Abstract
We investigate the role of recombination in transposable element (TE) proliferation in the cyclical parthenogen, Daphnia pulex. Recombination provides a mechanism by which the rate of both TE gain and loss can be accelerated, a duality that has long intrigued many biologists interested in the influence of sex on mutation accumulation. We compared TE loads among populations of D. pulex where sex occurs regularly (cyclical parthenogens or ‘sexuals’) with those in which the ability to reproduce sexually has been completely lost (obligate ‘asexuals’) for six different families of DNA transposons. Transposon display assays showed that sexuals have more TEs than asexuals, contrary to the expectations under Muller's ratchet but consistent with the idea that sex facilitates TE spread. Sexuals also exhibit higher insertion site polymorphism among lineages, as predicted because recombination accelerates rates of loss and gain. Asexuals, however, have proportionally more singletons (loci occupied in a single isolate), which differs from previous studies where selfing and outcrossing were used as a proxy for high and low recombination. Our multi-element survey reveals that the impact of sex on TE proliferation is consistent among different Class II TE families and we discuss the genomic consequences of different reproductive strategies over long time periods.
Keywords: transposable elements, recombination, Daphnia pulex, cyclical parthenogenesis, sex, mutation
1. Introduction
The evolutionary dynamics of transposable elements (TEs) are shaped by intrinsic properties of TEs, host genome environments and features of the natural history and demography of the populations in which the TE is found. One feature of the host genome thought to be especially important for the dynamics of TE evolution is recombination via sexual reproduction (Wright & Finnegan 2001). The mechanisms of both TE gain and loss are different in genomic backgrounds with and without meiotic recombination, ultimately affecting both TE load and distribution. For example, independent assortment of chromosomes during meiosis can lead to the loss of heterozygous TE copies, a mechanism of loss that does not occur in asexual lineages. At the same time, sex provides the chief mechanism through which TEs can multiply among lineages in a population, a means of spread unavailable in asexuals. These effects of meiotic recombination on TE loads within individual lineages may have long-term consequences for the success of TEs at the population level.
Interest in the impact of reproductive strategies on patterns of TE accumulation in host genomes has motivated comparative studies of TE loads based on patterns of recombination (Charlesworth et al. 1992; Hoogland & Biemont 1996). These studies, especially in Drosophila, have also been used to infer the nature of the cost of new insertions, a topic of considerable debate in the literature (Biemont et al. 1997; Charlesworth et al. 1997). Some argue that the primary cost of increased copy number is indirect (i.e. higher risk of non-homologous, or ectopic, recombination (Montgomery et al. 1987)), as opposed to the more direct costs of insertion (Finnegan 1992) or transposition itself (Brookfield 1991). To evaluate these hypotheses, researchers examine correlations between rates of recombination and TE density (e.g. Rizzon et al. 2002; Song & Boissinot 2007). If, for example, the primary cost of new insertions is an increased risk of deletion via ectopic recombination (indirect), one would predict that selection would lead to a negative relationship between the frequency of recombination and the abundance of TEs or, more generally, lower loads in sexuals than in asexual lineages. This pattern is indistinguishable, however, from that which would be observed because of an increase in the efficacy of selection in sexuals. This underscores the challenge of distinguishing among the factors controlling the distribution of TEs and highlights the importance of understanding their distribution within the genome and among populations.
Surveys of TEs and the long-term costs and benefits associated with sexual versus asexual reproductive strategies have also been performed by comparing inbred versus outcrossing species (inbreeding is used as a proxy for low recombination because recombination without genetic variation offers little advantage; Wright et al. 2001; Dolgin et al. 2008). These studies, however, evaluate the impact of selfing on TE dynamics rather than the effects of recombination itself and, although informative, unavoidably introduce the confounding effects of species differences to the comparison. Intraspecific studies of TE dynamics take advantage of known differences in recombination rates within and among chromosomes in classic model organisms (e.g. Caenorhabditis elegans, Duret et al. 2000; Drosophila melanogaster, Bartolome et al. 2002; Bartolome & Maside 2004). Although these studies reveal patterns of TE distribution based on local rates of recombination, they do not explain the long-term consequences of different reproductive strategies, thereby helping to further elucidate reasons for the origin and maintenance of sex and the abundance of asexual clades in primarily sexual taxa (see Butlin 2002 for review).
