Abstract
The sustained transmission and spread of environmentally mediated infectious diseases is governed in part by the dispersal of parasites, disease vectors and intermediate hosts between sites of transmission. Functional geospatial models can be used to quantify and predict the degree to which environmental features facilitate or limit connectivity between target populations, yet typical models are limited in their geographical and analytical approach, providing simplistic, global measures of connectivity and lacking methods to assess the epidemiological implications of fine-scale heterogeneous landscapes. Here, functional spatial models are applied to problems of surveillance and control of the parasitic blood fluke Schistosoma japonicum and its intermediate snail host Oncomelania haupensis in western China. We advance functional connectivity methods by providing an analytical framework to (i) identify nodes of transmission where the degree of connectedness to other villages, and thus the potential for disease spread, is higher than is estimated using Euclidean distance alone and (ii) (re)organize transmission sites into disease surveillance units based on second-order relationships among nodes using non-Euclidean distance measures, termed effective geographical distance (EGD). Functional environmental models are parametrized using ecological information on the target organisms, and pair-wise distributions of inter-node EGD are estimated. A Monte Carlo rank product analysis is presented to identify nearby nodes under alternative distance models. Nodes are then iteratively embedded into EGD space and clustered using a k-means algorithm to group villages into ecologically meaningful surveillance groups. A consensus clustering approach is taken to derive the most stable cluster structure. The results indicate that novel relationships between nodes are revealed when non-Euclidean, ecologically determined distance measures are used to quantify connectivity in heterogeneous landscapes. These connections are not evident when analysing nodes in Euclidean space, and thus surveillance and control activities planned using Euclidean distance measures may be suboptimal. The methods developed here provide a quantitative framework for assessing the effectiveness of ecologically grounded surveillance systems and of control and prevention strategies for environmentally mediated diseases.
Keywords: geospatial connectivity, environmental transport, infectious disease spread, Schistosoma japonicum, network epidemiology, graph theory
1. Introduction
Infectious disease transmission involves connections between susceptible and infected hosts—connections which, in the case of pathogens with environmental stages or those carried by vectors and intermediate hosts, are strongly mediated by the intervening environmental features between hosts. These features can regulate disease transmission, where, for example, rivers mediate the spread of rabies (Smith et al. 2002), or animal or human hosts migrating across heterogeneous landscapes govern the onset of diseases such as measles or foot-and-mouth disease (Ferguson et al. 2001; Grenfell et al. 2001).
Generally, strong linkages between subpopulations result in strong synchrony in transmission among subpopulations, and persistence, establishment and other effects are known to be modified by network connectivity (Adler 1993; Hess 1996; Ruxton & Rohani 1999; Bjornstad 2001; Koopman et al. 2002). Important practical implications for interventions emerge from these studies, where, for example, focusing antihelminthic treatment on highly infected villages may be inefficient when compared with a regional approach involving careful exploration of network topology to identify key nodes that contribute to downstream infection and targeting those (Gurarie & Seto 2008).
This raises an important opportunity for public health decision-making. If disease persistence and establishment, and intervention optimization, are dependent on connectivity in a system, and that connectivity is strongly environmentally determined, then environmental datasets have the potential to inform public health decisions such as where to focus surveillance efforts, or identifying clusters of related transmission loci. This is especially true for changing environments, where phenomena such as climate change can effectively bring some hosts (or vectors) closer to (and push some further from) vectors (or hosts) than they have been historically, a context where environmental data may be crucial (Sutherst 2004).
Metapopulation models are commonly used to conceptualize the role of connectivity in infectious disease systems (Hanski 2001), and the popularity of these approaches has led to calls for rigorous quantification of host, vector and parasite migration between patches (Ferguson et al. 2001; Hanski 2001). Both graph and spatial network models are limited in their applications to systems with complex responses to environmental heterogeneity. For instance, typical graph network models applied to directly transmitted diseases are constructed such that only certain pairs of nodes are connected (Keeling & Eames 2005). Yet, in the context of environmentally mediated diseases at the community scale, where distant transport of vectors or free-living stages is possible, there is rarely a definitive basis for excluding edges between node pairs altogether. Methods that account for the strength of connections (rather than treating edges as either present or absent; Keeling 1999) and connection asymmetries (instead of simple symmetric linkages between nodes; Jeger et al. 2007) are needed for organisms with dispersive environmental stages subject to directional environmental flows. In those rare cases where the strength of interaction between nodes has been evaluated, graph theory measures of network properties (degree distribution, triples, etc. which ignore edge weights) are ineffective (Brooks et al. 2008), and edge weights are often dropped prior to analysis (Urban & Keitt 2001; Brooks et al. 2008).
Spatial network models used to simulate spread across continuous space have similar limitations. They are typically based on radially symmetric, monotonically decreasing functions of distance that define a spatial sphere of influence (e.g. an exponential kernel) assuming spatial homogeneity and isotropy (Reluga et al. 2006; Brooks et al. 2008; Parham et al. 2008). Where directional forces (anisotropy) have been considered for spatial point processes, they have been applied as a constant force across the spatial domain (Soubeyrand et al. 2008). Such models are of limited value when transmission processes exhibit variable, directional dispersal rates mediated by heterogeneous landscape features, as in the case of waterborne- or habitat-mediated, vector-borne transmission.
New analytical tools are thus needed that account for complex environmental heterogeneity, network asymmetries and stochastic dispersal modes, while providing measures of local and global network properties. These methods are especially desirable for disease agents with free-living stages, where the geometry and magnitude of connections between patches are highly sensitive to environmental factors, including land cover and hydrology (Gurarie & Seto 2008). Here, we provide a rigorous approach for just such a system, focusing on human schistosomes, which exhibit multiple free-living stages as well as transmission by intermediate hosts, and are thus model organisms for exploring the influence of environmental resistance to (or facilitation of) disease spread through heterogeneous landscapes. The re-emerging schistosomiasis context in Sichuan, China, offers a prime example of where decision-making tools for response and surveillance are badly needed.
In China, schistosomiasis re-emergence is defined as the incidence of new cases of infection where there had been previous attainment of transmission control criteria (prevalence of human and cattle infections less than 1%, no new infections in children less than 12 years old and in cattle less than 2 years old, no acute human cases and snail habitat reduced by 98% from pre-control levels; Liang et al. 2006). New acute cases over the last decade in Sichuan have signalled re-emergence of the disease and have triggered investigations by local disease control agencies as to whether these cases were the result of local transmission or importation. Infection examinations of potential human and cattle hosts were carried out throughout the geographical region, yet at the time, no systematic method was available to inform the selection of villages to include in surveys.
Environmental or social distances that define the degree of connection (or disconnection) between populations, hosts, vectors and pathogens can be useful in this context, and methods for estimating these distance metrics are greatly needed. Euclidean distance is clearly a good candidate for fulfilling this role; it is simple to estimate, easy to interpret and supported by well-developed statistical methods. Yet, there is evidence that Euclidean metrics alone fail to measure epidemiological distance when environmental pathways lie on heterogeneous landscapes (Ferguson et al. 2001; Grenfell et al. 2001). What is more, simple Euclidean distance may not sufficiently represent social distances that mediate transmission processes (Miller & Wentz 2003).
