Abstract
During the Late Pleistocene, the woolly mammoth (Mammuthus primigenius) experienced a series of local extinctions generally attributed to human predation or environmental change. Some small and isolated populations did however survive far into the Holocene. Here, we investigated the genetic consequences of the isolation of the last remaining mammoth population on Wrangel Island. We analysed 741 bp of the mitochondrial DNA and found a loss of genetic variation in relation to the isolation event, probably caused by a demographic bottleneck or a founder event. However, in spite of ca 5000 years of isolation, we did not detect any further loss of genetic variation. Together with the relatively high number of mitochondrial haplotypes on Wrangel Island near the final disappearance, this suggests a sudden extinction of a rather stable population.
Keywords: Wrangel Island, ancient DNA, mitochondrial DNA, genetic variation, Mammuthus primigenius
1. Introduction
The extinction of the once widespread woolly mammoth has fascinated naturalists for almost 300 years (Breyne et al. 1737). It was long thought that mammoths had become extinct by the Pleistocene–Holocene transition around 12 000–10 000 14C years before present (yr BP). However, recent radiocarbon data indicate a prolonged survival in some areas, including a few islands where mammoths persisted far into the Holocene (Vartanyan et al. 1993, 2008; Guthrie 2004; Yesner et al. 2005). The youngest radiocarbon date (3685 ± 60 yr BP) comes from a mammoth fossil discovered on Wrangel Island off the north-eastern Siberian coast (Vartanyan et al. 2008).
Wrangel Island was part of western Beringia during the Late Pleistocene, but became separated from the mainland ca 9000 yr BP owing to rising sea levels (Fairbanks 1989; Vartanyan et al. 2008). As a consequence, the mammoths on Wrangel Island became isolated and the population survived more than 5000 years on the island before it finally died out (Vartanyan et al. 1993). In a recent study, Vartanyan et al. (2008) presented 124 radiocarbon dates of randomly collected mammoth samples from Wrangel Island. Since 85 per cent of the dated samples fell within a time interval of 8980–3685 yr BP, Vartanyan et al. (2008) suggested a higher mammoth density in the area during the Holocene than during the Pleistocene, when Wrangel Island was still connected to the mainland. A gap in radiocarbon dates between 12 000 and 9000 yr BP was further suggested to indicate a period when mammoths were rare in, or absent from, the area (Vartanyan et al. 2008).
It is still unclear what caused the final extinction of mammoths on Wrangel Island. The two main hypotheses for the extinction of woolly mammoths on the mainland, environmental change and/or human impact (Barnosky et al. 2004), have been difficult to attribute to the extinction on Wrangel Island. The mammoths on Wrangel Island survived the climate change associated with the Pleistocene–Holocene transition, and eventually went extinct during a period of relatively stable climate (Lozhkin et al. 2001; Vartanyan 2007). Further, to date, there is no direct evidence for human arrival on Wrangel Island prior to the extinction of the woolly mammoth (Dikov 1988).
The carrying capacities of large mammals on islands are often limited (Alcover et al. 1998), which means that the risk of extinction through demographic stochasticity increases (Lande 1993). Further, small isolated populations are likely to be negatively affected by genetic drift and inbreeding (Frankham 2003, 2005). An alternative explanation for the extinction on Wrangel Island could thus be that the island was simply too small to support a long-term viable mammoth population.
In this study, we wanted (i) to examine whether mammoths were absent from Wrangel Island between 12 000 and 9000 yr BP, which would influence the genetic composition and also have a bearing on our interpretation of the viability of the last population. Further, we wanted (ii) to investigate whether changes in genetic variation through time could help explain why the mammoth population on Wrangel Island became extinct. To address these questions, we analysed mitochondrial DNA from radiocarbon-dated mammoth fossils, ranging from before the isolation of Wrangel Island up until the final extinction (greater than 38 000–3685 ± 60 yr BP, table 1). If the genetic composition of the population inhabiting Wrangel prior to the gap in the fossil record (12 000–9000 yr BP) is found to be highly different from that of the post-gap population, this would support the hypothesis that mammoths were absent on Wrangel Island between 12 000 and 9000 yr BP. For the second question regarding the final extinction on Wrangel Island, we see two alternative scenarios. If the extinction was a delayed response to deteriorating environmental conditions associated with the Pleistocene–Holocene transition, or a consequence of a reduced carrying capacity, we would expect to find a gradual loss of genetic variation in the population. On the other hand, a relatively constant level of genetic variation through time, followed by a rapid loss of variation near the extinction event, would indicate a sudden disappearance of a stable population, more likely caused by an abrupt environmental change or the arrival of humans on Wrangel Island.