We investigated the impact of recombination on TE dynamics in natural populations by surveying six families of DNA transposons among populations of Daphnia pulex, an aquatic microcrustacean. These families are class II ‘cut-and-paste’ elements that transpose via a DNA intermediate, unlike class I retroelements which have an RNA intermediate and are known to ‘copy-and-paste’. Daphnia pulex is a cyclical parthenogen and most populations reproduce clonally during the growing season and facultatively switch to sex in response to seasonal environmental cues. Some populations have lost the ability to switch to sex; these obligate asexuals can produce resting eggs (in the absence of males), which can overwinter similar to meiotically produced resting eggs of their cyclically parthenogenetic counterparts. Two previous studies have examined patterns for Pokey, the first TE described in Daphnia, among cyclically parthenogenetic and obligately asexual natural populations (Sullender & Crease 2001; Valizadeh & Crease 2008). Their results showed that obligate asexuals have fewer Pokey insertions and cyclically parthenogenetic populations exhibit higher levels of insertion presence–absence polymorphism than those observed among asexual isolates. In order to assess whether this is a general trend among families and superfamilies of TEs, we present data comparing the abundance and distribution of five recently discovered TE families, in addition to Pokey, among D. pulex isolates collected from throughout North America.
2. Material and methods
(a). Collection and reproductive mode determination
Natural populations of D. pulex1 were collected from 56 permanent and ephemeral ponds from 12 different states and provinces throughout North America (IL, IN, WI, OH, MI, OR, MN, ME, NY, ON, NB, QU; see electronic supplementary material, table S2). Animals were collected using mesh-bottomed cups and transported in containers of pond water on ice. Single individual females were used to establish multiple clonal lines from each population, hereafter referred to as isolates. Isolates were confirmed to be D. pulex using allozymes according to previously published methods (see Hebert et al. 1988). Each isolate was maintained in the laboratory under common conditions: they were maintained in Percival environmental chambers at 20°C, fed chemostat-grown Scenedesmus obliquus three times per week, and cultured in filtered lake water collected primarily from Lake Lemon, Bloomington, IN, USA.
In order to establish whether isolates were cyclical parthenogens or obligate asexuals, approximately five individual females from each were raised in 250 ml beakers in the absence of males. Water from separate, high-density cultures was used to provide a crowding cue to the females to induce ephippial production (Stross & Hill 1965). Any live young or males were removed from the beakers, and all ephippia produced were collected, opened and scored for the presence of eggs. Cyclical parthenogens cannot produce egg-bearing ephippia in the absence of males, unlike obligate asexuals (Innes et al. 1986). Females were categorized as obligate asexuals once they produced egg-bearing ephippia in the absence of males a minimum of 10 times. A single isolate was used for each population determined to be obligately asexual because previous studies indicated no polymorphism among isolates collected from such populations, whereas multiple isolates were used to represent cyclically parthenogenetic populations because polymorphism among isolates in TE distribution has been previously demonstrated (see electronic supplementary material, table S2; Sullender & Crease 2001).
(b). Transposon display
Transposon display (TD; Van den Broeck et al. 1998) was performed on six families (representing four superfamilies and two subclasses) of DNA transposons previously identified in the D. pulex genome (Penton et al. 2002; Schaack et al. in press). TE families assayed included two families of Tc1-like elements, two Helitron families, a hAT homologue and the previously described piggyBac element, Pokey. Approximately 20 clonally produced individuals from each isolate were used to perform DNA extractions using the CTAB protocol developed by Doyle & Doyle (1987). Transposon display was performed using EcoRI to digest genomic DNA from each isolate (total n = 84; cyclical parthenogens = 41 and obligate asexuals = 43). Digests were performed for 6 h at 37°C followed by 22 min at 80°C. Adaptors consisting of approximately 20 bp oligonucleotide pairs with a non-complementary mid-portion were ligated on to the ends of each fragment after the digest (16 h ligation using T4 ligase at room temperature; see electronic supplementary material, table S1 for oligosequences). The DNA concentration of the restriction–ligation reaction was checked in a subset of samples using a Nanodrop ND-1000 and was found to range from 30 to 70 ng µl−1. Element-containing fragments were amplified via nested PCR using approximately 50 ng of DNA as template and an element-specific primer (forward) and a reverse primer complementary to the mid-portion of one strand of the oligos used to construct the ligated adaptors, which include an internal region of non-complementarity (see electronic supplementary material, table S1). Only fragments of the genome containing copies of a given element were amplified during PCR because the reverse primer cannot anneal unless the element-specific primer binds and elongates.
