Abstract
Bacterial biofilms have an enormous impact on medicine, industry and ecology. These microbial communities are generally considered to adhere to surfaces or interfaces. Nevertheless, suspended filamentous biofilms, or streamers, are frequently observed in natural ecosystems where they play crucial roles by enhancing transport of nutrients and retention of suspended particles. Recent studies in streamside flumes and laboratory flow cells have hypothesized a link with a turbulent flow environment. However, the coupling between the hydrodynamics and complex biofilm structures remains poorly understood. Here, we report the formation of biofilm streamers suspended in the middle plane of curved microchannels under conditions of laminar flow. Experiments with different mutant strains allow us to identify a link between the accumulation of extracellular matrix and the development of these structures. Numerical simulations of the flow in curved channels highlight the presence of a secondary vortical motion in the proximity of the corners, which suggests an underlying hydrodynamic mechanism responsible for the formation of the streamers. Our findings should be relevant to the design of all liquid-carrying systems where biofilms are potentially present and provide new insights on the origins of microbial streamers in natural and industrial environments.
Keywords: biofilms, bacterial streamers, Pseudomonas aeruginosa, microfluidics, secondary flow
1. Introduction
Biofilms are generally identified as microbial communities embedded in a self-secreted matrix of polymeric substances (Costerton et al. 1995). Due to their increased resistance to antimicrobial agents (Mah & O'Toole 2001; Drenkard & Ausubel 2002), bacterial biofilms are involved in many health-related problems, ranging from infections and disease transmission (Costerton et al. 1999; Parsek & Singh 2003) to wound healing and medical implants (Edwards & Harding 2004; Zimmerli et al. 2004). Biofilms also have a significant impact on many manufacturing processes, as in the food industry (Brooks & Flint 2008) and basic industrial quality control.
Extensive research has shown that biofilms are dynamic systems that can develop into highly differentiated and complex architectures (O'Toole et al. 2000; Stoodley et al. 2002). A remarkable example is given by filamentous flow-shaped biofilms, called ‘streamers’, which generally stem from a surface-attached biofilm base while a long downstream ‘tail’ freely oscillates in the bulk fluid. Streamers are frequently found in rivers (Hall-Stoodley et al. 2004), acidic metal-rich waters (Edwards et al. 2000; Hallberg et al. 2006), hydrothermal hot springs (Reysenbach & Cady 2001), and have been shown to substantially contribute to ecosystem processes (Battin et al. 2003) and fouling of membranes (Vrouwenvelder et al. 2009). Streamers have also been reported to grow from mixed bacterial communities in streamside flumes (Besemer et al. 2009) and monospecies biofilms in laboratory flow cells (Stoodley et al. 1998) under conditions of turbulent flow.
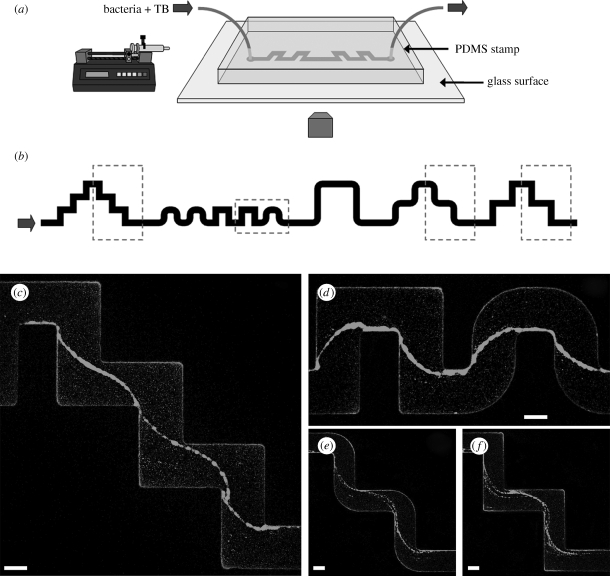
In our work, we exploited microfluidic technologies (Stone et al. 2004) in order to characterize the development of biofilms in both space and time while controlling the chemical and hydrodynamic environments (figure 1a). A further advantage of using microfluidics is the flexibility in the design of devices: we therefore prepared channels with curved sections (figure 1b), including both sharp and rounded corners, to investigate the role of geometric features, commonly present in water-handling systems and natural environments, on the evolution of a biofilm. Besides the growth of a biofilm on the walls of the channel, we discovered the striking, rapid formation of long, suspended bacterial streamers, which, to the best of our knowledge, have not been reported in straight channels under laminar flow conditions.
