Abstract
Oceanic islands are well known for harboring diverse species assemblages and are frequently the basis of research on adaptive radiation and neoendemism. However, a commonly overlooked role of some islands is their function in preserving ancient lineages that have become extinct everywhere else (paleoendemism). The island archipelago of Bermuda is home to a single species of extant terrestrial vertebrate, the endemic skink Plestiodon (formerly Eumeces) longirostris. The presence of this species is surprising because Bermuda is an isolated, relatively young oceanic island approximately 1000 km from the eastern United States. Here, we apply Bayesian phylogenetic analyses using a relaxed molecular clock to demonstrate that the island of Bermuda, although no older than two million years, is home to the only extant representative of one of the earliest mainland North American Plestiodon lineages, which diverged from its closest living relatives 11.5 to 19.8 million years ago. This implies that, within a short geological time frame, mainland North American ancestors of P. longirostris colonized the recently emergent Bermuda and the entire lineage subsequently vanished from the mainland. Thus, our analyses reveal that Bermuda is an example of a “life raft” preserving millions of years of unique evolutionary history, now at the brink of extinction. Threats such as habitat destruction, littering, and non-native species have severely reduced the population size of this highly endangered lizard.
Introduction
“…it appears that the true history of the colonization of the land, now Bermuda, is lost forever in oblivion” [1]
Studies of island biodiversity have focused largely on adaptive radiations associated with neoendemism (i.e., “cradles” of diversity) [2]–[4]. There are myriad factors that promote spectacular biodiversity on islands, but the factors that contribute to neoendemism ultimately derive from the fact that the islands formed de novo with no connection to a larger landmass and thus have abundant “empty” ecological niche space. However, the isolation that defines islands can also preserve genetic diversity of relict lineages, a pattern known as paleoendemism [3], [5]. The most prominent example of this phenomenon among vertebrates is the tuatara (Sphenodon) that represents a clade of reptiles once widespread, but now restricted to two remaining species found only on the offshore islands of New Zealand. Moreover, given numerous threats such as climate change, introduced species, and habitat destruction, insular fauna are subject to increased risk of extinction [6]. This vulnerability is of particular concern for paleoendemics, as these taxa represent a disproportionately high amount of phylogenetic diversity [7]–[9].
The islands of Bermuda (32.33°N, 64.75°W) are an isolated, 54 km2 archipelago (referred to as “island”, hereafter) approximately 1000 km east of the United States (Fig. 1a). The island is currently home to a single endemic terrestrial vertebrate, the scincid lizard Plestiodon longirostris (formerly Eumeces longirostris [10], [11]), although several other terrestrial vertebrates, including a tortoise (Hesperotestudo) and multiple species of birds, inhabited the island until from the Middle to Late Pleistocene [12]–[19]. Plestiodon longriostris is currently considered critically endangered [20] and faces continuing threats of extinction through human-caused habitat loss, competition with and predation from introduced species, and entrapment in discarded bottles [21]. Once abundant [22], [23], the species' range has dwindled to several small sub-populations, the largest of which occurs on Southampton Island and contains only an estimated 400 individuals [21]. Preliminary microsatellite analysis indicates that the current genetic diversity of P. longirostris is low [24].
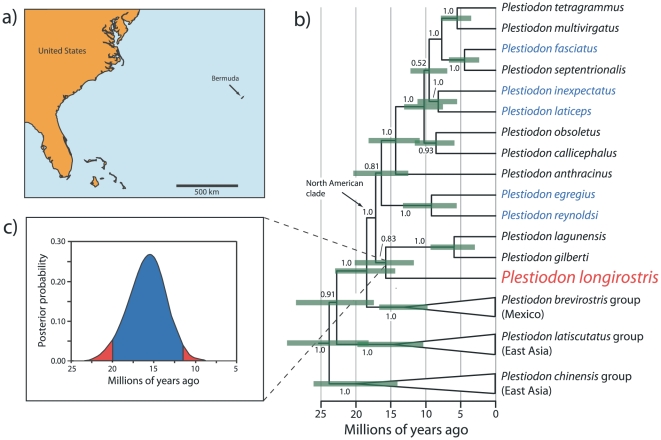
Figure 1. The location of Bermuda, phylogeny of Plestodon, and molecular estimates of divergence times.