Primary reactions were performed using Eppendorf Mastermix in a total volume of 10 µl (4 µl mastermix, 1 µl 25 mM MgCl2, 0.3 µl F primer (10 mmol), 0.3 µl R primer (10 mmol), 4 µl H2O and 1 µl of restriction–ligation reaction described above). Products from the primary reaction were diluted 1 : 10 in ddH2O, and 2.5 µl of the dilution was used as the template for the secondary reaction. The second round of PCR used a fluorescently labelled element-specific primer slightly more towards the 3′-end of the conserved region of the element and primer concentrations were adjusted (0.3 µl (10 mmol) F fluorescent primer and 0.03 µl (10 mmol) R primer). Reactions were 10 µl total in volume (same as previous, with adjusted H2O), and were diluted 1 : 75 in ddH2O after amplification. Conditions for the first and second round of PCR were as follows: initial denaturation at 94°C for 3 min, followed by 24 cycles of denaturation at 94°C for 30 s, annealing at 5°C below the melting temperature for each element-specific primer and elongation at 72°C for 1 min, ending with a 5 min elongation step at 72°C. The diluted secondary PCR product (1 µl) from each sample was added to a 96-well plate in which each well contained 9 µl of a ladder dilution (prepared for 96 reactions by diluting 5 µl LIZ 1200 size standard in 895 µl ddH2O). The mixture was spun, denatured for 5 min at 93°C and placed on ice for 5–10 min. Reactions were run on an ABI 3730 Genotyper and analysed using Genemapper.
Beginning at the PCR stage, all samples were run in triplicate and data were scored in cyclical parthenogens and obligate asexuals using a consistent rubric for each family (for details see electronic supplementary material, S2). Transposon display is sensitive but provides lower-bound estimates because long fragments may not amplify owing to PCR bias. In addition, because divergence among populations may have occurred, it is possible that element-specific primers may not have equivalent binding affinities at all loci in all populations. Non-fluorescent primers were used to run additional PCR reactions under the same conditions for each TE family in a small subset of populations (both sexuals and asexuals) to confirm that the assay was amplifying TE-bearing fragments. Fragments were cloned using the Invitrogen TOPO cloning kit following manufacturer's protocols. Cloned fragments were PCR-amplified using the reverse primer from the initial secondary PCR reaction (complementary to the adaptor) and the successful amplicons were sequenced using ABI BigDye sequencing mix on the ABI 3730. Sequences were aligned to verify that they contained the 3′-end of the TE for each family using CodonCode Aligner (CodonCode Corporation) and MEGA (Tamura et al. 2007).
(c). Data analysis
The mean number of occupied sites was compared between the two treatments, cyclical parthenogens and obligate asexuals, for each TE family separately and combining across all families using the Mann–Whitney U-test. The number of singletons (loci occupied in a single isolate) was compared between cyclical parthenogens and obligate asexuals (all families combined) using Fisher's exact test.
The frequency of occupation at any given site was compared between cyclical parthenogens and obligate asexuals by comparing across all families using analysis of covariance (ANCOVA) with total number of occupied sites as a covariate. Similarly, the number of polymorphic loci among isolates within each treatment (with the number of polymorphic loci across treatments per family as a covariate) was compared (with a Poisson distribution for count data; SPSS 16.0). In this comparison, one random representative isolate was selected from each of the 13 sexual populations and these were compared with a randomly chosen subset of obligate asexuals (n = 13 of 43 total isolates collected) to make sample sizes even.
Within the cyclical parthenogens, polymorphism patterns were analysed using analysis of molecular variance (AMOVA) to assess between-population and between-region (state or province) differences (cyclical isolates only (n = 41); AMOVA, Arlequin 3.11; Excoffier et al. 2005). In addition, to detect evidence for clustering based on breeding system or region, a neighbour-joining tree was generated based on a distance matrix of concatenated presence–absence data for all TE families using Arlequin 3.11. Limitations of the TD technique (the inability to distinguish between heterozygous and homozygous insertions and the technical constraints of PCR and fragment analysis) result in a lower-bound estimate of TE load and polymorphism based on the number and distribution of occupied loci, not based on copy number; however, because these caveats are applicable to both sexuals and asexuals, a comparison of reproductive strategies is reasonable.