Figure 1.
(a) Schematic of the microfluidic set-up. (b) Layout of a portion of the channel used in the experiments. The width is 200 μm and the typical height is 100 μm. The round corners have an inner radius of curvature of 50 μm and an outer of 250 μm. The turns are spaced differently all along the channel by 400 μm (c,d), 600 μm (e,f) and 1200 μm. The direction of the flow is from left to right. (c,d) Streamers with wild-type GFP-labelled PA14 (OD600 = 0.25) after 12 h of constant flow at 0.75 μl min−1. (e,f) Streamers with wild-type GFP-labelled PA14 (OD600 = 0.4) after 10 h of constant flow at 0.75 μl min−1. The images are taken in the middle horizontal plane of the channel with a confocal microscope (scale bars, 100 μm).
2. Material and methods
2.1. Microfluidic experiments
The microfluidic channels were prepared from polydimethylsiloxane (PDMS, Sylgard 184 silicone elastomer kit, Dow Corning) following conventional soft-lithography techniques (Duffy et al. 1998). Each channel was sealed to a glass microscope slide (25 × 60 × 0.17 mm, VWR) after 1 min exposure in a plasma chamber. Solutions were infused in the channel by means of a syringe pump (Harvard Apparatus). Images were collected by conventional fluorescence and phase-contrast microscopies (Leica DM IRB) and confocal microscopy (Leica TCS SP5).
2.2. Bacterial strains and growth conditions
Colonies of Pseudomonas aeruginosa strain PA14 were cultured at 37°C overnight on a Luria–Bertani (LB) agar plate. From these cultures, 5 ml of tryptone broth (TB, 10 g l−1 tryptone) medium was inoculated and incubated on a rotary shaker at 220 rpm for approximately 3 h at 37°C. Growth was measured as the optical density at 600 nm (OD600) in a spectrophotometer. We also measured the bacterial concentration in the solution left in the syringe at the end of the experiments and did not observe a major change in the optical density (about 30% over 24 h). In most of the experiments, solutions were continuously infused into the channels for less than 12 h. In addition, we performed two different sets of experiments in which, after 3 h, we replaced the bacterial solution with a newly incubated solution or with only TB medium. All the experiments were performed at room temperature. The following strains were used in this paper: P. aeruginosa PA14 wild-type, pilC::Tn5(Tet), flgK::Tn5(Tet) (O'Toole & Kolter 1998), ΔpelA (Friedman & Kolter 2004a), P. aeruginosa PA01 wild-type.
3. Results
3.1. Biofilm streamers: spatial localization and temporal development
After flowing the bacterial solution at constant flow rate for a few hours, many small colonies uniformly covered the top and bottom surfaces of the channel. More significantly, we noted the presence of thread-like structures suspended in the flow all along the length of the channel (figure 1c–f; see the electronic supplementary material, figure S1a–c and movies S1 and S2). The observed streamers have several distinguishing features: (i) They are located exactly in a plane at half the channel height. (ii) They connect only inner corners (where the angle spanned by the fluid flow is about 270°) and are typically attached to the wall directly after each turn. (iii) There is no significant difference between sharp and round corners. (iv) The streamers are found in all of the curved patterns, but never in the straight sections of the channel. (v) These structures are reproducible and appear in every experiment performed in a range of flow rates of 0.2–1 μl min−1 (which correspond to a range of average velocities of about 0.2–1 mm s−1). Moreover, the streamers can connect several consecutive turns with an overall length of few millimetres.