a., Map showing the location of Bermuda relative to North America. b., Phylogeny of the genus Plestiodon (outgroups not shown; see Brandley et al, 2010). Branch lengths are in units of time and represent the means of the posterior distribution. Numbers above or below the nodes indicate posterior probabilities. Triangles indicate groups for which multiple species were sampled, but are not shown. Taxa in blue are those that inhabit eastern North America. Green bars indicate the 95% credible interval for estimated divergence dates for that node. c., Posterior probability distribution of the age of the divergence between Plestiodon longirostris and its sister lineage. Areas shaded in red are values that exceed the 95% credible interval.
Pre-plate tectonics biogeographic hypotheses assumed that P. longirostris was a relict of an ancient lineage that became isolated on Bermuda after the closing of a land bridge [1]. This “relict” hypothesis was also influenced by the species' unique morphological characteristics (including an elongated snout). However, recent geologic studies have demonstrated that, although part of an ancient, volcanic sea-mount, most of the exposed, terrestrially habitable portion of Bermuda has never been connected to a larger landmass and instead consists of limestone deposited in the Pleistocene [25]–[28], the maximum age of which is no more than two million years [29]. Moreover, all known Bermudan vertebrate fossils are limited to sediments from the latter half of the Pleistocene [13]–[19]. A more reasonable alternative hypothesis is that the species is quite young, at most as old as the maximum age of Bermuda (1–2 million years), and is a descendent of one of the several species of Plestiodon inhabiting the eastern United States that dispersed over water to Bermuda following the emergence of the island. Another reptile (indeed, the only other potentially native reptile), the diamondback terrapin (Malaclemys terrapin), is a likely very recent immigrant that descended from populations of the same species that currently inhabit the eastern United States [30], [31].
To evaluate the evolutionary history of this unique skink, and specifically to test the “relict” and “recent immigrant” hypotheses, we conducted Bayesian relaxed molecular clock divergence dating analyses of an extensive, multi-locus DNA data set including all Plestiodon species from the eastern United States and the Bermudan P. longirostris. We demonstrate that the oceanic island of Bermuda, despite its young age (no more than two million years) and history of extreme changes in available habitat, harbors the last representative of one of the earliest North American lineages of Plestiodon that diverged ∼16 million years ago.
Results and Discussion
The results of the phylogenetic analysis (pruned to show relevant taxa only; Fig. 1b) show that the lineage that includes modern P. longirostris does not descend from any of the extant Plestiodon lineages that inhabit eastern North America. In fact, Bayesian divergence dating analyses using a relaxed molecular clock indicate that it is one of the earliest diverging lineages of the entire North American clade (Figs. 1b,c). Furthermore, this divergence occurred ∼16 million years ago (Ma) (95% credible interval = 11.5 to 19.8 Ma), well before any modern species, and the lineage contains no extant representative other than P. longirostris. We are therefore left with the remarkable conclusion that a two million-year-old island contains the sole survivor of an ancient lineage that predates the existence of Bermuda by well over 10 million years.
Although no fossils of the P. longirostris lineage are known from mainland North America [1], the phylogenetic and divergence date data clearly indicate that ancestors once existed on the mainland. However, P. longirostris is present in Bermudan fossil beds dated to approximately 400,000 years ago. Given the young age of Bermuda, these results support the hypothesis that within the past 400,000 to two million years, individuals from the North American mainland P. longirostris lineage dispersed to a presumably recently emerged Bermuda 1000 km offshore, yet subsequently became extinct on the mainland. Although we can only speculate how these colonizing individuals dispersed over water, we note that both hurricanes and ocean currents are known to transport living lizards and debris to and from islands [32]–[33], and that the powerful Gulf Stream ocean current runs along eastern North America to the mid-Atlantic Ocean [see 31].