3. Results
Mean number of occupied sites for each TE family in cyclically parthenogenetic and obligately asexual isolates of D. pulex is presented in table 1. Overall, the number of occupied sites per isolate is higher in cyclicals than in asexuals (all families combined; n = 84, Mann–Whitney U = 614.50, p = 0.017; figure 1). Specifically, this difference stems from higher numbers of occupied sites in three of the six families assayed (Tc1-2, Helitron 1 and Helitron 2; see table 1 and electronic supplementary material, table S2 for means and total numbers for each family, respectively). Across the four superfamilies assayed, the two families of Tc1 elements have the highest mean number of occupied sites per isolate and the hAT family has the lowest. The two non-autonomous Helitron families differ, with the shorter element (Helitron 1, 351 bp) being more abundant than the longer members of the Helitron 2 family (1359 bp). Our results are consistent with previous work by Valizadeh & Crease (2008) showing higher Pokey insertion numbers in cyclical parthenogens, although the difference observed here is not significant and the mean number of occupied sites for Pokey in our populations (both treatments) is lower overall. This is likely because we used a less-frequent cutter to perform the initial digest of genomic DNA. However, we were able to score fragments ranging up to 800 bp for most elements (versus an upper bound of 500 bp), which makes it possible to detect copies located further upstream from cut sites. Overall, the frequency of insertion at any given locus was generally low and similar among families, except Tc1-2 where the average frequency of insertion was high (table 1; analysis of variance, d.f. = 5, F = 5.04, p < 0.0001). Although asexual isolates had lower numbers of insertions overall, the number of singletons (number of sites occupied in only one isolate) was higher in asexuals in five out of six families (table 1 and electronic supplementary material, table S2; for concatenated dataset of all six families combined, Fisher's exact test, p = 0.012).
Table 1.
For each TE family assayed, mean occupation frequency per locus, proportion of singletons (loci only occupied in a single isolate), range (number of occupied sites per isolate), mean number of occupied sites for each family of DNA transposons surveyed in cyclically parthenogenetic (cyclicals; n = 41) and obligately asexual populations (obligates; n = 43) of D. pulex, Mann–Whitney U statistics from comparison of mean rank number, and associated p-values.
| singletons |
mean number (±s.e.) |
|||||||
|---|---|---|---|---|---|---|---|---|
| TE family | mean occupation frequency (per locus) | cyclicals | obligates | range | cyclicals | obligates | U | p |
| Tc1-1 | 0.05 | 0.44 | 0.51 | 1–37 | 18.05 (1.12) | 18.12 (1.18) | 875.5 | 0.957 |
| Tc1-2 | 0.10 | 0.38 | 0.45 | 2–27 | 19.78 (0.82) | 14.98 (0.88) | 451.0 | <0.0001 |
| Pokey | 0.04 | 0.58 | 0.60 | 1–8 | 4.56 (0.23) | 3.83 (0.30) | 702.5 | 0.104 |
| hAT | 0.04 | 0.29 | 0.20 | 0–3 | 0.463 (0.12) | 0.349 (0.08) | 860.0 | 0.814 |
| Helitron 1 | 0.06 | 0.42 | 0.54 | 2–23 | 15.39 (0.64) | 12.40 (0.90) | 630.0 | 0.024 |
| Helitron 2 | 0.06 | 0.45 | 0.67 | 1–7 | 3.63 (0.19) | 2.02 (0.17) | 308.5 | <0.0001 |
Figure 1.
Frequency distribution of transposable element loads (all six DNA transposon families combined) for D. pulex from throughout North America showing (a) cyclically parthenogenetic isolates have higher loads (black bars; mean = 61.9, n = 41, s.d. = 14.02) than (b) obligately asexual isolates white bars; mean = 51.7, n = 43, s.d. = 18.26).