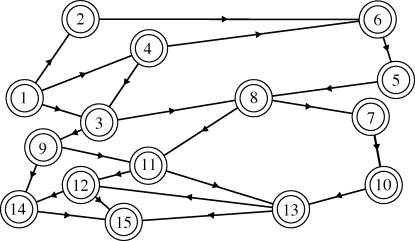
We present methods for estimating and analysing the influence of environmental or social distances that define the degree of connection (or disconnection) between hosts, vectors and pathogens. We make use of simple environmental models in this study in order to emphasize methods that convert model output into public health decision-making tools. A technique is presented for prioritizing treatment or surveillance in neighbour nodes when responding to an acute case, as is a method for re-clustering nodes, based on connectivity information, to form new spatial units of surveillance or coordination. We apply these approaches to Schistosoma japonicum, the parasite that causes schistosomiasis in East and Southeast Asia. The larval stages are fully aquatic, as are the juvenile stages of the intermediate host, thus hydrology plays a major role in determining ease of larval and snail host dispersal between villages. Adult intermediate hosts are amphibious, and their dispersal is determined by a set of terrestrial land cover, soil moisture and other environmental parameters. Key environmental determinants of schistosome infection in western China have been identified (Spear et al. 2004; Remais et al. 2007), including dispersive flows such as hydrology and snail movement that modify S. japonicum transmission along paths between villages in rural Sichuan Province, China, illustrated diagrammatically in figure 1. Parasites enter the environment as eggs that hatch in water into a free-swimming miracidium that seeks a specific species of snail, Oncomelania hupensis, to infect. The snails are amphibious and largely inhabit the margins of irrigation canals where they are subject to advective transport as well as active dispersal. Asexual reproduction of the parasite within the snail produces cercariae, another free-swimming aquatic stage with a lifespan of the order of a day. These larvae penetrate the intact skin of a definitive host (human or other mammal) and mature into adult worms. Eggs are excreted in faeces, which find their way into the environment in the absence of basic sanitation, through alternative mammalian hosts, or through the use of human waste as fertilizer, and begin the cycle again. We present methods that, accounting for connectivity, can be used to plan surveillance efforts or identify clusters of environmentally related loci in the context of S. japonicum transmission.
Figure 1.
Diagrammatic model of connected village nodes where land cover and hydrology-dependent diffusion of the parasite and intermediate host is estimated on the paths between nodes.
2. Material and methods
2.1. Study sites
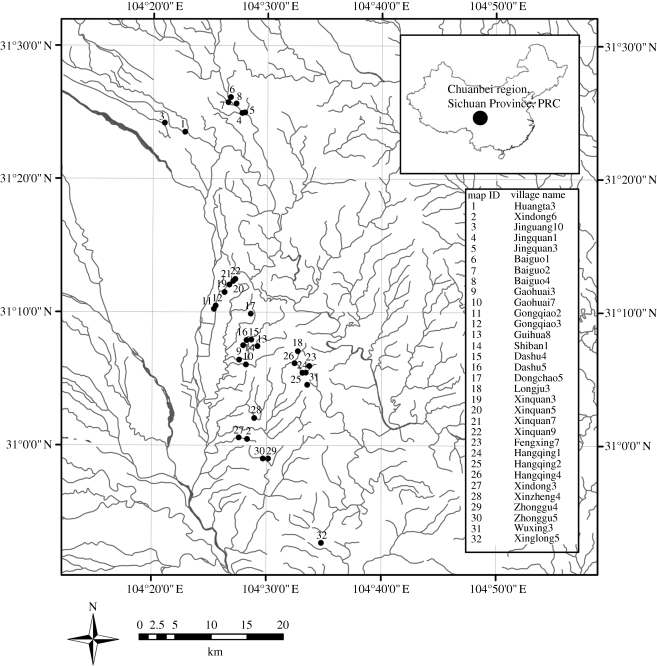
The study was conducted in 32 villages (figure 2) within three counties in the Chuanbei region of Sichuan Province, People's Republic of China (104°29′ E, 31°06′ N). Schistosoma japonicum has re-emerged in this region in areas that had previously attained transmission control according to Chinese Ministry of Health guidelines (Liang et al. 2006). The villages lie on the mountainous areas surrounding the city of Deyang. The region is characterized by a subtropical climate with an annual average temperature of 17°C and annual rainfall greater than 1100 mm. The landscape is dominated by intense, irrigated agricultural cultivation, especially rice, corn, peanuts and vegetables. Use of human waste, termed nightsoil, for crop fertilization is pervasive in this region, leading to the release of parasitic ova into the environment and sustaining schistosomiasis transmission. Villages were selected from a related study on social–environmental factors associated with re-emergence (defined elsewhere; Liang et al. 2006). The villages were not selected at random, but focus was first placed on villages with data on the presence of schistosomiasis infection in snails, acute human cases or infected children (less than 12 years old) since control status was attained in each county. These villages were paired with villages in the same township (an administrative unit of organization, approx. 3 km2) for which historical data existed, but infections had not been found. The extensive human and intermediate host data, and protocols for their collection, used as a basis for these classifications, are described in detail elsewhere (Liang et al. 2006).
Figure 2.
Map of study region with inset table showing 32 participating villages across three counties in Sichuan Province, People's Republic of China. Filled circle, study villages; solid line, river.
2.2. Functional environmental models
Functional environmental models quantify the degree to which the intervening landscape facilitates or impedes the movement of a focal organism between geographically defined nodes (Hansson 1991). The approach has been widely applied in conservation biology and landscape ecology as a means to identify paths that support gene flow between populations of organisms that require high-quality habitat linkages (Macdonald & Johnson 2001). Indeed, a common outcome of these studies is that distances defined by functional environmental models better explain observed dispersal, population dynamics or genetic differentiation than Euclidean distance, reinforcing the need for improved geographical measures of ecological connectivity to predict rates and likelihoods of spread (Michels et al. 2001; Adriaensen et al. 2003; Stevens et al. 2006; Epps et al. 2007).
The functional connectivity approach models the dispersal of a focal organism across a habitat matrix by calculating a weighted or ‘effective’ geographical distance (EGD) between each pair of nodes. Ecologically relevant resistance values are defined on each cell in the matrix, which is then parsed using an algorithm that sums the effective distance experienced while moving from source to destination node. Most functional connectivity studies summarize the EGD between a pair of nodes by taking the single minimum resistance path, assuming the cost of this path to be the most informative measure of node connectivity. Clearly, the distribution of EGDs (along all possible paths) between a pair of nodes provides a more complete representation of the diversity of connectivity modes (Boone & Hunter 1996; Schippers et al. 1996), including those that pass through contiguous corridors, fragmented habitat patches and indirect paths (Theobald 2006). EGD distributions are estimated for S. japonicum and O. hupensis using a series of environmental resistance models parametrized with ecological, experimental and behavioural data.