Thus, despite its young age, the island preserves the last representative of one of the oldest lineages of mainland North American Plestiodon – it is essentially an evolutionary “life raft”. This “life raft” role is remarkable considering that extreme fluctuations in sea level during the Pleistocene have intermittently decreased the available terrestrial habitat on Bermuda by orders of magnitude [18]. This extreme contraction in habitat was the likely cause of the extinction of several endemic birds [13]–[19]. Bermuda's only other native reptile, Malaclemys terrapin, only colonized the island in the past 3000 to 400 years from populations that currently inhabit the Eastern United States [30].
These results are extremely unlikely to be the result of error in divergence date estimates or phylogenetic uncertainty. One advantage of Bayesian methods of divergence dating is their ability to incorporate error in both the calibration age constraint and phylogeny, and to infer posterior probability distributions of estimated ages. The 95% credible interval of the date of divergence between P. longirostris and other North American Plestiodon species ranges from 11.5 to 19.8 Ma and excludes the earliest date that Bermuda may have emerged (2 Ma). Although the relationship between P. longirostris and its sister lineage is not well supported (posterior probability<0.95), this species is nonetheless excluded from any other younger clades with strong statistical support (Fig. 1b). In other words, the lineage cannot be any younger than the other major clades that all predate the emergence of Bermuda. Furthermore, if this lack of resolution represents a rapid radiation at the base of the North American Plestiodon phylogeny, then the age of the P. longirostris lineage is even older (∼13–21 Ma; Fig. 1b). Finally, extensive analyses of this data set have demonstrated that the use of models that account for rate heterogeneity among subsets of the DNA dramatically improve divergence date estimates and help mitigate potential problems inherent in using distantly related age calibrations [34].
We also note that the history of P. longirostris somewhat parallels that of the extinct Bermudan turtle, Hesperotestudo bermudae. Fossil evidence indicates this species became extinct on Bermuda 300,000 years ago, yet was the last representative of a genus of tortoise that inhabited North America from the Oligocene to Pleistocene [13], [18]. With the caveat that more recent fossils of North American Hesperotestudo may yet to be discovered, these data currently suggest that Bermuda also served as an evolutionary life raft for this genus after extinction on the mainland.
Although we certainly do not discount the profoundly important role of islands in generating biodiversity, our results highlight the frequently overlooked role of islands in preserving diversity (acting as evolutionary “museums”). This role is of particular importance given that preservation of phylogenetic diversity has been an increasingly important goal of conservation biology as the extinction of “old” species would result in a greater loss of genetic diversity than that of a “young” species with close phylogenetic relatives [7]–[9]. Therefore, because the Bermuda skink, Plestiodon longirostris, represents the sole representative of one of the earliest diverging lineages among North American Plestiodon, efforts to preserve this species are also preserving ∼12 to 20 millions of years of unique evolutionary history at the risk of extinction.
Materials and Methods
Taxon and character sampling
DNA for 62 individuals representing 37 of ∼43 recognized species of Plestiodon and 25 outgroups was isolated from tissue using Qiagen DNeasy™ columns (see [34] for detailed specimen information and methods). We amplified BDNF, MKL, mtDNA [ND1+tRNAs], PRLR, PTGER4, R35, RAG1, and SNCAIP loci using standard PCR techniques (Genbank numbers upon acceptance). In few cases, we were unable to obtain reliable sequences for some species; in this case, another species from the same family was used. PCR products were cleaned using ExoSap-IT (USB Corp.). Purified templates were dye-labeled using BigDye™ (ABI) and sequenced on an ABI 3077™ automated DNA sequencer. Nucleotide sequences were examined and aligned by eye. This process was relatively straightforward for the protein-coding genes (BDNF, MKL, mtDNA ND1, PRLR, PTGER4, R35, RAG1, and SNCAIP) due to their codon reading frames. MtDNA tRNAs were aligned according to their secondary structure, and regions in which homology was uncertain due to multiple insertions and deletions were excluded from subsequent analysis. The size of the final data set for phylogenetic analysis was 7667 bp.
Phylogenetic analyses
Brandley et al. [34] demonstrated that accommodating different rates of evolution among subsets of DNA data may improve divergence time estimation, especially when different subsets of the data evolve at different rates. We therefore partitioned the data a priori by locus and codon position (and a single partition for the tRNAs) for a total of 28 partitions. For each partition, we determined the appropriate model of nucleotide substitution using the Bayesian information criterion (BIC) [35].