Table 1.
The specimens used in the study. (Specimens are listed with associated radiocarbon number and dates before present (Vartanyan et al. 2008), type of material used for DNA analysis and mitochondrial DNA haplotype and haplogroup (sensu Debruyne et al. 2008).)
| 14C lab. no. | 14C age BP | material | haplotype | haplogroup |
|---|---|---|---|---|
| Ua-13365 | >38 000 | tooth | W1 | E |
| LU-3511 | 37 080 ± 1650 | bone | W2 | D |
| GIN-8257 | 22 400 ± 200 | bone | W3 | D |
| LU-2807 | 20 000 ± 110 | tooth | W4 | E |
| LU-3510a | 18 030 ± 130 | bone | — | — |
| LU-2792 | 12 980 ± 80 | tooth | W5 | E |
| LU-2823 | 12 010 ± 110 | tooth | W6 | D |
| AA60048 | 8980 ± 90 | bone | W1 | E |
| AA60049 | 8870 ± 100 | bone | W1 | E |
| Ua-13377 | 8030 ± 75 | tooth | W1 | E |
| Ua-13370 | 7910 ± 80 | tooth | W1 | E |
| LU-4473 | 7850 ± 80 | tusk | W1 | E |
| GIN-6995 | 7710 ± 40 | tusk | W1 | E |
| GIN-6996a | 7620 ± 30 | tusk | — | — |
| Ua-13372 | 7510 ± 80 | tooth | W1 | E |
| LU-2809 | 7250 ± 60 | tooth | W7 | E |
| LU-2746 | 7040 ± 60 | tusk | W1 | E |
| LU-3515 | 6830 ± 40 | bone | W1 | E |
| GIN-6997 | 6690 ± 60 | tusk | W1 | E |
| Ua-13361 | 6560 ± 75 | tooth | W1 | E |
| LU-4449 | 6560 ± 60 | tusk | W1 | E |
| Ua-13360 | 6530 ± 70 | tooth | W1 | E |
| AA40666 | 6499 ± 66 | bone | W1 | E |
| LU-2799 | 6260 ± 50 | tooth | W1 | E |
| LU-4474 | 5910 ± 50 | tusk | W1 | E |
| GIN-6988 | 5610 ± 40 | tusk | W1 | E |
| LU-2742 | 5310 ± 90 | tusk | W8 | E |
| Ua-13368 | 4730 ± 65 | tooth | W1 | E |
| Ua-13373 | 4675 ± 75 | tooth | W1 | E |
| Ua-13378 | 4475 ± 60 | tooth | W9 | E |
| LU-2756 | 4400 ± 40 | tusk | W1 | E |
| AA40667 | 4389 ± 46 | bone | W10 | E |
| GIN-6989 | 4370 ± 40 | tusk | W1 | E |
| LU-3513 | 4350 ± 60 | bone | W1 | E |
| Ua-13362 | 4260 ± 75 | tooth | W10 | E |
| Ua-13375 | 4210 ± 70 | tooth | W1 | E |
| LU-4448 | 4120 ± 110 | tusk | W1 | E |
| Ua-13369 | 4085 ± 65 | tooth | W1 | E |
| GIN-6985 | 3920 ± 40 | tusk | W1 | E |
| AA40665 | 3905 ± 47 | tooth | W1 | E |
| LU-2741 | 3730 ± 40 | tusk | W1 | E |
| Ua-13366 | 3685 ± 60 | tooth | W1 | E |
aNo amplification product or not amplified for all 741 bp.