2.3. Model development
Functional models rely on experimental data, literature sources and expert opinion on the habitat requirements, relative mobility and dispersal characteristics of an organism of interest. Each functional model can be viewed as a hypothesis that asserts that a specific set of landscape features governs dispersal. Ultimately, these models must be confronted by objective data (observed dispersal from mark–recapture experiments, multilocus genotype data, etc.), which we discuss further below. Here, we focus on relatively simple functional models in order to emphasize the methods we have developed for analysing functional model output in the presence of uncertainty in such data. Oncomelania hupensis snails have an affinity for perennially wet environments, and S. japonicum larvae are fully aquatic (Fan et al. 1998; Xu et al. 2000). Thus, we define at each cell a property that influences snail or larval dispersal, based on literature data indicating the varying resistance of hydrological classes that impede, have no effect on or facilitate dispersal, as outlined below. We limit the scope of this paper to three very simple functional models in order to focus on the methodological advances presented below; models can of course be extended to include additional land cover and feature data, as well as anisotropy.
2.3.1. Euclidean (null model)
The Euclidean model gives equal resistance to all cells, thus generating straight line distances between points.
2.3.2. Overland distance
The overland distance resistance model is a Euclidean distance model corrected for the distance travelled when moving over sloped topography.
2.3.3. Watershed
The watershed model considers the distance along flow paths to streams, where the cost of movement through a given cell increases with distance from the closest stream cell. This model represents the hypothesis that the position in the watershed in relation to the nearest stream cell will determine the likelihood that a dispersing snail or larval stage will encounter a stream corridor. Watershed boundaries, being the furthest from streams, were considered barriers to dispersal and are thus assigned the highest resistance values. Snails or larvae originating from villages in higher reaches of a watershed were assumed to be less likely to encounter a stream during active dispersal or passive transport, such as during a rain event, in line with observed associations between O. hupensis and S. japonicum and waterways (Xu et al. 2000; Li & Fu 2006). Following other work (Chardon et al. 2003; Driezen et al. 2007), the resistance value of a cell, Ci,j, was defined by an exponential function of distance from the nearest stream cell, 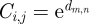 , where d is overland distance from (m, n), the location of the nearest stream pixel, to (i, j).
, where d is overland distance from (m, n), the location of the nearest stream pixel, to (i, j).
2.4. Cost distributions
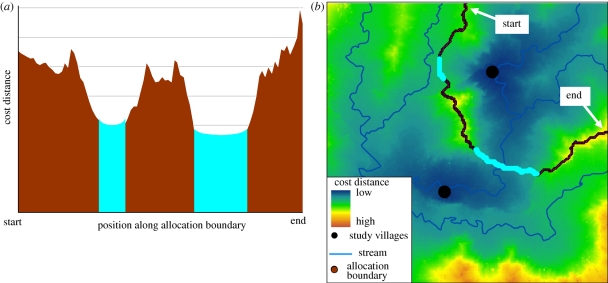
For each environmental model, the distribution of EGD values between each pair of the 32 villages (n = 496 possible pairs) was generated using an iterative walk procedure, with path EGD values recorded, for convenience, at points along the weighted-distance mid-point isoclines (termed allocation boundary) between nodes as described elsewhere (Theobald 2006). The isocline provides a simple accounting scheme for recording path costs between a pair of nodes, and while other approaches are possible, the methods discussed below are not sensitive to how the distribution of costs is recorded or stored. The allocation boundary approach is useful in that it allows an analysis of the cross-sectional distribution of cost distance values, revealing the presence of contiguous low-cost corridors and other features (figure 3). The full distributions of EGD values extracted from the allocation boundary are included in the analysis below, with the extent of the allocation boundary, which stretches infinitely in both directions, set proportional to the straight line distance between each node pair. Functional environmental models were coded in ArcGIS ModelBuilder (ESRI 2008) and Python (van Rossum 2008), and run iteratively.
Figure 3.
Example distribution of cost distances displayed (a) ordered along the allocation boundary and (b) spatially over a cost distance surface. For illustration, blue highlighted values show lowest 20 cost distances in each representation.
2.5. Statistical analysis
The cost distribution between a pair of nodes represents the aggregate available paths for travel between that pair. Where traditional methods ignore all but the lowest cost path, methods presented here account for the full distribution of paths using Monte Carlo techniques. The approach is as follows: consider a set of n nodes S = (p1, p2, …, pn) in Euclidean space ℝN where all inter-node distances are known and define a distance matrix, D = [dij] in 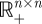 . Matrix D is translation and rotation invariant and satisfies the Euclidean metric properties of non-negativity (dij > 0), identity (dij = 0⇔i = j), symmetry (dij = dji) and subadditivity (dij ≤ dik + dkj). Applying a functional cost model deforms Euclidean space, resulting in D′, a modified distance matrix. Each informative element dij in D′ can be described by the probability density πij defined from the set of cost-weighted distances along the allocation boundary between pi and pj.
. Matrix D is translation and rotation invariant and satisfies the Euclidean metric properties of non-negativity (dij > 0), identity (dij = 0⇔i = j), symmetry (dij = dji) and subadditivity (dij ≤ dik + dkj). Applying a functional cost model deforms Euclidean space, resulting in D′, a modified distance matrix. Each informative element dij in D′ can be described by the probability density πij defined from the set of cost-weighted distances along the allocation boundary between pi and pj.
Our interest in D′ is in the information it contains about relative positions (and configurations) of nodes under alternative models of environmental distance. In the public health context, we are interested in connectivity measures that can be derived from D′ and in the decision-making tools these measures can provide, such as the identification of neighbouring nodes and alternative visualizations of connected epidemic landscapes.
2.5.1. Identifying key neighbouring nodes: surveillance response to an acute case
Designing a surveillance response to an acute case detected in network node pa depends on a range of factors, including the population at risk in other nodes, node characteristics such as availability of clinical care, availability of trained surveillance personnel and other resource constraints. Here, we focus on the information a functional environmental model can provide in prioritizing surveillance of nodes proximal to pa. The elements of D′ can provide a ranked listing of nodes near pa as measured by EGD. A Monte Carlo approach is used to generate different realizations of D′, where values of elements dij are drawn repeatedly from the probability density πij. For each Monte Carlo simulation, nodes pn (where n ≠ a) are ranked as to their distance (daj) from pa. Rankings are then analysed using a rank product approach (Breitling et al. 2004), a non-parametric statistic that detects items that are consistently highly ranked in replicated lists. Our interest is in nodes that rank consistently closer under EGD measures when compared with Euclidean distance; these would be nodes worthy of surveillance consideration that simple inspection of Euclidean proximity would not reveal. This approach is compatible with models accounting for anisotropic effects, although for simplicity we use an isotropic environmental model, the watershed model. Because the large number of comparisons among ranked lists could inflate the rate of falsely significant rank changes, the rank product method accounts for multiple testing by allowing for the flexible control of the false discovery rate (FDR). Cost distances were subject to quantile normalization (Bolstad et al. 2003), and variance was stabilized using a generalized logarithm (glog) procedure (Durbin et al. 2002). Rank product comparisons for both increasing and decreasing proximity were made between Euclidean and EGD distance models at all nodes. We report the FDR level for a given node calculated using a permutation-based procedure (Breitling et al. 2004), which represents the expected proportion of true null hypotheses that are erroneously rejected, out of the total number of hypotheses rejected (i.e. the proportion of type I errors among all significant results).