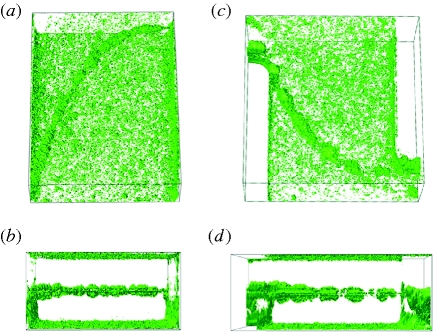
To illustrate the three-dimensional structure of the streamers, we created software reconstructions of confocal z-scan images (figure 2a–d; see the electronic supplementary material, figure S2a,b). The cell distribution on the top and the bottom surfaces consists of a large number of bacterial clusters, which slightly increases only in the proximity of the lateral walls and forms layers that are a few microns in thickness. The streamer, in contrast, is located in the middle of the channel, connected only to the lateral walls at the corners, and is otherwise freely suspended in the flow. The average thickness, or diameter, of the streamer is, after 12 h of flow, typically 10–20 μm. In addition, the measurement of the total light intensity gives an estimate of the percentage of cells constituting a single streamer, which, by the end of the experiment, is more than 25 per cent of the total number of bacteria present in that portion of the channel.
Figure 2.
(a–d) Three-dimensional reconstruction of two streamers from confocal z-scan images. Above perspective (a,c) and horizontal views (b,d) are shown.
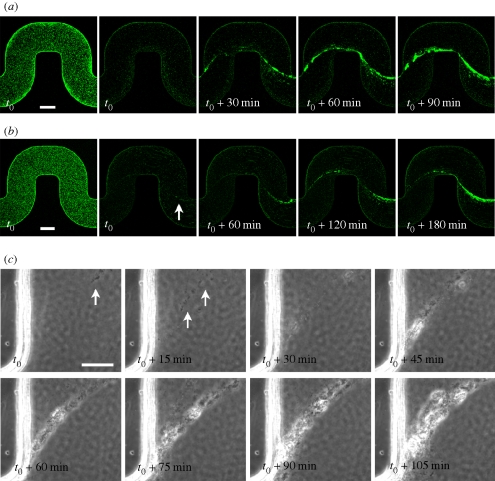
Time-lapse images provide useful information about the development of the streamers. The image sequences shown in figure 3a,b were obtained from confocal z-scans of the channel every 15 min for two different concentrations of bacteria at the same flow rate. In both cases, the streamer appeared at first as a very thin filament, already connecting the corners of the channel, which progressively became thicker with time. We note that the growth of the streamer in the first experiment (OD600 = 0.4) was faster than in the second experiment in which there was a lower concentration of bacteria in solution (OD600 = 0.17). A further idea of the early formation of the streamer is given by the sequence shown in figure 3c (see also the electronic supplementary material, movie S3). These phase-contrast images, taken with higher magnification right after a corner, show that the initial thread is made purely of extracellular matrix, recognizable only through a series of single cells or small clusters of bacteria suspended across the channel in a synchronous oscillating motion. These early filaments are clearly connected to the endpoint of the corner, and progressively grow to form a single bigger streamer as more bacteria stick and more polymeric material accrues. In addition, the timing of the early formation of the streamers is reproducible: in experiments with approximately the same flow rate we observed the first appearance of streamers approximately 6–7 h after the beginning of the experiment.
Figure 3.
(a,b) Time evolution of the streamers: the first image in the sequences is taken at the bottom of the channel, the next four at the middle-height plane. The optical densities are (a) OD600 = 0.4 and (b) OD600 = 0.17. The time t0 corresponds to 6 h (a) and 7 h (b) of constant flow at 0.75 μl min−1. The white arrow indicates a very thin initial streamer. (c) Time sequence of phase-contrast microscopy images acquired every 1 min at 40× magnification (the focus is roughly in the middle of the channel). Time t0 corresponds to 5 h and 45 min of constant flow at 1 μl min−1. The initial concentration of PA14 is equivalent to OD600 = 0.5. White arrows point to single cells or isolated small clusters that are not moving with the mainstream flow but, instead, seem to be attached to very thin filaments of extracellular matrix (not visible; scale bars, (a,b) 100 μm; (c) 25 μm).
Furthermore, in an attempt to visualize the extracellular polymeric substances (EPS) in the developing streamers, we used fluorescently labelled lectins (Strathmann et al. 2002): although this approach only worked in a test with a different strain (P. aeruginosa PA01) and in the presence of a sufficient amount of extracellular matrix, it confirmed the occurrence of aggregates of EPS in the proximity of the corners and in the middle-height plane of the channel. These aggregates were connected by a filament of EPS suspended across the channel (see the electronic supplementary material, figure S3).