All phylogenetic analyses of the combined data set were conducted using BEAST v1.4.8 [36] assuming an uncorrelated lognormal relaxed molecular clock [37]. A total of seven analyses were performed. Each analysis used a coalescent starting tree and was run for 108 generations, sampled every 10,000th generation. We used the program's default prior distributions with the exception of GTR substitution rates in which we used a uniform (0,100) distribution, and the date distributions of the most recent common ancestor of the three clades used for calibration (see below). To determine convergence, we constructed cumulative posterior probability plots for each clade using the cumulative and compare function in AWTY [38]. Posterior probabilities≥0.95 are considered statistically significant clade support [39].
Because the Plestiodon fossil record, and the record of fossil skinks in general, are extremely poor, we used three fossil calibration age prior distributions from non-scincid fossil taxa whose phylogenetic placement in the squamate tree was recently inferred [40]. The age of crown Episquamata (represented here as Anniella, Aspidoscelis, Basiliscus, and Bipes) was calibrated using the age of the earliest stem “anguimorph” fossils, Becklesius, Dorsetisaurus, Paramacellodus, and Pseudosaurilius [34], [40]. We chose a lognormal distribution so that the earliest possible sampled age corresponds to 148 Ma and the older 97.5% credible interval (CI) encompasses the earliest age of crown Squamata (180 Ma: standard deviation = 1.769; [40], [41]). The age of the divergence between Amphisbaenia (Bipes biporus) and Teiidae (Aspidoscelis) was calibrated using the age (Albian - Cenomanian boundary) of the earliest teiioid (Polyglyphanodontidae) fossils, (e.g., Bicuspidon [40], [42]). We chose a lognormal distribution so that the earliest possible sampled age corresponds to 96 Ma and the older 97.5% credible interval (CI) encompasses the earliest age of crown Episquamata (148 Ma; standard deviation = 2.016). The age of Scinciformata (represented here by skinks, Gerrhosauridae, and Xantusiidae) was calibrated using the age (Berriasian) of the fossil Sakurasaurus [40], [43]. We chose a lognormal distribution so that the earliest possible sampled age corresponds to 138 Ma and the older 97.5% credible interval (CI) encompasses the earliest age of the root (151 Ma; standard deviation = 1.309). We therefore enforced the monophyly of these clades in accordance with recent phylogenetic analyses that have inferred these relationships [44], [45]. The full phylogeny, including all outgroups, is provided in Fig. S1.
Supporting Information
Full phylogeny including fossil calibrations (in red). Boxes indicate 95%CIs of node ages.
(1.48 MB EPS)
Acknowledgments
We thank J. Davenport, A. Dornburg, J. McGuire, A. Seago, and one anonymous reviewer for advice and/or comments on the manuscript; J. Aguilar, K. Ashton, J. Campbell, S. Daniels, U. García, L. Kitson, J. Lazell, I. López, R. Macey, A. Mendoza, M. Mociño, R. Murphy, E. Pérez, T. Reeder, J. Richmond, CAS, FMNH, KUZ, LSUMZ, MVZ, UMMZ, and YPM for providing tissue samples; and J. Ichikawa, I. Katsube, A. Mendoza Hernández, E. Pérez Ramos, A. Seago, and Y. Yamamoto for fieldwork assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was funded the National Natural Science Foundation of China (30700062) awarded to X.G.; and the the National Science Foundation East Asia and Pacific Program grants (OISE 0513295 and OISE 0611646, in conjunction with the National Natural Science Foundation of China and the Japan Society for the Promotion of Science), National Science Foundation Doctoral Dissertation Improvement grant (DEB 0709885), the Linnean Society Systematics Fund, and multiple grants from the Museum of Vertebrate Zoology awarded to M.C.B. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Taylor EH. A taxonomic study of the cosmopolitan scincoid lizards of the genus Eumeces with an account of the distribution and relationships of its species. Kansas Univ Sci Bull. 1935;23:1–641. [Google Scholar]