2. Material and methods
(a). DNA analysis
Forty-two radiocarbon-dated specimens were included in this study (details are provided in table 1). DNA was extracted from about 0.1 g bone, tooth or tusk powder according to protocol C in Yang et al. (1998). A region comprising 741 bp of the mitochondrial DNA, including the 3′ end of the cytochrome b gene, the tRNA threonine and proline genes, and the first part of the control region, was amplified using six overlapping primer pairs (set A inner primer 22F, mammoth_15530_R, set A inner primer 22R, set B inner primer 22F, mammoth_15780_R and mammoth_15038_F from Krause et al. (2005); mammoth_15183_R, mammoth_15175_F, mammoth_15311_R and mammoth_15270_F from Barnes et al. (2007); forward primer 5′-CATAGACCATACTATGTATAATC-3′ and reverse primer 5′-CATTATGTATGGGGACGAGCAT-3′ from this study). Polymerase chain reactions (PCRs) were performed in 25 µl volumes with 5 µl of DNA extract, 0.2 µM of each primer, 0.4 mM dNTPs, 0.25 U UNG (Sigma-Aldrich), 2.5 mM MgCl2 (Qiagen), 1× PCR buffer (Qiagen) and 1.5 U HotStar Taq (Qiagen). PCR thermal cycling conditions were initiated with a 10 min pre-incubation step at 37°C to activate UNG, followed by 15 min denaturation at 95°C, 40 cycles of 30 s denaturation at 94°C, 40 s annealing at 54°C, 1 min extension at 72°C and a single 10 min extension step at 72°C. PCR products were cleaned using ExoSAP-IT (USB Corporation) and both strands were sequenced using either a CEQ 8000 (Beckman Coulter) or an ABI3130 (Applied Biosystems) automated sequencer according to the manufacturers' instructions.
(b). Precaution and authentication
All samples in this study had previously been radiocarbon dated (Vartanyan et al. 2008) and were thus likely to contain preserved DNA. Since ancient DNA (aDNA) is usually degraded and exists in few copy numbers (Hofreiter et al. 2001), general precautions and standards of authentication for aDNA work were performed. Extractions and PCR reactions were set up in a laboratory dedicated for analysis of aDNA at the Archaeological Research Laboratory, Stockholm University (Sweden). PCRs and post-PCR work were performed at the Department of Zoology and the Department of Genetics, Microbiology and Toxicology, Stockholm University (Sweden). Working areas, equipment and reagents were decontaminated using bleach, hydrochloric acid and/or ultraviolet irradiation. Negative controls were used in all extractions and PCRs. Pleistocene horse samples were also used as negative controls. DNA was extracted twice for several of the specimens, and all extracts were amplified at least two times for each fragment to monitor the effect of misincorporated bases during the PCR. Six of the samples were also independently replicated at Centro UCM-ISCII de Evolución y Comportamiento Humanos, Madrid (Spain). All controls were negative during the course of the study and independently derived replicates as well as overlapping sequences consistently matched, lending support to the authenticity of the results.
(c). Data analysis
The overlapping sequences were aligned and assigned to haplotypes using BioEdit v. 7.0.5.2 (Hall 1999). After supplementing our dataset with 41 sequences from GenBank (see electronic supplementary material, table S1), we constructed a haplotype network based on statistical parsimony using the program TCS v. 1.21 (Clement et al. 2000). The added sequences shared 705–741 bp with our sequence and represented all haplotypes comprising haplogroups D and E (Debruyne et al. 2008) within mammoth mitochondrial DNA clade 1 (Barnes et al. 2007; Gilbert et al. 2008).
Genetic variation in the population on Wrangel Island before 12 000 yr BP and after 9000 yr BP was measured as the number of haplotypes (nh), haplotype diversity (Hd) and nucleotide diversity (nd). Haplotype and nucleotide diversities, with standard errors, were estimated using Arlequin v. 3.01 (Excoffier et al. 2005), and differences between samples were evaluated using t-tests. To test whether there was a difference in genetic composition on Wrangel Island before and after 9000 yr BP, we performed an exact contingency test with 1000 dememorization steps and 100 000 iterations on a contingency table of haplotype frequencies, using the program STRUC (Raymond & Rousset 1995) in Genepop v. 3.1c.