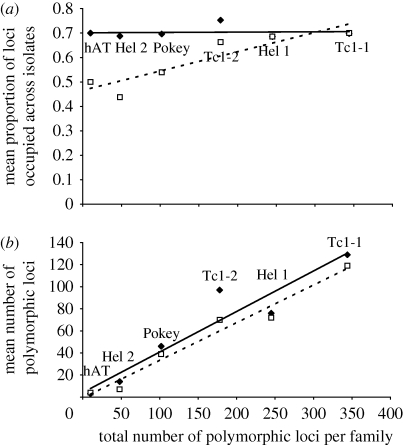
Although the number of potentially occupied sites across isolates (total number of polymorphic loci) ranged widely among the six TE families surveyed (from 10 (hAT) to 344 (Tc1-1)), the proportion of those sites that were occupied in at least one isolate did not vary greatly among cyclical parthenogens (figure 2a; also see distribution of occupied loci among isolates for each family in electronic supplementary material, S1a–f). For obligate asexuals, however, the proportion of total sites observed that were occupied was lower, but increased as a function of TE family size (interaction between reproductive mode and family size; d.f. = 1, F = 14.23, p = 0.005). There were more loci occupied by at least a single isolate among cyclical parthenogens relative to their asexual counterparts across all families assayed, when total number of polymorphic loci per TE family was included as a covariate (figure 2b; ANCOVA: n=28, slope, d.f. = 1, n.s.; intercept, d.f. = 1, F = 3.8, p = 0.038; estimated marginal means are 49.5 and 42.2 (±2.9, 2.6 s.e.) among cyclicals and obligates, respectively). AMOVA for each TE family indicates variation attributable to locale (variance partitioned by state; electronic supplementary material, table S3), as does a neighbour-joining tree constructed based on a distance matrix of pairwise differences using concatenated data from all six families (data not shown). This tree did not, however, reveal any clustering of isolates based on reproductive strategy alone, indicating that asexuality has likely arisen independently more than once (see also Paland et al. 2005).
Figure 2.
(a) Mean proportion of loci occupied by a transposable element in at least one isolate for each TE family (n = 84) compared between cyclical parthenogens and obligate asexuals and (b) mean number of polymorphic loci among populations of cyclical parthenogens and obligate asexuals based on a random subsample of the data (n = 13 for each treatment) with total number of polymorphic loci for each family as a covariate. Cyclical parthenogens are denoted as black diamonds/solid line and obligate asexuals are denoted as white squares/dashed line.
4. Discussion
The dual role of recombination in TE proliferation has generated great interest in the effects of sexual versus asexual reproduction on TE loads in the genome over time (Arkhipova & Meselson 2000; Wright & Finnegan 2001; Dolgin & Charlesworth 2006). Sex can facilitate the spread of a new TE throughout a population, but it also provides a mechanism through which new copies can be lost. Thus, sex can lead to an accelerated rate of increase and decrease in TEs over time relative to asexuals, and also impact the distribution of TEs among individuals within a population where sex has been lost (Schaack et al. in press). We surveyed six families of transposable elements in populations of D. pulex, which reproduce either with or without sex and find that both the number and distribution of TEs differ between cyclical parthenogens and obligate asexuals, despite the fact that obligately asexual populations in this species are thought to be relatively young (Lynch et al. 2008). Even though purging deleterious mutations and increased efficiency of selection are major arguments for the evolution of sex, we find that obligate asexuals have lower TE loads than sexuals. Overall, sexual populations bear higher numbers of occupied loci and, despite fewer singletons, exhibit higher levels of presence–absence polymorphism at individual loci (figures 1 and 2b).
A simple explanation for higher TE loads in sexuals is the opportunity sex provides for multiplicative spread (copies located at different loci can be united in the genome of sexually produced offspring; Hickey 1982). In addition, sexuals can become homozygous at a given locus through meiotic and mitotic recombination, whereas asexuals can only become homozygous by mitotic recombination. Homozygous insertions are more difficult to eliminate from the genome because homologue-dependent DNA repair can reconstitute a copy of a homozygous element after excision. A second explanation for why sexuals appear to have higher TE loads than asexuals is unavoidable sampling bias. Asexual lineages have repeatedly been spun-off from sexual progenitors, and those derived asexual lineages with low TE loads may have a better chance of surviving in nature and the laboratory relative to high-load asexual isolates suffering a greater mutational ‘hangover’ from sex (Nuzhdin & Petrov 2003).
