similar to many cancers(13). In mouse ES cells, the Cdk4/Cdk6-Cyclin D complex is not present (14), while the Cdk2–Cyclin E complex is constitutively active throughout the cell cycle (15). During differentiation, the restriction or check point pathway is established. Accordingly Cdk4/Cdk6–Cyclin D and Cdk2-Cyclin E activity becomes cell cycle regulated and responsive to external cues. These events lead to an elongated G1 phase and slower proliferation of the resulting somatic cells (16). A central role for miRNAs in the ES cell cycle was initially suggested by the analysis of ES cell models involving the deletion of either Dicer (17) or Dgcr8 (18), two key components in the miRNA biogenesis pathway. The loss of either resulted in slower proliferation. Careful analysis of the proliferation phenotype in Dgcr8 knock out cells uncovered a relative accumulation of cells in the G1 phase of the cell cycle. This finding suggested that miRNAs play a role in promoting the G1/S transition in ES cells. However, to prove this hypothesis, it was essential to uncover the specific miRNAs responsible.
Members of the miR-290 cluster regulates the G1/S transition in ES cells
The reintroduction of wild-type Dgcr8 into Dgcr8 knockout ES cells rescued the proliferation and cell cycle defects, proving these defects are not due to the secondary and irreversible cellular events. In addition, the reversibility suggested that it may be possible to rescue these defects by reintroducing individual miRNAs. To this end, a screening strategy was established where chemically synthesized miRNA duplexes, called miRNA mimics, were individually transfected into the Dgcr8 knockout cells (19). The transfected cells were then evaluated for changes in their rate of cell proliferation. Using a colorimetric based assay in 96 well plates, it was possible to expand this screen to hundreds of miRNA mimics. This unbiased screening approach identified multiple miRNAs that partially rescued the proliferation defect. Most of these miRNAs shared a common seed sequence “AAGUGCU”. These miRNAs include members of the miR-290 cluster (miR-291-3p, miR-294, and mir-295) and the miR-302 cluster, and those with the slightly different seed sequence “AAAGUGC” including miR-20, miR-93 and miR-106 belonging to the miR-17/20/106 family. Similar seed sequences suggest similar sets of genes are regulated by these miRNAs and, therefore, there is a high degree of redundancy among these miRNAs. All these miRNAs are expressed in wild type ES cells. The mir-290 cluster alone makes up greater than 70% of the total quantity of miRNAs in ES cells (20). Members of this cluster are co-transcribed as a single transcript (21), suggesting synergistic regulation by these miRNAs. Furthermore, expression of this cluster is rapidly downregulated upon differentiation, coincident with the elongation of the cell cycle. Members from the miR-17/20/106 family are analogously highly expressed in many cancers promoting their growth (22).
Further characterization showed that the miR-291-3p, miR-294 and miR-295 fully rescued the G1 accumulation phenotype suggesting they were acting to promote the G1/S transition (Fig. 1). To confirm this hypothesis it was essential to identify the targets of these miRNAs. The previous work on the ES cell cycle had provided important hints to what these targets may be (14-16, 23). Specifically, the miRNAs are presumably acting through the Cdk2–Cyclin E pathway as this is the key G1/S promoting pathway in ES cells. Moreover, a previous report showed p21 which inhibits the Cdk2-Cyclin E activity is post-transcriptionally regulated during ES cell differentiation with p21 protein, but not mRNA, levels dramatically increasing as the G1 phase extended (24). Indeed, p21 levels were elevated in the Dgcr8 knockout cells, while introduction of the rescuing miRNAs reduced p21 to the wild-type level. p21 was then confirmed as a direct target of the miRNAs by luciferase assays. However, miRNAs are unlikely to function through a single target as they are known to influence the levels of numerous proteins(25, 26). Indeed, the overexpression of p21 in wild-type ES cells only partially phenocopied the G1 accumulation seen in the Dgcr8 knockout cells. Further analysis using mRNA profiling data, target prediction, and luciferase assays identified additional inhibitors of the Cdk2–Cyclin E pathway including p130 (Rbl2) and Lats2 as direct targets of these miRNAs (Fig. 1). These results show that the members of the miR-290 cluster act to suppress several well known inhibitors of the G1/S transition thereby modulating the cell cycle structure of ES cells. Interestingly, mir-106b has recently been reported to promote cell cycle progression in a breast cancer cell line (27) by mechanisms very similar to the mir-290 cluster in ES cells reflecting parallels in the molecular control of the cell cycle of embryonic and cancer cells.