2.5.2. Embedding and cluster analysis
Presuming that transmission across the network is governed both by properties of individual nodes (patch properties) and by environmental features that provide linkages among a group(s) of nodes (corridor properties), it is logical to identify network groupings that arise by considering environmental connectivity, groupings that may form epidemiological units useful for surveillance or control activities. To approach this question, we turn to traditional second-order analyses such as clustering, first embedding nodes in non-Euclidean, EGD space and then carrying out a consensus cluster analysis.
To relocalize nodes under alternative distance measures, each realization of D′ was used to embed nodes in ℝe EGD space using a semi-definite programming relaxation approach. Methods for choosing the number of dimensions e are discussed elsewhere (Loland & Host 2003), and as in other analyses, scree plots in this study (data not shown) showed marginal improvement in embedding for e > 2, and thus we localize on a ℝ2 plane. Anchor-free node embedding is an optimization problem, where inter-node distances are treated as constraints and the coordinate space is systematically searched to find node coordinates that satisfy these constraints. An intuitive description of the problem is as follows: for every node pair a random draw is performed from πij until a full set D′ is assembled. Then, a nonlinear global minimization problem is solved with the objective function
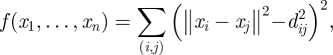 |
2.1 |
where xn is the position of node pn, and a set of coordinates x1,…, xn is a solution if and only if it is the global minimizer of f, with the global minimum being zero. Numerous multidimensional scaling algorithms have been developed. Here, we achieve efficient optimization over large samples of D′ using a semi-definite programming relaxation solution recently proposed for sensor network localization (Biswas et al. 2006; Kim et al. 2008), undertaken in Matlab (Mathworks Inc. 2008) using the SeDuMi toolbox for optimization over symmetric cones (Strum 1999). The result for each realization of D′ is an embedding of each pn in EGD coordinate space, denoted S′ = (p′1, p′2, … , p′n), with localization error estimated by a stress function given by (Golub & Van Loan 1996)
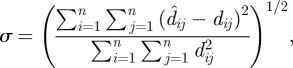 |
2.2 |
which captures the distance error between D′ and the distances resulting from the embedding technique (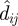 ).
).
The resulting representation of nodes, S′, is useful in that we can compare clustering of sites under Euclidean and EGD domains. We use a simple k-means clustering approach here, although alternative clustering approaches (Gaussian mixture models, c-means, etc.) could easily be substituted. The domain of each S′ is partitioned into clusters of k nodes, which can be represented as a vector of labels over S′. Let λ(q)∈{1, 2, …, k(q)}n denote the label vector of the clustering of qth realization of S′; i.e. 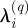 is the membership label of p′i in the partitioning of the qth realization of S′. A set of m such partitions λ(1, 2, …, m) from Monte Carlo relocalizations forms a cluster ensemble, where the goal now is to seek a consensus function that combines the information in the m partitions into a single clustering. The cluster labels are symbolic and thus we must simultaneously solve a correspondence problem, as well as the optimization problem where, given a set of m clusterings, we seek the consensus clustering that minimizes the disagreement with the m clusterings. Here, we follow previous work (Strehl & Ghosh 2002) and define the optimal combined clustering as the one that shares the most mutual information with the m original clusterings, as estimated by the normalized mutual information (NMI). We note that the consensus function is capable of combining multiple partitionings without accessing the original partitioning features. This approach is useful for combining results from diverse partitionings generated using multiple algorithms and applied to differing subsets of the network. Consensus clustering was performed in Matlab (Mathworks Inc. 2008) using three previously developed algorithms (Strehl & Ghosh 2002): Cluster-Based Similarity Partitioning, HyperGraph Partitioning and Meta-Clustering, with the final consensus clustering determined by the maximum average NMI.
is the membership label of p′i in the partitioning of the qth realization of S′. A set of m such partitions λ(1, 2, …, m) from Monte Carlo relocalizations forms a cluster ensemble, where the goal now is to seek a consensus function that combines the information in the m partitions into a single clustering. The cluster labels are symbolic and thus we must simultaneously solve a correspondence problem, as well as the optimization problem where, given a set of m clusterings, we seek the consensus clustering that minimizes the disagreement with the m clusterings. Here, we follow previous work (Strehl & Ghosh 2002) and define the optimal combined clustering as the one that shares the most mutual information with the m original clusterings, as estimated by the normalized mutual information (NMI). We note that the consensus function is capable of combining multiple partitionings without accessing the original partitioning features. This approach is useful for combining results from diverse partitionings generated using multiple algorithms and applied to differing subsets of the network. Consensus clustering was performed in Matlab (Mathworks Inc. 2008) using three previously developed algorithms (Strehl & Ghosh 2002): Cluster-Based Similarity Partitioning, HyperGraph Partitioning and Meta-Clustering, with the final consensus clustering determined by the maximum average NMI.
3. Results
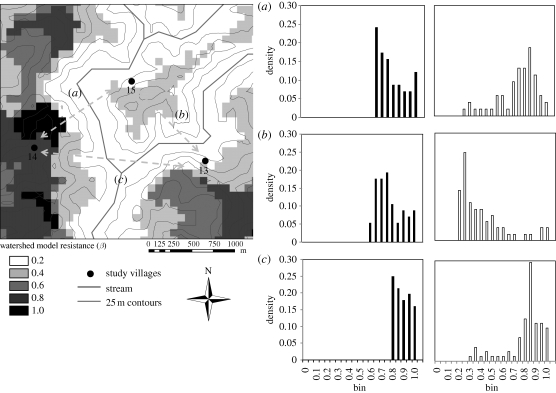
Cost distributions for inter-node paths between the 32 villages using environmental models differed depending on the underlying environmental inputs. In particular, when models incorporate directed paths, such as streams, narrow, low-cost corridors are pronounced for nodes that lie along those paths. Figure 4 shows a subset of village nodes with insets showing distributions of path costs dividing three node pairs (13,14), (13,15) and (14,15), estimated using the overland distance and watershed models. Histograms corresponding to the simple overland distance model (filled bars) reflect the distribution of unweighted paths between villages, showing a greater frequency of low-cost paths between closer village pairs (13,15) and (14,15) than those farther apart (13,14). By contrast, the watershed model (unfilled bars) shows that village 14 is isolated from the two other villages by a topographic boundary, resulting in a right-skewed distribution of path costs for (13,14) and (14,15), whereas villages 13 and 15 are linked by stream corridors, resulting in a left-skewed distribution between (13,15).