Thirdly, although our data provide no evidence for Muller's ratchet (Muller 1964; Felsenstein 1974) operating in asexual lineages, Paland & Lynch (2006) found an excess of replacement site substitutions in populations of D. pulex, where sex has been permanently lost. If the fitness consequences of new TE insertions are sufficiently high, it may not be possible to observe their accumulation prior to extinction. Finally, it is possible that TEs in sexually reproducing host genomes garner certain direct advantages from sex that allow them to persist and spread more effectively. Presumably, when TEs invade a host genome, selective pressure favours the coevolution of some type of host suppression machinery to minimize mutations resulting from TE activity, much like an immune response. TEs can evade such machinery if, like their hosts, they are able to recombine, exchange genetic material and produce novel genetic combinations undetectable to the host (Sharma et al. 2008). If true, one would predict TEs to be more active, and potentially more abundant, in sexually reproducing populations of D. pulex relative to asexuals where inter-element exchange is far less likely.
As has been seen in several TE families surveyed in Drosophila melanogaster (Bartolome et al. 2002), most sites are occupied at low frequency across isolates for each family surveyed in D. pulex (table 1). The differences between cyclical parthenogens and obligate asexuals in mean occupation frequencies (figure 2a) indicate that such patterns are consistent across families in sexuals but vary with TE family size in asexuals. This pattern may be expected because TEs can spread and reach higher occupation frequencies among sexual isolates, whereas in asexuals no such spread is possible and singletons are more common. The overall higher levels of polymorphism among cyclical parthenogens is similar to that observed in other studies looking at the relationship between recombination and TE load, even when inbreeding and outcrossing were used as a proxy for low and high recombination, respectively (Wright et al. 2001; Dolgin et al. 2008). One important difference between the patterns observed in those studies and that found here is the frequency of singletons; in interspecific comparisons of Arabidopsis thaliana and its outcrossing relative A. lyrata, for example, the excess singletons observed in A. lyrata was interpreted to be the evidence for stronger purifying selection on new insertions in high recombination environments. In contrast, here we see an excess of singletons in asexual isolates in four of the six families examined, which would be expected because any new insertions in asexual lineages cannot spread between populations.
Valizadeh & Crease (2008) suggest that Pokey may, indeed, be active based on patterns of polymorphism observed among natural populations (i.e. the presence of novel insertion sites and an inability to group isolates based on breeding system or region). Our data also indicate no clustering based on reproductive strategy, but do show variation based on region (electronic supplementary material, table S3). This does not eliminate the possibility that these TE families are active, but rather indicates that there may also be natural variation segregating locally in populations. Additionally, lab-reared mutation accumulation lines of D. pulex provide evidence for somatic insertions for all six of the TE families assayed in this study, as well as support for a germline transposition event for a hAT-like element (Schaack et al. in press). This family is found at very low copy numbers in D. pulex, making it difficult to ascertain whether reproductive mode has impacted patterns of proliferation for this element (e.g. there was no difference in load or polymorphism between cyclical parthenogens and obligate asexuals within this family; table 1 and figure 2b). The frequency of occupation across sites, however, was most different between cyclical parthenogens and asexuals for this element family (figure 2a), a pattern that indicates that the impact of breeding system may vary between low and high copy number element families.
5. Conclusions
Our survey of one previously identified and five recently discovered families of DNA transposons in natural populations of both cyclically parthenogenetic and obligately asexual isolates of D. pulex revealed that sexually reproducing D. pulex generally exhibit higher TE loads and increased polymorphism among individuals. These results corroborate previous research demonstrating higher numbers of Pokey insertions in isolates where sex occurs (Valizadeh & Crease 2008) and suggest that this pattern is more general within class II elements because it was observed in three additional superfamilies, including two non-autonomous elements from the recently discovered Helitron subclass of the DNA transposons (see Feschotte & Pritham 2007 for review of DNA transposon diversity). The same pattern is also observed, although less robustly, in two families of retroelements surveyed in D. pulex (S. Schaack 2010, unpublished data). Differences in patterns of polymorphism are similar to the previously reported trends from other model systems in which outcrossers and selfers were used as proxies for high and low rates of recombination, respectively (Caenorhabditis spp. (Dolgin et al. 2008), Arabidopsis spp. (Wright et al. 2001)), although the source of the increased polymorphism among sexual isolates is not an excess of singletons in sexuals as in these previous reports. Instead, singletons are more common in obligate asexuals, as expected based on the limitations of this reproductive strategy for transposable element proliferation. Despite this excess, overall TE loads were lower in asexuals which counters the predicted build-up of mutations that accompanies loss of sex. Lack of recombination may, instead, represent an advantage for the host in asexual lineages because TE copies cannot multiply via sex, rarely become homozygous, and are less likely to recombine, thereby avoiding coevolved host suppression machinery.