- 2.Schluter D. Oxford: Oxford University Press; 2000. The Ecology of Adaptive Radiation.296 [Google Scholar]
- 3.Gillespie RG, Roderick GK. Arthropods on islands: colonization, speciation, and conservation. Ann Rev Entomol. 2002;47:595–632. doi: 10.1146/annurev.ento.47.091201.145244. [DOI] [PubMed] [Google Scholar]
- 4.Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature. 2009;457:830–835. doi: 10.1038/nature07893. [DOI] [PubMed] [Google Scholar]
- 5.Stebbins GL, Major J. Endemism and speciation in the California flora. Ecol Monogr. 1965;35:1–35. [Google Scholar]
- 6.Jamieson IG. Has the debate over genetics and extinction of island endemics truly been resolved? Animal Conserv. 2007;10:139–144. [Google Scholar]
- 7.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv. 1992;61:1–10. [Google Scholar]
- 8.Redding DW, Mooers AØ. Incorporating evolutionary measures into conservation prioritization. Conserv Biol. 2006;20:1970–1978. doi: 10.1111/j.1523-1739.2006.00555.x. [DOI] [PubMed] [Google Scholar]
- 9.Isaac NJB, Turvey ST, Collen B, Waterman C, Baillie JEM. Mammals on the EDGE: conservation priorities based on threat and phylogeny. PLoS One. 2007;2:e296. doi: 10.1371/journal.pone.0000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandley MC, Schmitz A, Reeder TW. Partitioned Bayesian analyses, partition choice, and the phylogenetic relationships of scincid lizards. Syst Biol. 2005;54:373–390. doi: 10.1080/10635150590946808. [DOI] [PubMed] [Google Scholar]
- 11.Smith HM. Plestiodon: a replacement name for most members of the genus Eumeces in North America. J Kan Herpetol Soc. 2005;14:15–16. [Google Scholar]
- 12.Sterrer W. How many species are there in Bermuda? Bull Mar Sci. 1998;62:809–840. [Google Scholar]
- 13.Meylan PA, Sterrer W. Hesperotestudo (Testudines: Testudinidae) from the Pleistocene of Bermuda, with comments on the phylogenetic position of the genus. Zool J Linn Soc. 2000;128:51–76. [Google Scholar]
- 14.Olson SL, Wingate DB. Two new species of flightless rails (Aves: Rallidae) from the Middle Pleistocene “crane fauna” of Bermuda. Proc Biol Soc Wash. 2000;113:356–368. [Google Scholar]
- 15.Olson SL, Wingate DB. A new species of large flightless rail of the Rallus longirostris/elegans complex (Aves: Rallidae) from the late Pleistocene of Bermuda. Proc Biol Soc Wash. 2001;114:509–516. [Google Scholar]
- 16.Olson SL, Hearty PJ. Probable extirpation of a breeding colony of short-tailed albatross (Phoebastria albatrus) on Bermuda by Pleistocene sea-level rise. Proc Nat Acad Sci U S A. 2003;100:12825–12829. doi: 10.1073/pnas.1934576100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olson SL, Wingate DB, Hearty PJ, Grady FV. Prodromus of Vertebrate Palaeontology and Geochronology of Bermuda. Monografies de la Societat d'Historia Natural de les Balears. 2005;12:219–232. [Google Scholar]
- 18.Olson SL, Hearty PJ, Pregill GK. Geological constraints on evolution and survival in endemic reptiles on Bermuda. J Herpetol. 2006;40:394–398. [Google Scholar]
- 19.Olson SL. A new genus and species of buteonine hawk from Quaternary deposits in Bermuda (Aves: Accipitridae). Proc Biol Soc Wash. 2008;121:130–141. [Google Scholar]
- 20.IUCN. 2008. 2008 IUCN Red List of Threatened Species. IUCN, Gland, Switzerland. http://www.iucnredlist.org [accessed 07 April 2008].