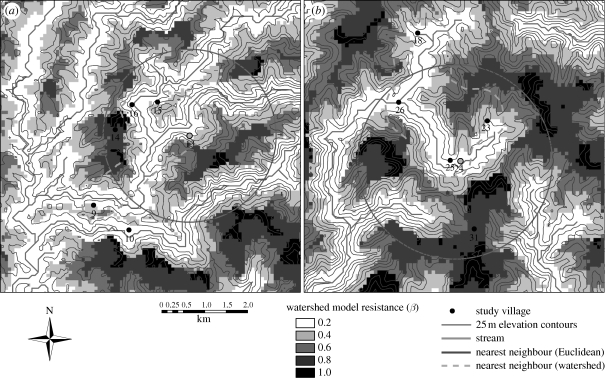
Figure 4.
Map (left) showing locations of three study villages over a watershed resistance surface. Darker cells indicate positions higher in the watershed (further from streams along flow paths) and are assigned higher resistance values. Cost distance histograms (right) correspond to these three village pairs for two models: overland distance (filled bars) and watershed (unfilled bars). Here, resistance (β) represents Ci,j values normalized by the maximum Ci,j. Note in the watershed model, the greater frequency of high-cost paths linking villages 14 and 15 (a) and 13 and 14 (c) compared with the greater number of low-cost paths between villages 13 and 15 (b).
3.1. Identifying key neighbouring nodes
The watershed functional model aids in prioritizing surveillance of nodes along flow paths to a node with an acute case, pa. Table 1 shows the results of ranking the closest 10 nodes for two representative nodes, pa = 13 and pa = 24, generated using 1000 realizations of D′. Rankings of proximal nodes are significantly altered under the watershed model when compared with the Euclidean null. In table 1, the FDR level is reported as a measure of the significance of the change in proximity of the village. Here, the FDR can be interpreted as a conservative estimate of the percentage false positives if this village and all other villages, with rank product values smaller than this village, were considered as significantly more or less proximate. Essentially, the FDR level for a village is the proportion of significant shifts in rank that are truly null if the shift in rank of the current village is taken as significant.
Table 1.
Selected villages where ranking of proximal nodes is significantly altered in the watershed model when compared with the Euclidean null model, as estimated using 1000 realizations of D′. The median rank of nearest neighbour nodes under the watershed model is compared with the Euclidean model, with changes in rank noted, and the false discovery rate (FDR) reported as described in the text.
| village 13 |
village 24 |
|||||||
|---|---|---|---|---|---|---|---|---|
| nodes |
nodes |
|||||||
| ranking | Euclidean | watershed | change in ranking | FDR | Euclidean | watershed | change in ranking | FDR |
| 1 | 15 | 16 | ↑2 | 0.01 | 25 | 25 | — | 0.98 |
| 2 | 14 | 15 | ↓1 | 0.06 | 23 | 23 | — | 0.99 |
| 3 | 16 | 9 | ↑2 | 0 | 31 | 26 | ↑1 | 0.03 |
| 4 | 10 | 14 | ↓2 | 0.04 | 26 | 18 | ↑1 | 0.03 |
| 5 | 9 | 10 | ↓1 | 0 | 18 | 31 | ↓2 | 0.26 |
| 6 | 17 | 17 | — | 0.63 | 13 | 28 | ↑4 | 0.04 |
| 7 | 26 | 19 | ↑7 | 0.10 | 10 | 2 | ↑7 | 0 |
| 8 | 18 | 20 | ↑6 | 0.04 | 9 | 27 | ↑7 | 0.15 |
| 9 | 24 | 26 | ↓2 | 0.58 | 15 | 10 | ↓2 | 0.05 |
| 10 | 25 | 25 | — | 0.66 | 28 | 9 | ↓2 | 0.26 |
When pa = 13, key shifts in ranks occur among neighbouring village nodes. Figure 5a shows how the top five ranked nodes proximal to pa = 13 change under the watershed model, whereas figure 5b shows a similar phenomenon for pa = 24.
Figure 5.
Maps showing clustering of proximal villages around village 13 (a) and village 24 (b) in the watershed model (dashed grey line) when compared with the Euclidean null model (solid circle). Resistance (β) is defined as in figure 4.
3.2. Embedding and cluster analysis
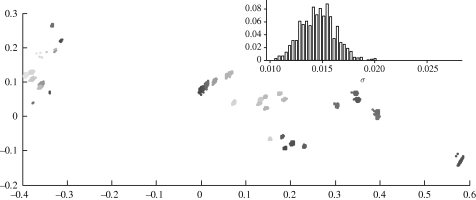
Embedding of multiple realizations of S′ in EGD space produces a cloud of potential coordinates for each p′n. Figure 6 shows these clouds for the embedding of 1000 realizations of the overland functional model, which exhibits strong resemblance to the Euclidean model because elevation gradients in this region are modest. Embedding error was low across models, as seen in the inset in figure 6, which shows the distribution of σ for the ℝ2 embedding of 1000 watershed model realizations.
Figure 6.
Multiple realizations of S′ localizations generated from 1000 Monte Carlo samples of overland model inter-node cost distributions, forming for each node a cloud of potential localizations in a two-dimensional, normalized EGD domain (axes units are normalized overland distance). For visualization, plotted realizations were aligned by Procrustes transformations to minimize sum of squared error. Inset shows the distribution of error, σ, for the ℝ2 embedding of 1000 realizations of all nodes using the watershed model.
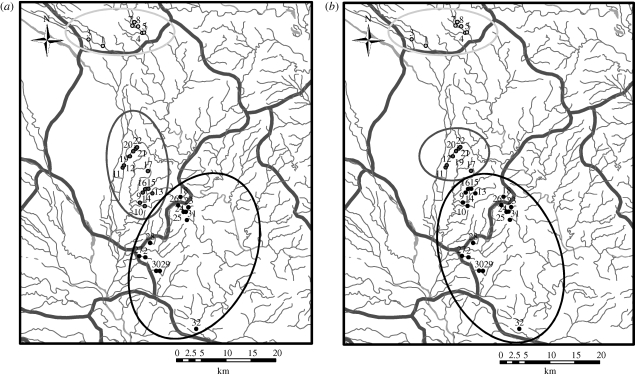
As expected, non-Euclidean models strongly altered clustering patterns of nodes as explored using a k-means analysis. In the watershed model for instance, the consensus cluster analysis revealed nodes which, while separated by short Euclidean distances, bridge drainage divides and are therefore separated by long effective distances. Figure 7 shows how some of these nodes (such as 16 and 17) are clustered together in Euclidean space but are clustered with other nodes when embedded in EGD space. The nodes in this study were (non-randomly) sampled from three counties, and k-means analysis shows that within-county nodes are indeed members of the same cluster when k = 3. The watershed model embedding shows, however, that village nodes in one county may naturally group with nodes in other counties when watershed features are accounted for. In figure 7, consensus cluster membership of several nodes in Jinyang county shift to the cluster formed by a majority of Zhongjiang county nodes. Surveillance and control activities are organized at the county level in this region, and sometimes in a manner uncoordinated with neighbouring counties. A simple k-means analysis can reveal the limited relevance of county boundaries for effectively grouping villages.