Acknowledgements
The authors would like to thank E. Williams, A. Omilian, A. Seyfert for assistance with field collection and processing samples. L. Weider provided D. pulex samples, and T. Crease provided D. pulex samples and helpful comments on performing transposon display. L. Washington and L. Blackwell provided technical assistance. T. Doak, F. Catania, J. Lucas, X. Gao, C. Lively, J. Bever, M. Hahn, B. Koskella, C. Gilbert and J. Meik improved the manuscript with their comments and discussion. This project was possible because of financial support from the National Science Foundation (award DDIG-0608254 to S.S. and grant EF-0328516 to M.L.) and the National Institutes of Health (fellowship to S.S.).
Endnote
A note on the species: The D. pulex species complex is globally distributed and includes several distinct mitochondrial lineages. This has led to a still unresolved debate on both nomenclature and taxonomy for the species. The populations used in the study belong to the fresh-water dwelling group of D. pulex distributed throughout North America, which includes a nested subclade, sometimes referred to as D. arenata, located primarily in western Oregon, USA.
References
- Arkhipova I., Meselson M.2000Transposable elements in sexual and ancient asexual taxa. Proc. Natl Acad. Sci. USA 97, 14 473–14 477 (doi:10.1073/pnas.97.26.14473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome C., Maside X.2004The lack of recombination drives the fixation of transposable elements on the fourth chromosome of Drosophila melanogaster. Genet. Res. 83, 91–100 (doi:10.1017/S0016672304006755) [DOI] [PubMed] [Google Scholar]
- Bartolome C., Maside X., Charlesworth B.2002On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19, 926–937 [DOI] [PubMed] [Google Scholar]
- Biemont C., Tsitrone A., Vieira C., Hoogland C.1997Transposable element distribution in Drosophila. Genetics 147, 1997–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookfield J. F. Y.1991Models of repression of transposition in P–M hybrid dysgenesis by P-cytotype and by zygotically encoded repressor proteins. Genetics 128, 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butlin R.2002The costs and benefits of sex: new insights from old asexual lineages. Nat. Rev. Genet. 3, 311–317 (doi:10.1038/nrg749) [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Lapid A., Canada D.1992The distribution of transposable elements within and between chromosomes in a population of Drosophila-melanogaster. 2. Inference on the nature of selection against elements. Genet. Res. 60, 115–130 (doi:10.1017/S0016672300030809) [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Langley C. H., Sniegowski P. D.1997Transposable element distributions in Drosophila. Genetics 147, 1993–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. S., Charlesworth B.2006The fate of transposable elements in asexual populations. Genetics 174, 817–827 (doi:10.1534/genetics.106.060434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolgin E. S., Charlesworth B., Cutter A. D.2008Population frequencies of transposable elements in selfing and outcrossing Caenorhabditis nematodes. Genet. Res. 90, 317–329 (doi:10.1017/S0016672308009440) [DOI] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L.1987A rapid DNA isolation procedure from small quantities of leaf tissues. Phytochem. Bull. 19, 11–15 [Google Scholar]
- Duret L., Marais G., Biemont C.2000Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 156, 1661–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S.2005Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J.1974Evolutionary advantage of recombination. Genetics 78, 737–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C., Pritham E. J.2007DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 (doi:10.1146/annurev.genet.40.110405.090448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan D. J.1992Transposable elements. Curr. Opin. Genet. Dev. 2, 861–867 (doi:10.1016/S0959-437X(05)80108-X) [DOI] [PubMed] [Google Scholar]