3.2. Mutant strains
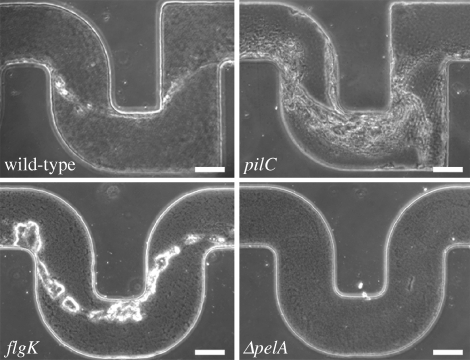
The crucial role of the extracellular matrix in the formation process of the streamers is further confirmed when we compare results obtained using different mutants of P. aeruginosa PA14. We used mutants (pilC) defective in the biogenesis of type IV pili (i.e. extremely thin surface appendages that are a few microns long), non-motile mutants (flgK) defective in the synthesis of a functional flagellum (O'Toole & Kolter 1998) and mutants (ΔpelA) defective in the production of PEL, a glucose-rich polymer that is one of the main components of the EPS in PA14 biofilms (Friedman & Kolter 2004a). In four identical channels we injected four different solutions (three mutants and the wild-type) at the same concentration (OD600 = 0.5) and the same flow rate (1 μl min−1). After approximately 12 h, we observed the presence of streamers for pili-deficient and non-motile mutants (as well as the wild-type) but not in the matrix-deficient mutants (as shown in figure 4), with the only exception of a few tiny filaments observed in the proximity of the corners, probably made from polymeric substances whose synthesis was not prevented by the pelA mutation (Friedman & Kolter 2004b). Moreover, although they both build biofilm streamers, the different architectures shown by pilC and flgK mutant strains are noteworthy (figure 4; see the electronic supplementary material, figure S4): the streamers grown with the pili-deficient mutants appear to be formed by multiple thin filaments rather than a more massive single thread. On the other hand, the flagellum-deficient bacteria made more clustered, clumpy streamers compared with the smooth wild-type morphology, which suggests a role of motility in the structure of the streamers. A similar diversity in architectures has been reported for surface-attached biofilms formed by wild-type and motility mutants in flow chambers (Klausen et al. 2003). Future experiments will be required to better elucidate the role played by pili and flagellum in the formation process of biofilm streamers.
Figure 4.
Phase-contrast images (the focus is approximately in the middle of the channel) after 12 h of continuous flow at 1 μl min−1. Comparison between wild-type, type IV pili defective mutant (pilC), flagellar-mediated motility defective mutant (flgK) and mutant (ΔpelA) defective in the synthesis of one of the main components of the extracellular matrix in P. aeruginosa PA14 biofilms (scale bars, 100 μm).
4. Numerical simulations
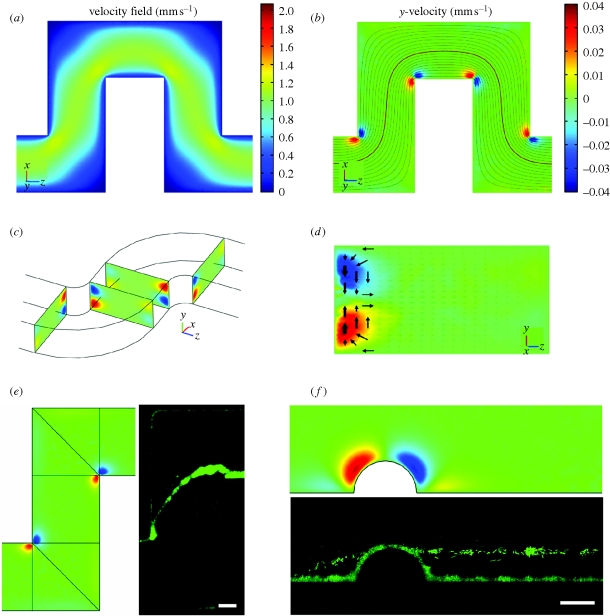
What is the link between the observed streamers and the geometric features of the channel? Since the Reynolds number (i.e. a dimensionless number expressing the ratio of inertial to viscous forces) is relatively small (in the range 0.02–0.1), the flow in the microchannels is everywhere laminar. Hence, we used a commercial finite-element software (COMSOL) to perform three-dimensional numerical simulations of the flow in curved channels with the same geometry and physical parameters as used in the experiments. The main features of the results are shown in figure 5a–d. The primary flow matches the flow pattern predicted in a two-dimensional planar geometry: a contour plot of the velocity field and the associated streamlines, in a plane at a quarter of the channel height from the upper surface, are displayed, respectively, in figure 5a,b. However, an additional velocity field superimposed on the primary flow, usually called a ‘secondary flow’, develops only in the proximity of the corners, both rounded and sharp (figure 5b–d). This secondary flow consists of two symmetrical counter-rotating vortices of length scale comparable to half the channel height. These two open eddies rotate from the middle plane towards the bottom and the top walls at the beginning of the curve, while the motion is