Figure 7.
Results of k-means clustering (k = 3) performed on Euclidean space (a) and the watershed model embedded into EGD space (b). Note that for clarity both maps are drawn using the Euclidean coordinate system. Node colours represent cluster membership (also indicated by ellipses), which shifts when clusters are formed under EGD space. Light grey circle, 1; dark grey circle, 2; black circle, 3; light grey line, stream; dark grey line, county boundary.
4. Discussion
Meaningful measures of environmental distance are key to conceptualizing and quantifying transport processes that underlie the spread of environmentally mediated infectious diseases and are critically important to the planning of disease surveillance and control. The methods presented here allow us to explore alternative models of connectivity and determine the value of adding additional model complexity. Inter-node cost distributions are robust representations of the degree of connectivity between nodes. Previous work has frequently relied on unweighted, binary edges, isotropic kernel functions, or has used a single path cost as the measure of inter-node distance, which others have identified as a problematic representation (Adriaensen et al. 2003; Stevens et al. 2006). The cost distribution approach presented here is one alternative to least cost path approaches; other alternatives have been proposed (McRae et al. 2008), yet are limited in application to isotropic systems. We note that the analysis presented here does not make use of the order of path costs along the allocation boundaries because for the system under study here, we have no ecological basis to consider contiguous paths more favourable than discontiguous paths. But in the context of other species where these effects are known, path costs can be weighted based on the costs of adjacent paths (using a simple moving average function across the allocation boundary, for instance), rewarding paths that are members of a contiguous, high-value corridor.
Two novel methods were presented here to take advantage of information on the distribution of paths of varying quality across a heterogeneous environment. First, the cost distribution approach can be used to quantitatively identify nodes proximal to a node of interest, such as the location of an acute case. Combined with other sources of data, these results could be used to prioritize resources for disease control and surveillance following an index case at the beginning of an epidemic, or resurgence of a previously controlled disease. We demonstrated that rank product statistics are a straightforward approach to analysing the Monte Carlo output described in this study, where our interest is in detecting items that are consistently highly ranked in replicated lists. We found that rankings of proximal nodes are significantly altered under the assumptions of functional environmental models when compared with the Euclidean null, and the approach is compatible with more complex environmental models, including anisotropic models. While this approach is valuable for analysing relationships between a given node and nearby nodes, an alternative approach is necessary to explore the global implications of a functional environmental model across all points.
To achieve global insights across all nodes, a second method was presented that positioned a set of nodes so that the Euclidean distance between these nodes in EGD space matches the set of non-Euclidean EGD distances specified by the functional environmental model. The analysis then proceeded using the distances resulting from the embedding, avoiding issues of invalid variogram and covariance functions that arise with non-Euclidean distance measures (Loland & Host 2003). Embedding results were stable when nodes were added or deleted. As a simple demonstration of the approach, nodes were clustered under Euclidean and non-Euclidean embeddings, showing that functional environmental models can suggest alternative groupings of nodes that may have relevance for surveillance and control. For instance, stable groupings of nodes may be used to establish sentinel sites for ongoing surveillance, optimizing the allocation of public health resources. Here, we used a simple k-means approach, but more sophisticated techniques are amenable. We deliberately proposed a cluster consensus procedure that ignores the clustering algorithm that generated the partition; this can be advantageous if multiple clustering algorithms were employed or subsets of nodes are clustered.
As a simple measure of the utility of non-Euclidean metrics for defining epidemiological distance in this system, we examined the official Chinese Centers for Disease Control classification of each study village (and its neighbours) as re-emergent (R) or not re-emergent (NR) based on the criteria presented above. We inquired for each functional model as to whether the nearest neighbours of R villages are generally other R villages as opposed to NR villages, and whether NR villages were generally closer neighbours with other NR villages. For each node pm, nodes pn (where n ≠ m) were ranked as to their distance from pm, using Euclidean distance and then watershed distance. Under the Euclidean distance model, like-classified neighbours were ranked no higher (closer) than non-like-classified neighbours using a non-parametric test comparing the distances of like and unlike neighbours (Mann–Whitney–Wilcoxon test: Z = −0.62, p = 0.53). The watershed model, however, ranked like-classified neighbours higher (closer) than unlike neighbours (Mann–Whitney–Wilcoxon test: Z = −3.70, p < 0.001), indicating that watershed distance may be an improved measure of epidemiological distance when compared with Euclidean distance in this system. Much additional work is needed to rigorously confirm this finding; an ongoing study being conducted by the authors is collecting extensive epidemiological and molecular genetic data in these villages over a 5-year interval to confirm the levels of infection in both R and NR villages and to directly estimate parasite dispersal coefficients. Combining these data with mathematical modelling approaches will be instrumental in gaining insight from, and extending, the approach described here. Indeed, the methods presented are valuable in that they can reveal in which systems non-Euclidean distance measures may be of value. Even confirmation of the null, that Euclidean distances are sufficient, is useful in that it provides a test of a commonly untested assumption: that Euclidean distance is the most suitable metric for geographical analyses in a particular system.
Two considerable challenges face the application of these methods to infectious disease transmission. First, assigning relative mobility coefficients to environmental features and surfaces requires detailed data on the habitat preferences, occurrence and movement rates of the organism of interest through diverse land cover classes. Where multiple species or life stages are involved, the data requirements are large and future work should address the need to synthesize connectivity models parametrized for multiple organisms of interest. Strong habitat preferences do not guarantee use of that habitat for dispersal (Schippers et al. 1996; Vignieri 2005), and an organism's decisions along the way may be more important in determining the overall path than the cumulative resistance over the entire trajectory (Brooker et al. 1999). Here, we explicitly consider the uncertainty associated with typical, least cost path techniques by developing simulation approaches to examine the implications of alternative connectivity models in two infectious disease applications.
Even having parametrized several competing functional models, a second major challenge is determining which of these best represents the processes that govern host/vector/pathogen spread; that is, which model is ‘best’. This requires additional, objective data for model selection. Contemporary measures of migration from multilocus genotypes (Wilson & Rannala 2003) will be estimated in our ongoing study in this region, but traditional epidemiologic surveillance datasets may be equally valuable for this purpose, although reporting, classification and selection issues certainly arise with these data. The utility of the competing model approach is that we are able to determine the value of adding additional model complexity, that is, which models provide explanatory power that significantly exceeds simple Euclidean distance. This information can then be used to prioritize the type and quantity of data collection necessary to validate an operational model of connectivity for a given transmission system.