- Hebert P. D. N., Ward R. D., Weider L. J.1988Clonal-diversity patterns and breeding-system variation in Daphnia pulex, an asexual–sexual complex. Evolution 42, 147–159 (doi:10.2307/2409123) [DOI] [PubMed] [Google Scholar]
- Hickey D. A.1982Selfish DNA: a sexually-transmitted nuclear parasite. Genetics 101, 519–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogland C., Biemont C.1996Chromosomal distribution of transposable elements in Drosophila melanogaster: test of the ectopic recombination model for maintenance of insertion site number. Genetics 144, 197–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes D. J., Schwartz S. S., Hebert P. D. N.1986Genotypic diversity and variation in mode of reproduction among populations in the Daphnia pulex group. Heredity 57, 345–355 (doi:10.1038/hdy.1986.134) [Google Scholar]
- Lynch M., Seyfert A., Eads B., Williams E.2008Localization of the genetic determinants of meiosis suppression in Daphnia pulex. Genetics 180, 317–327 (doi:10.1534/genetics.107.084657) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery E., Charlesworth B., Langley C. H.1987A test for the role of natural-selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet. Res. 49, 31–41 (doi:10.1017/S0016672300026707) [DOI] [PubMed] [Google Scholar]
- Muller H. J.1964The relation of recombination to mutational advance. Mut. Res. 1, 2–9 (doi:10.1016/0027-5107(64)90047-8) [DOI] [PubMed] [Google Scholar]
- Nuzhdin S. V., Petrov D. A.2003Transposable elements in clonal lineages: lethal hangover from sex. Biol. J. Linn. Soc. 79, 33–41 (doi:10.1046/j.1095-8312.2003.00188.x) [Google Scholar]
- Paland S., Lynch M.2006Transitions to asexuality result in excess amino acid substitutions. Science 311, 990–992 (doi:10.1126/science.1118152) [DOI] [PubMed] [Google Scholar]
- Paland S., Colbourne J. K., Lynch M.2005Evolutionary history of contagious asexuality in Daphnia pulex. Evolution 59, 800–813 [PubMed] [Google Scholar]
- Penton E., Sullender B., Crease T.2002Pokey, a new DNA transposon in Daphnia (Cladocera: Crustacea). J. Mol. Evol. 55, 664–673 (doi:10.1007/s00239-002-2362-9) [DOI] [PubMed] [Google Scholar]
- Rizzon C., Marais G., Gouy M., Biemont C.2002Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaack S., Choi E., Lynch M., Pritham E. J.In press DNA transposons and the role of recombination in mutation accumulation in Daphnia pulex. Genome Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A., Schneider K. L., Presting G. G.2008Sustained retrotransposition is mediated by nucleotide deletions and interelement recombinations. Proc. Natl Acad. Sci. USA 105, 15 470–15 474 (doi:10.1073/pnas.0805694105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. Z., Boissinot S.2007Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene 390, 206–213 (doi:10.1016/j.gene.2006.09.033) [DOI] [PubMed] [Google Scholar]
- Stross R. G., Hill J. C.1965Diapause induction in Daphnia requires two stimuli. Science 150, 1462–1464 (doi:10.1126/science.150.3702.1462) [DOI] [PubMed] [Google Scholar]
- Sullender B. W., Crease T. J.2001The behavior of a Daphnia pulex transposable element in cyclically and obligately parthenogenetic populations. J. Mol. Evol. 53, 63–69 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S.2007MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- Valizadeh P., Crease T. J.2008The association between breeding system and transposable element dynamics in Daphnia pulex. J. Mol. Evol. 66, 643–654 (doi:10.1007/s00239-008-9118-0) [DOI] [PubMed] [Google Scholar]
- Van den Broeck D., Maes T., Sauer M., Zethof J., De Keukeleire P., D'Hauw M., Van Montagu M., Gerats T.1998Transposon display identifies individual transposable elements in high copy number lines. Plant J. 13, 121–129 (doi:10.1046/j.1365-313X.1998.00004.x) [DOI] [PubMed] [Google Scholar]
- Wright S., Finnegan D.2001Genome evolution: sex and the transposable element. Curr. Biol. 11, R296–R299 (doi:10.1016/S0960-9822(01)00168-3) [DOI] [PubMed] [Google Scholar]
- Wright S. I., Le Q. H., Schoen D. J., Bureau T. E.2001Population dynamics of an Ac-like transposable element in self- and cross-pollinating Arabidopsis. Genetics 158, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]