The evolutionary rate for this region of the mitochondrial genome has previously been estimated as 2.47 × 10−7 mutations site−1 yr−1 (Barnes et al. 2007). For the whole 741 bp sequence analysed in this study, this is equivalent to a haplotype mutation rate (i.e. the probability that a new haplotype is formed) of 0.0002 per year. Assuming a generation time of 15 years, as suggested for Asian elephants (Sukumar 1989), this corresponds to a haplotype mutation rate of 0.003 per generation. To examine the probability of obtaining the observed number of haplotypes after 9000 yr BP through mutation from a single haplotype, we used a slightly modified version of the program Easypop (Balloux 2001). The modification allowed us to sample one individual from the population at specified generations corresponding to the radiocarbon dates for the post-isolation samples used in this study (i.e. one individual from each of generations 0, 7, 63, 71, … etc. assuming a generation time of 15 years), and to store data over repeated runs. A time span of 353 generations was used in the simulations, which represents the isolation period on Wrangel Island. We ran the simulation for seven different haploid population sizes (harmonic means: 10, 50, 100, 500, 1000, 5000 and 10 000) and four haplotype mutation rates per generation (0.01, 0.003, 0.001 and 0.0001). We used the simulation settings: single locus, haploid data, no migration, the K-allele model (KAM) of mutation and an initial variation of zero. After each run, the number of allelic variants (i.e. haplotypes) was calculated in the sample. All simulation combinations were replicated 999 times, and the probability of obtaining the observed number of haplotypes in our real dataset (five or more; see below) was calculated for each mutation rate/population size combination. To investigate the effect of different generation times, we also repeated the simulations for generation times of 10, 20 and 30 years (electronic supplementary material, table S2).
We estimated the carrying capacity of mammoths on an island the size of Wrangel Island (ca 8000 km2), using Damuth's (1981) equation on the relationship between body size and population density for herbivores in an arctic environment. Since it has been suggested that the mammoths on Wrangel Island were of smaller size (Vartanyan et al. 1993), we assumed an average body weight of between 1000 and 6000 kg in the equation.
3. Results
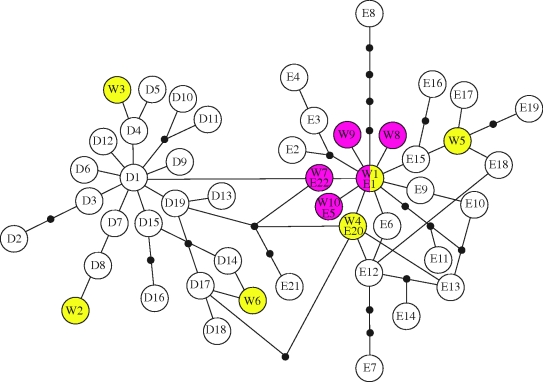
All 741 bp were successfully amplified from 40 of the 42 individuals (table 1). We identified 10 haplotypes on Wrangel Island of which all belonged to clade I, previously described by Barnes et al. (2007) (the haplotypes have been deposited in GenBank with accession numbers GU984769-GU984778). Six of the haplotypes were present before 12 000 yr BP and five after 9000 yr BP. Whereas the haplotypes from before 12 000 yr BP were spread out in the haplotype network (figure 1), the haplotypes from after 9000 yr BP formed a star-like shape around the dominant haplotype (W1/E1) in the sample. When compared with previously published sequences (Krause et al. 2005; Rogaev et al. 2006; Barnes et al. 2007; Debruyne et al. 2008; Gilbert et al. 2008; electronic supplementary material, table S1), the haplotypes from before 12 000 yr BP were found within both haplogroups D and E (sensu Debruyne et al. 2008), whereas all haplotypes from after 9000 yr BP clustered within haplogroup E (table 1 and figure 1).
Figure 1.
A network showing the relationship between mitochondrial DNA haplotypes on Wrangel Island and previously published haplotypes within haplogroups D and E (sensu Debruyne et al. 2008). The network is constructed from sequences of 705 bp. Each branch represents a mutational step and missing haplotypes are marked with black dots. The yellow dots are haplotypes present on Wrangel Island before 12 000 yr BP, and the magenta dots are haplotypes present after 9000 yr BP.