A final point of utility for the methods presented herein is that they allow us to explore the potential health effects of environmental change, which can alter natural groupings of populations. The clustering methods presented here can be applied to the questions: what are natural groupings in the network under current conditions, and how does environmental change alter these groupings? Proximity to hazards (infectious or otherwise) is highly relevant to the epidemiology of environmental change. Distance, linear and social, is a component of key variables of interest, such as access to care, proximity to environmental hazards (e.g. disease vectors, contaminated water sources, exposure to airborne disease, etc.) and mobility of environmental hazards themselves. In essence, environmental change can pull (push) some hazards ‘closer’ (‘further’) than they have been historically, while also changing the magnitude of certain hazardous exposures. There is a great need, then, to design interventions to increase the effective distance between hazards and susceptible populations. The methods presented above provide a framework for carrying out such an analysis.
Acknowledgements
The authors wish to thank Kang Junxin, Director of the Sichuan Center for Disease Control and Prevention (Chengdu, People's Republic of China), and our colleagues at the Anxian, Zhongjiang and Jinyang County Anti-Schistosomiasis Stations for their continued support and collaboration. This work was supported in part by the NIH/NSF Ecology of Infectious Disease Program (grant no. 0622743), the National Institute for Allergy and Infectious Disease (grant no. R01AI068854) and the Emory Global Health Institute Faculty Distinction Fund.
References
- Adler F. 1993. Migration alone can produce persistence of host-parasitoid models. Am. Nat. 141, 642 ( 10.1086/285496) [DOI] [Google Scholar]
- Adriaensen F., Chardon J. P., De Blust G., Swinnen D., Villalba D., Gulinck H., Matthysen E. 2003. The application of ‘least-cost’ modelling as a functional landscape model. Landscape Urban Plann. 64, 233–247. ( 10.1016/S0169-2046(02)00242-6) [DOI] [Google Scholar]
- Biswas P., Lian T.-C., Wang T.-C., Ye Y. 2006. Semidefinite programming based algorithms for sensor network localization. ACM Trans. Sen. Netw. 2, 188–220. ( 10.1145/1149283.1149286) [DOI] [Google Scholar]
- Bjornstad O. N. 2001. Cycles and synchrony: two historical ‘experiments’ and one experience. J. Anim. Ecol. 69, 869–873. ( 10.1046/j.1365-2656.2000.00444.x) [DOI] [PubMed] [Google Scholar]
- Bolstad B. M., Irizarry R. A., Åstrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. ( 10.1093/bioinformatics/19.2.185) [DOI] [PubMed] [Google Scholar]
- Boone R., Hunter M. 1996. Using diffusion models to simulate the effects of land use on grizzly bear dispersal in the Rocky Mountains. Landscape Ecol. 11, 51–64. ( 10.1007/BF02087113) [DOI] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573, 83–92. ( 10.1016/j.febslet.2004.07.055) [DOI] [PubMed] [Google Scholar]
- Brooker L., Brooker M., Cale P. 1999. Animal dispersal in fragmented habitat: measuring habitat connectivity, corridor use and dispersal mortality. Conserv. Ecol. (online) 3, 4. [Google Scholar]
- Brooks C. P., Antonovics J., Keitt T. H. 2008. Spatial and temporal heterogeneity explain disease dynamics in a spatially explicit network model. Am. Nat. 172, 149–159. ( 10.1086/589451) [DOI] [PubMed] [Google Scholar]
- Chardon J. P., Adriaensen F., Matthysen E. 2003. Incorporating landscape elements into a connectivity measure: a case study for the Speckled wood butterfly (Pararge aegeria L.). Landscape Ecol. 18, 561–573. ( 10.1023/A:1026062530600) [DOI] [Google Scholar]
- Driezen K., Adriaensen F., Rondinini C., Doncaster C. P., Matthysen E. 2007. Evaluating least-cost model predictions with empirical dispersal data: a case-study using radiotracking data of hedgehogs (Erinaceus europaeus). Ecol. Model. 209, 314–322. ( 10.1016/j.ecolmodel.2007.07.002) [DOI] [Google Scholar]
- Durbin B. P., Hardin J. S., Hawkins D. M., Rocke D. M. 2002. A variance-stabilizing transformation for gene-expression microarray data. Bioinformatics 18(Suppl. 1), S105–S110. [DOI] [PubMed] [Google Scholar]
- Epps C. W., Wehausen J. D., Bleich V. C., Torres S. G., Brashares J. S. 2007. Optimizing dispersal and corridor models using landscape genetics. J. Appl. Ecol. 44, 714–724. ( 10.1111/j.1365-2664.2007.01325.x) [DOI] [Google Scholar]
- ESRI. 2008. ArcGIS Model Builder. Redlands, CA. [Google Scholar]
- Fan J., Minchella D. J., Day S. R., McManus D. P., Tiu W. U., Brindley P. J. 1998. Generation, identification, and evaluation of expressed sequence tags from different developmental stages of the Asian blood fluke Schistosoma japonicum. Biochem. Biophys. Res. Commun. 252, 348–356. ( 10.1006/bbrc.1998.9491) [DOI] [PubMed] [Google Scholar]
- Ferguson N. M., Donnelly C. A., Anderson R. M. 2001. The foot-and-mouth epidemic in Great Britain: pattern of spread and impact of interventions. Science 292, 1155–1160. ( 10.1126/science.1061020) [DOI] [PubMed] [Google Scholar]
- Golub G. H., Van Loan C. F. 1996. Matrix computations. Baltimore, MD: Johns Hopkins Press. [Google Scholar]
- Grenfell B. T., Bjornstad O. N., Kappey J. 2001. Travelling waves and spatial hierarchies in measles epidemics. Nature 414, 716–723. ( 10.1038/414716a) [DOI] [PubMed] [Google Scholar]
- Gurarie D., Seto E. Y. 2008. Connectivity sustains disease transmission in environments with low potential for endemicity: modelling schistosomiasis with hydrologic and social connectivities. J. R. Soc. Interface 6, 495–508. ( 10.1098/rsif.2008.0265) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski I. 2001. Spatially realistic theory of metapopulation ecology. Naturwissenschaften 88, 372–381. ( 10.1007/s001140100246) [DOI] [PubMed] [Google Scholar]
- Hansson L. 1991. Dispersal and connectivity in metapopulations. Biol. J. Linn. Soc. 42, 89–103. ( 10.1111/j.1095-8312.1991.tb00553.x) [DOI] [Google Scholar]
- Hess G. 1996. Disease in metapopulation models: implications for conservation. Ecology 77, 1617–1632. ( 10.2307/2265556) [DOI] [Google Scholar]