Figure 1.
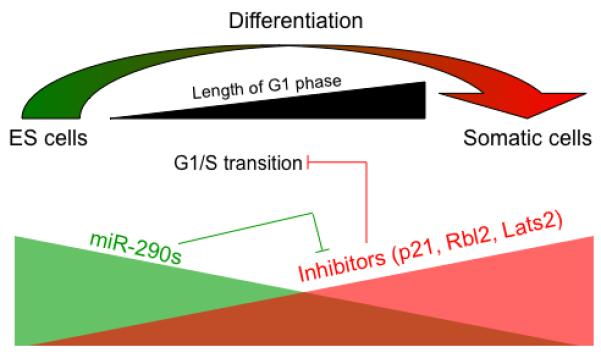
The miR-290 family suppresses multiple inhibitors of the G1/S transition to enable a short G1 phase in ES cells. During differentiation, the expression level of the miR-290 family is downregulated and the G1 phase of the cell cycle is elongated.
The screening approach in the miRNA deficient background provides a powerful strategy to identify functions of individual miRNAs. Widespread redundancy of miRNAs often impedes assigning biological functions to specific miRNAs by reverse genetic approaches. Starting with cells deficient for all miRNAs and reintroducing individual miRNAs overcomes this problem. For example, besides the miR-290 cluster, miRNAs from the miR-302 cluster and the miR-17/20/106 family are also expressed in ES cells (20). All together more than ten miRNAs containing the seed sequence “AAGUGCU” or “AAAGUGC” make up a majority of the total miRNA population in ES cells. This degree of redundancy makes it very difficult to use a reverse knockdown approach. Indeed, knockdown of a number of these miRNAs individually or even in combination did not produce a significant phenotype (19).
Future directions
miRNAs have a broad impact on both the transcriptome and proteome of cells. The three targets p21, Rbl2 and Lats2 are certainly not the sole targets of the miR-290 cluster. Only systems approaches can generate a global image of the miRNA regulation. Indeed, our preliminary studies suggest that dozens of cell cycle related genes are regulated by miR-294. It will be a challenge to study the relative contribution of each of the targets. However, dissection of such complex regulation is critical, as it is key to understanding the mechanisms governing fundamental cellular processes in early embryonic development.
Homologs of miR-291-3p, miR-294 and miR-295 are highly expressed in early embryos of other species including the miR-430 family in Zebrafish embryos (28) and the miR-371/2/3 and mir-302 clusters in human ES cells (29). It will be important to determine if these miRNAs regulate cell cycle progression in these systems in a way analogous to the miR-290 cluster in mouse ES cells. Human ES cells, like mouse ES cells, have an expedited cell cycle progression with a shortened G1 phase. However, many components involved in cell cycle regulation are differentially expressed between human and mouse ES cells (30). It was recently reported that the miR-302 cluster promotes cell cycle progression in human ES cells (31). This report identified Cyclin D1 as the target, suggesting yet another potential means for these miRNAS to promote the ES cell cycle, although how repression of cyclin D1 could promote cell cycle progression is unclear. In mouse, the miR-290 and mir-17 clusters are also highly expressed in primordial germ cells (32). It will be interesting to determine whether they perform similar functions in these cells as they do in ES cells. Furthermore, potential roles for these or related miRNAs in regulating the cell cycle of organ resident stem/ progenitor cells should be an exciting avenue of inquiry.
Another important question is how the expression of these miRNAs is regulated. What are the cis-elements (promoters and enhancers), the transcription factors and the epigenetic regulators that sustain extremely high levels of expression of these miRNAs during early development and then silenced in differentiated somatic cells? A recent study has begun to gain insights into the transcriptional regulation of these miRNAs, at least in ES cells, by showing the binding of pluripotency factors Oct4, Sox2, Nanog and Tcf3 on the promoter of the mir-290 and miR-302 clusters (20, 31). Manipulating the expression or function of this large family of cell cycle promoting miRNAs may provide an important therapeutic avenue. For example, inhibiting their expression in cancer cells may suppress tumor growth in vivo. Alternatively, ectopic expression may be useful in the expansion of adult stem cells, which then can be harnessed to replace damaged tissues.