reversed after the turn (figure 5c,d). The numerical results also show that the magnitude of the secondary vortical flow is directly proportional to the average velocity in the channel and about 5 per cent of the velocity in the primary flow direction (see colour scales in figure 5a,b). Moreover, for increasing values of the Reynolds number, only a small asymmetry between the vortices at the beginning and the end of the turn begins to appear, in which the latter are stronger than the former eddies. The phenomenon of eddies associated with a three-dimensional flow around a corner, in the limit of small Reynolds numbers (Stokes flow), is not very well known, but has been shown to stem from a change in the curvature of the boundary (Balsa 1998). This result means that in confined Stokes flows where the geometry has a constant curvature (e.g. in the annular space between two concentric cylinders) there is no secondary flow (as also shown in Lauga et al. 2004), whereas the vortical motions we found develop as soon as the streamlines deviate from a rectilinear to a curvilinear path.
Figure 5.
(a) Numerical results of the modulus of the velocity field in a plane at ¼ of the channel height from the upper surface. Coordinate system (x, y, z) and colour scale are shown. (b) Flow components perpendicular to the primary flow (represented here with streamlines) in the same plane as (a). Red and blue colours indicate here velocity components orthogonal to the plane of the channel, and directing, respectively, upwards (positive y) and downwards (negative y). (c) Prospective view of the channel showing pairs of counter-rotating vertices in cross-sectional planes right before and after the turns. (d) A cross-section of the channel, right after a turn. Arrows show the motion of fluid elements in the secondary flow. (e,f) Comparisons between numerical simulations of the secondary flow and experimental observations of the streamers, in a curved channel (e) and in a straight channel with a lateral hemi-cylindrical bump (f) (scale bars, (e) 50 μm; (f) 25 μm).
5. Discussion
We hypothesize a connection between the experimental demonstration of the streamers and the features of the flow highlighted by the numerical simulations. First of all, the locations where the streamers are connected to the lateral walls of the channel correspond exactly to the positions of secondary vortical motion (see figure 5e). In particular, the streamers are linked to the ends of the turns rather than the beginning (as also shown by the time-lapse images), which is where the fluid elements move towards the middle of the channel, as represented by the arrows in figure 5d. The direction of these two rolls of the secondary flow indeed suggests an accrual of biomass in the horizontal plane of the channel, which is where the streamers develop. However, while at the time of the initial development of the streamers the bottom and top surfaces of the channel are already covered by microcolonies, as shown, for instance, by the first image in the sequences of figure 3a,b, we did not observe any increase of cells on the side walls at half the channel height. Thus, a probable mechanism for the formation of the streamers is that the vortical flow at the corner accumulates polymeric substances (in solution or, more probably, secreted from the bacteria adhered to the surfaces) on the lateral walls at half the channel height and forms one or more precursor threads, which are then stretched by the primary flow until they reach the other corner. We believe this process can happen simultaneously at different corners all along the channel, and the subsequent time evolution probably involves the collection of cells and matrix along the streamers, which eventually connect together to form the long structures that we observe after a few hours of continuous flow (as shown in figure 3).