- Jeger M. J., Pautasso M., Holdenrieder O., Shaw M. W. 2007. Modelling disease spread and control in networks: implications for plant sciences. New Phytol. 174, 279–297. ( 10.1111/j.1469-8137.2007.02028.x) [DOI] [PubMed] [Google Scholar]
- Keeling M. J. 1999. The effects of local spatial structure on epidemiological invasions. Proc. Biol. Sci. 266, 859–867. ( 10.1098/rspb.1999.0716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling M. J., Eames K. T. 2005. Networks and epidemic models. J. R. Soc. Interface 2, 295–307. ( 10.1098/rsif.2005.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kojima M., Waki H. 2008. Exploiting sparsity in SDP relaxation for sensor network localization. Tokyo, Japan: Department of Mathematical and Computing Sciences, Tokyo Institute of Technology. [Google Scholar]
- Koopman J. S., Chick S. E., Simon C. P., Riolo C. S., Jacquez G. 2002. Stochastic effects on endemic infection levels of disseminating versus local contacts. Math. Biosci. 180, 49–71. ( 10.1016/S0025-5564(02)00124-4) [DOI] [PubMed] [Google Scholar]
- Li D., Fu X. 2006. Design of hydraulic structures to prevent the spread of intermediate snails hosts of schistosomiasis. Irrigation Drainage Syst. 20, 69–82. ( 10.1007/s10795-006-2252-1) [DOI] [Google Scholar]
- Liang S., Yang C., Zhong B., Qiu D. 2006. Re-emerging schistosomiasis in hilly and mountainous areas of Sichuan, China. Bull. World Health Organ. 84, 139–144. ( 10.2471/BLT.05.025031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loland A., Host G. 2003. Spatial covariance modelling in a complex coastal domain by multidimensional scaling. Environmetrics 14, 307–321. ( 10.1002/env.588) [DOI] [Google Scholar]
- Macdonald D., Johnson D. 2001. Dispersal in theory and practice: consequences for conservation biology. In Dispersal (eds Colbert J., Danchin E., Dhondt A., Nichols J.). New York, NY: Oxford University Press. [Google Scholar]
- Mathworks Inc. 2008. Matlab, v. 2007b. Natick, MA. [Google Scholar]
- McRae B., Dickson B., Keitt T. H., Shah V. B. 2008. Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89, 2712–2724. ( 10.1890/07-1861.1) [DOI] [PubMed] [Google Scholar]
- Michels E., Cottenie K., Neys L., De Gelas K., Coppin P., De Meester L. 2001. Geographical and genetic distances among zooplankton populations in a set of interconnected ponds: a plea for using GIS modelling of the effective geographical distance. Mol. Ecol. 10, 1929–1938. ( 10.1046/j.1365-294X.2001.01340.x) [DOI] [PubMed] [Google Scholar]
- Miller H., Wentz E. 2003. Representation and spatial analysis in geographic information systems. Ann. Assoc. Am. Geogr. 93, 574–594. ( 10.1111/1467-8306.9303004) [DOI] [Google Scholar]
- Parham P. E., Singh B. K., Ferguson N. M. 2008. Analytic approximation of spatial epidemic models of foot and mouth disease. Theor. Popul. Biol. 73, 349–368. ( 10.1016/j.tpb.2007.12.010) [DOI] [PubMed] [Google Scholar]
- Reluga T. C., Medlock J., Galvani A. P. 2006. A model of spatial epidemic spread when individuals move within overlapping home ranges. Bull. Math. Biol. 68, 401–416. ( 10.1007/s11538-005-9027-y) [DOI] [PubMed] [Google Scholar]
- Remais J., Hubbard A., Zisong W., Spear R. C. 2007. Weather-driven dynamics of an intermediate host: mechanistic and statistical population modelling of Oncomelania hupensis. J. Appl. Ecol. 44, 781–791. ( 10.1111/j.1365-2664.2007.01305.x) [DOI] [Google Scholar]
- Ruxton G. D., Rohani P. 1999. Fitness-dependent dispersal in metapopulations and its consequences for persistence and synchrony. J. Anim. Ecol. 68, 530–539. ( 10.1046/j.1365-2656.1999.00300.x) [DOI] [Google Scholar]
- Schippers P., Verboom J., Knaapen J. P., van Apeldoorn R. C. 1996. Dispersal and habitat connectivity in complex heterogeneous landscapes: an analysis with a GIS-based random walk model. Ecography 19, 97–106. ( 10.1111/j.1600-0587.1996.tb00160.x) [DOI] [Google Scholar]
- Smith D. L., Lucey B., Waller L. A., Childs J. E., Real L. A. 2002. Predicting the spatial dynamics of rabies epidemics on heterogeneous landscapes. Proc. Natl Acad. Sci. USA 99, 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soubeyrand S., Held L., Höhle M., Sache I. 2008. Modelling the spread in space and time of an airborne plant disease. J. R. Statist. Soc. Ser. C Appl. Stat. 57, 253–272. ( 10.1111/j.1467-9876.2007.00612.x) [DOI] [Google Scholar]
- Spear R. C., et al. 2004. Factors influencing the transmission of Schistosoma japonicum in the mountains of Sichuan Province. Am. J. Trop. Med. Hyg. 70, 48–56. [PubMed] [Google Scholar]
- Stevens V., Verkenne C., Vandewoestijne S., Wesselingh R. A., Baguette M. 2006. Gene flow and functional connectivity in the natterjack toad. Mol. Ecol. 15, 2333–2344. ( 10.1111/j.1365-294X.2006.02936.x) [DOI] [PubMed] [Google Scholar]
- Strehl A., Ghosh J. 2002. Cluster ensembles: a knowledge reuse framework for combining multiple partitions. J. Mach. Learn. Res. 3, 583–617. ( 10.1162/153244303321897735) [DOI] [Google Scholar]
- Strum J. 1999. SeDuMi 1.02, a MATLAB toolbox for optimization over symmetric cones. Optim. Methods Softw. 11, 625–653. [Google Scholar]
- Sutherst R. W. 2004. Global change and human vulnerability to vector-borne diseases. Clin. Microbiol. Rev. 17, 136–173. ( 10.1128/CMR.17.1.136-173.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald D. 2006. Exploring the functional connectivity of landscapes using landscape networks. In Connectivity conservation (eds Crooks K., Sanjayan M.). Cambridge, UK: Cambridge University Press. [Google Scholar]
- Urban D., Keitt T. 2001. Landscape connectivity: a graph-theoretic perspective. Ecology 82, 1205–1218. ( 10.1890/0012-9658(2001)082[1205:LCAGTP]2.0.CO;2) [DOI] [Google Scholar]
- van Rossum G. 2008. Python computer language. See http://www.python.org/. [Google Scholar]
- Vignieri S. N. 2005. Streams over mountains: influence of riparian connectivity on gene flow in the Pacific jumping mouse (Zapus trinotatus). Mol. Ecol. 14, 1925–1937. ( 10.1111/j.1365-294X.2005.02568.x) [DOI] [PubMed] [Google Scholar]
- Wilson G. A., Rannala B. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. J., Wei F. H., Yang X. X., Dai Y. H., Yu G. Y., Chen L. Y., Su Z. M. 2000. Possible effects of the Three Gorges dam on the transmission of Schistosoma japonicum on the Jiang Han plain, China. Ann. Trop. Med. Parasitol. 94, 333–341. [DOI] [PubMed] [Google Scholar]