Acknowledgements
Grant support: National Institutes of Health K08 NS48118 and RO1 NS057221, California Institute of Regenerative Medicine Seed Grant RS1-0161-1, Stem Cell Research Foundation, and the Pew Foundation. YMW is supported by a California Institute of Regenerative Medicine training grant.
We thank members in Blelloch Lab for critical reading of the manuscript.
References
- 1.Sherr CJ, Roberts JM. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9(10):1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 2.Fukasawa K. P53, cyclin-dependent kinase and abnormal amplification of centrosomes. Biochim. Biophys. Acta. 2008;1786(1):15–23. doi: 10.1016/j.bbcan.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein GS, van Wijnen AJ, Stein JL, et al. An architectural perspective of cell-cycle control at the G1/S phase cell-cycle transition. J. Cell Physiol. 2006;209(3):706–710. doi: 10.1002/jcp.20843. [DOI] [PubMed] [Google Scholar]
- 4.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 5.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432(7015):298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 6.Deshpande A, Sicinski P, Hinds PW. Cyclins and cdks in development and cancer: a perspective. Oncogene. 2005;24(17):2909–2915. doi: 10.1038/sj.onc.1208618. [DOI] [PubMed] [Google Scholar]
- 7.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet. 2008;9(11):831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faller M, Guo F. MicroRNA biogenesis: there's more than one way to skin a cat. Biochim. Biophys. Acta. 2008;1779(11):663–667. doi: 10.1016/j.bbagrm.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gregory RI, Yan KP, Amuthan G, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432(7014):235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 10.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18(24):3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Bueno MJ, de Castro IP, Malumbres M. Control of cell proliferation pathways by microRNAs. Cell Cycle. 2008;7(20):3143–3148. doi: 10.4161/cc.7.20.6833. [DOI] [PubMed] [Google Scholar]
- 13.Berthet C, Kaldis P. Cell-specific responses to loss of cyclin-dependent kinases. Oncogene. 2007;26(31):4469–4477. doi: 10.1038/sj.onc.1210243. [DOI] [PubMed] [Google Scholar]
- 14.Savatier P, Lapillonne H, van Grunsven LA, Rudkin BB, Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12(2):309–322. [PubMed] [Google Scholar]
- 15.Stead E, White J, Faast R, Conn S, Goldstone S, Rathjen J, Dhingra U, Rathjen P, Walker D, Dalton S. Pluripotent cell division cycles are driven by ectopic Cdk2, cyclin A/E and E2F activities. Oncogene. 2002;21(54):8320–8333. doi: 10.1038/sj.onc.1206015. [DOI] [PubMed] [Google Scholar]
- 16.White J, Stead E, Faast R, Conn S, Cartwright P, Dalton S. Developmental activation of the Rb-E2F pathway and establishment of cell cycle-regulated cyclin-dependent kinase activity during embryonic stem cell differentiation. Mol Biol Cell. 2005;16(4):2018–2027. doi: 10.1091/mbc.E04-12-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. U S A. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R. Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat. Genet. 2008;40(12):1478–1483. doi: 10.1038/ng.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11(8):1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faast R, White J, Cartwright P, Crocker L, Sarcevic B, Dalton S. Cdk6-cyclin D3 activity in murine ES cells is resistant to inhibition by p16(INK4a) Oncogene. 2004;23(2):491–502. doi: 10.1038/sj.onc.1207133. [DOI] [PubMed] [Google Scholar]
- 24.Sabapathy K, Klemm M, Jaenisch R, Wagner EF. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 1997;16(20):6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 26.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanovska I, Ball AS, Diaz RL, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell Biol. 2008;28(7):2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 29.Bar M, Wyman SK, Fritz BR, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neganova I, Lako M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 2008;213(1):30–44. doi: 10.1111/j.1469-7580.2008.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Card DA, Hebbar PB, Li L, et al. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell Biol. 2008;28(20):6426–6438. doi: 10.1128/MCB.00359-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayashi K, Chuva de Sousa Lopes SM, Kaneda M, et al. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3(3):e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]