Several studies have shown that biofilms behave as viscoelastic polymer materials, with relaxation times on the order of seconds or minutes (Shaw et al. 2004; Hohne et al. 2009), which means that on a time scale of a few hours a streamer can be considered essentially as a viscous fluid. Then, an order of magnitude estimate for the ‘extrusion’ process exerted by the main flow can be provided. The rate of deformation (Δɛ/Δt) of an initial thin thread is given by Δɛ/Δt ≈ σ/η, where σ is the applied stress and η the viscosity of the material. In our case σ is the viscous stress exerted by the flow, which can be estimated (Hinch 1976) as σ ≈ (2πμV)/(d ln(2L/d)) ≈ μV/d ≈1 Pa, where μ is the dynamic viscosity of the medium (≈10−3 Pa s), V the velocity of the flow (≈10−3 m s−1), L the length (≈10−4 m) and d the diameter of the thread (≈10−6 m). If we suppose that the precursor streamer is stretched by about 10 times its original length (say, from about 25 μm, which is the size of the secondary eddy, up to 250 μm) in a few hours (≈104 s), we obtain η ≈ 103 Pa s, which is a value on the order of those reported in the literature for P. aeruginosa biofilms (Shaw et al. 2004).
Finally, to further validate the hypothesis of a hydrodynamic mechanism underlying the formation of the streamers, we considered a simple case of change of curvature in a channel, i.e. the presence of a hemi-cylindrical bump on the side wall of a straight channel. Numerical simulations for this confined geometry show the presence of a secondary flow located around the bump, similar to the vortical motion around the corners (figure 5f). Once again, we observed streamers (after approx. 20 h) attached to the bump at half the channel height and suspended in parallel to the side walls (figure 5f; see the electronic supplementary material, figure S5).
6. Conclusions
The discovery of streamers in laminar flows in curved channels offers new opportunities to study the mechanics and microbiological features relevant to the formation of these filamentous biofilms. Bacterial streamers, which can be detrimental or beneficial to a given system, develop in the bulk fluid and appear to be linked to inevitable three-dimensional flow features accompanying even the simplest geometries. Such suspended biofilms may therefore be much more widespread than previously thought in all manners of liquid distribution systems.
Acknowledgements
We thank W. Chai, D. Lopez and H. Vlamakis for providing strains, culture media and technical assistance. We thank R. Losick for critically reading the manuscript, A. Forsyth, U. Hees, J. Hinch, R. Kolter, R. Mitchell, J. Rieger and B. Sobotka for helpful discussions, and the BASF-Harvard working group for useful feedback. This work was supported by the BASF Advanced Research Initiative at Harvard University.
References
- Balsa T. F. 1998. Secondary flow in a Hele-Shaw cell. J. Fluid Mech. 372, 25–44. ( 10.1017/S0022112098002171) [DOI] [Google Scholar]
- Battin T. J., Kaplan L. A., Newbold J. D., Hansen C. M. E. 2003. Contributions of microbial biofilms to ecosystem processes in stream mesocosms. Nature 426, 439–442. ( 10.1038/nature02152) [DOI] [PubMed] [Google Scholar]
- Besemer K., Hödl I., Singer G., Battin T. J. 2009. Architectural differentiation reflects bacterial community structure in stream biofilms. ISME J. 3, 1318–1324 ( 10.1038/ismej.2009.73) [DOI] [PubMed] [Google Scholar]
- Brooks J. D., Flint S. H. 2008. Biofilms in the food industry: problems and potential solutions. Int. J. Food Sci. Tech. 43, 2163–2176. ( 10.1111/j.1365-2621.2008.01839) [DOI] [Google Scholar]
- Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. ( 10.1146/annurev.mi.49.100195.003431) [DOI] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. ( 10.1126/science.284.5418.1318) [DOI] [PubMed] [Google Scholar]
- Drenkard E., Ausubel F. M. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416, 740–743. ( 10.1038/416740a) [DOI] [PubMed] [Google Scholar]
- Duffy D. C., McDonald J. C., Schueller O. J. A., Whitesides G. M. 1998. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal. Chem. 70, 4974–4984. ( 10.1021/ac980656z) [DOI] [PubMed] [Google Scholar]