- 21.Davenport J, Hills J, Glasspool A, Ward J. Threats to the critically endangered endemic Bermudian skink Eumeces longirostris. Oryx. 2001;35:332–339. [Google Scholar]
- 22.Garman S. The reptiles of Bermuda. Bull US Nat Hist Mus. 1884;25:287–303. [Google Scholar]
- 23.Wingate DB. Terrestrial herpetofauna of Bermuda. Herpetologica. 1965;21:202–218. [Google Scholar]
- 24.Coughlan J, Kitson L, Dillane E, Cross TF, Davenport J. Characterisation of six polymorphic microsatellite loci in the Bermuda skink, Eumeces longirostris. Mol Ecol Notes. 2004;4:678–679. [Google Scholar]
- 25.Hearty PJ, Vacher HL, Mitterer RM. Aminostratigraphy and ages of Pleistocene limestones of Bermuda. Geol Soc Am Bull. 1992;104:471–480. [Google Scholar]
- 26.Hearty PJ, Kindler P, Cheng H, Edwards RL. +20 m middle Pleistocene sea-level highstand (Bermuda and the Bahamas) due to partial collapse of Antarctic ice. Geology. 1999;27:368–375. [Google Scholar]
- 27.Hearty PJ. Revision of the Late Pleistocene sratigraphy of Bermuda. Sed Geol. 2002;153:1–21. [Google Scholar]
- 28.Hearty PJ, Olson SL, Kaufman DS, Edwards RL, Cheng H. Stratigraphy and geochronology of pitfall accumulations in caves and fissures, Bermuda. Quat Sci Rev. 2004;23:1151–1171. [Google Scholar]
- 29.Vacher HL, Hearty PJ, Rowe MP. Stratigraphy of Bermuda: Nomenclature, concepts, and status of multiple systems of classification. Geol Soc Am Special Paper. 1995;300:269–294. [Google Scholar]
- 30.Parham JF, Outerbridge ME, Stuart BL, Wingate DB, Erlenkeuser H, Papenfuss TJ. Introduced delicacy or native species? A natural origin of Bermudian terrapins supported by fossil and genetic data. Biol Lett. 2008;4:216–219. doi: 10.1098/rsbl.2007.0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davenport J, Glasspool AF, Kitson L. Occurrence of diamondback terrapins, Malaclemys terrapin, on Bermuda: native or introduced? Chelonian Conserv Bi. 2005;4:956–959. [Google Scholar]
- 32.Calsbeek R, Smith TB. Ocean currents mediate evolution in island lizards. Nature. 2003;426:552–555. doi: 10.1038/nature02143. [DOI] [PubMed] [Google Scholar]
- 33.Censky EJ, Hodge K, Dudley J. Over-water dispersal of lizards due to hurricanes. Nature. 1998;395:556. [Google Scholar]
- 34.Brandley MC, Wang Y, Guo X, Nieto Montes de Oca A, Fería Ortíz M, Hikida T, Ota H. Accommodating high rates of evolution in molecular dating methods: an example using inter-continental dispersal of Plestiodon (Eumeces) lizards. 2010. Syst Biol, in press. [DOI] [PubMed]
- 35.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6:461–464. [Google Scholar]
- 36.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLOS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 39.Huelsenbeck JP, Rannala B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol. 2004;53:904–913. doi: 10.1080/10635150490522629. [DOI] [PubMed] [Google Scholar]
- 40.Conrad JL. Phylogeny and systematics of Squamata (Reptilia) based on morphology. Bull Am Mus Nat Hist. 2008;310:1–182. [Google Scholar]
- 41.Wiens J, Brandley MC, Reeder TW. Why does a trait evolve multiple times within a clade? Repeated evolution of snake-like body form in squamate reptiles. Evolution. 2006;60:123–141. [PubMed] [Google Scholar]
- 42.Nydam RL, Cifelli RL. A new teiid lizard from the Cedar Mountain Formation (Albian-Cenomanian boundary) of Utah. J Vert Paleo. 2002;22:276–285. [Google Scholar]
- 43.Evans SE, Manabe M. Early Cretaceous lizards from the Okurodani Formation of Japan. Geobios. 1999;32:889–899. [Google Scholar]
- 44.Townsend T, Larson A, Louis EJ, Macey JR. Molecular phylogenetics of Squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst Biol. 2004;53:735–757. doi: 10.1080/10635150490522340. [DOI] [PubMed] [Google Scholar]
- 45.Hugall AF, Foster R, Lee MSY. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full phylogeny including fossil calibrations (in red). Boxes indicate 95%CIs of node ages.
(1.48 MB EPS)