- Edwards R., Harding K. G. 2004. Bacteria and wound healing. Curr. Opin. Infect. Dis. 17, 91–96. ( 10.1097/00001432-200404000-00004) [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Bond P. L., Gihring T. M., Banfield J. F. 2000. An archaeal iron-oxidizing extreme acidophile important in acid mine drainage. Science 287, 1796–1799. ( 10.1126/science.287.5459.1796) [DOI] [PubMed] [Google Scholar]
- Friedman L., Kolter R. 2004a. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51, 675–690. ( 10.1046/j.1365-2958.2003.03877) [DOI] [PubMed] [Google Scholar]
- Friedman L., Kolter R. 2004b. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186, 4457–4465. ( 10.1128/JB.186.14.4457-4465.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg K. B., Coupland K., Kimura S., Johnson D. B. 2006. Macroscopic streamer growths in acidic, metal-rich mine waters in North Wales consist of novel and remarkably simple bacterial communities. Appl. Environ. Microbiol. 72, 2022–2030. ( 10.1128/AEM.72.3.2022-2030.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L., Costerton J. W., Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108. ( 10.1038/nrmicro821) [DOI] [PubMed] [Google Scholar]
- Hinch E. J. 1976. The distortion of a flexible inextensible thread in a shearing flow. J. Fluid Mech. 74, 317–333. ( 10.1017/S002211207600181X) [DOI] [Google Scholar]
- Hohne D. H., Younger J. G., Solomon M. J. 2009. Flexible microfluidic device for mechanical property characterization of soft viscoelastic solids such as bacterial biofilms. Langmuir 25, 7743–7751. ( 10.1021/1a803413x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jørgensen A., Molin S., Tolker-Nielsen T. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48, 1511–1524. ( 10.1046/j.1365-2958.2003.03525.x) [DOI] [PubMed] [Google Scholar]
- Lauga L., Stroock A. D., Stone H. A. 2004. Three-dimensional flows in slowly varying planar geometries. Phys. Fluid 16, 3051–3062. ( 10.1063/1.1760105) [DOI] [Google Scholar]
- Mah T.-F. C., O'Toole G. A. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9, 34–39. ( 10.1016/S0966-842X(00)01913-2) [DOI] [PubMed] [Google Scholar]
- O'Toole G. A., Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30, 295–304. ( 10.1046/j.1365-2958.1998.01062) [DOI] [PubMed] [Google Scholar]
- O'Toole G. A., Kaplan H. B., Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79. ( 10.1146/annurev.micro.54.1.49) [DOI] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701. ( 10.1146/annurev.micro.57.030502.090720) [DOI] [PubMed] [Google Scholar]
- Reysenbach A., Cady S. L. 2001. Microbiology of ancient and modern hydrothermal systems. Trends Microbiol. 9, 79–86. ( 10.1016/S0966-842X(00)01921-1) [DOI] [PubMed] [Google Scholar]
- Shaw T., Winston M., Rupp C. J., Klapper I., Stoodley P. 2004. Commonality of elastic relaxation times in biofilms. Phys. Rev. Lett. 93, 098102 ( 10.1103/PhysRevLett.93.098102) [DOI] [PubMed] [Google Scholar]
- Stone H. A., Stroock A. D., Ajdari A. 2004. Engineering flows in small devices: microfluidics towards a lab-on-a-chip. Annu. Rev. Fluid Mech. 36, 381–411. ( 10.1146/annurev.fluid.36.050802.122124) [DOI] [Google Scholar]
- Stoodley P., Lewandowski Z., Boyle J. D., Lappin-Scott H. M. 1998. Oscillation characteristics of biofilm streamers in turbulent flowing water as related to drag and pressure drop. Biotechnol. Bioeng. 57, 536–544. () [DOI] [PubMed] [Google Scholar]
- Stoodley P., Sauer K., Davies D. G., Costerton J. W. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209. ( 10.1146/annurev.micro.56.012302.160705) [DOI] [PubMed] [Google Scholar]
- Strathmann M., Wingender J., Flemming C. 2002. Application of fluorescently labelled lectins for the visualization and biochemical characterization of polysaccharides in biofilms of Pseudomonas aeruginosa. J. Microbiol. Meth. 50, 237–248. ( 10.1016/S0167-7012(02)00032-5) [DOI] [PubMed] [Google Scholar]
- Vrouwenvelder J. S., Graf von der Schulenburg D. A., Kruithof J. C., Johns M. L., van Loosdrecht M. C. M. 2009. Biofouling of spiral-wound nanofiltration and reverse osmosis membranes: a feed spacer problem. Water Res. 43, 583–594. ( 10.1016/j.watres.2008.11.019) [DOI] [PubMed] [Google Scholar]
- Zimmerli W., Trampuz A., Ochsner P. E. 2004. Prosthetic-joint infections. New Engl. J. Med. 351, 1645–1654. ( 10.1056/NEJMra040181) [DOI] [PubMed] [Google Scholar]