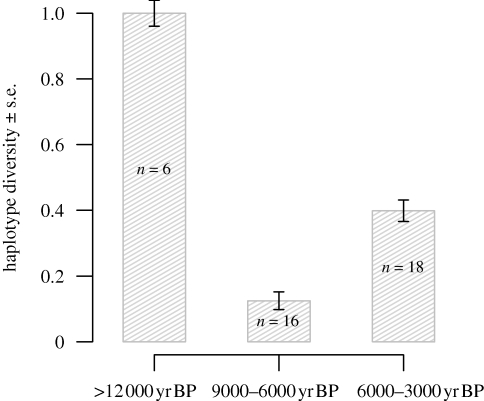
When comparing the genetic variation before and after the gap in the fossil record (12 000–9000 yr BP), we found higher haplotype and nucleotide diversities in the samples with radiocarbon dates older than 12 000 yr BP (t38 = 16.63, p < 0.0001 and t38 = 8.31, p < 0.0001, respectively, table 2). We also found a significant difference in haplotype frequencies (p < 0.0005) between samples older than 12 000 yr BP and those younger than 9000 yr BP. When dividing the samples with radiocarbon dates younger than 9000 yr BP into two groups (9000–6000 and 6000–3000 yr BP), haplotype and nucleotide diversities were higher in the samples within the time interval 6000–3000 yr BP than in samples within the time interval 9000–6000 yr BP (t32 = 6.42, p < 0.0001 and t32 = 2.43, p < 0.05, respectively; table 2 and figure 2). This pattern, with an initial high haplotype diversity, followed by a reduction and subsequent increase in diversity, was also observed when the samples were divided into shorter time periods (electronic supplementary material, figure S1).
Table 2.
Genetic variation in the mammoth population on Wrangel Island before 12 000 yr BP and after 9000 yr BP. (Sample size (n), number of haplotypes (nh), haplotype and nucleotide diversities with standard errors (Hd ± s.e., nd ± s.e.) and the number of transitions and transversions.)
| population | n | nh | Hd ± s.e. | nd ± s.e. | transitions/transversions |
|---|---|---|---|---|---|
| before 12 000 yr BP | 6 | 6 | 1 ± 0.0393 | 0.0058 ± 0.0016 | 10/0 |
| after 9000 yr BP | 34 | 5 | 0.2745 ± 0.0170 | 0.00039 ± 0.00008 | 4/0 |
| 9000–6000 yr BP | 16 | 2 | 0.1250 ± 0.0266 | 0.00017 ± 0.00008 | 1/0 |
| 6000–3000 yr BP | 18 | 4 | 0.3987 ± 0.0325 | 0.00058 ± 0.00015 | 3/0 |
Figure 2.
Haplotype diversity with standard error (s.e.) and sample size (n) on Wrangel Island before 12 000 yr BP, between 9000–6000 yr BP and 6000–3000 yr BP.
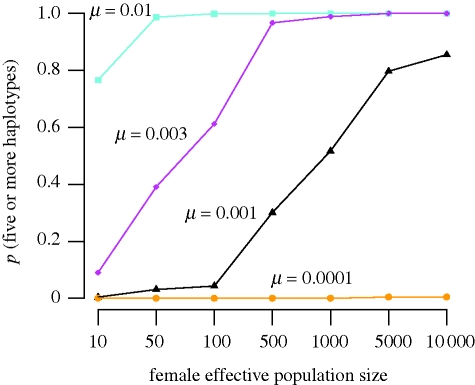
The computer simulations indicate that the observation of five haplotypes in our post-isolation sample (34 mammoths) is likely (p > 0.05) for a wide range of seemingly realistic mutation rate and population size combinations, also if the population was monomorphic immediately after isolation and all the subsequent variation was generated by mutation. As seen in figure 3, the probability of obtaining the observed result (five or more haplotypes) is less than 0.05 only at mutation rates substantially lower than that estimated in Barnes et al. (2007). At a mutation rate of one-third of that estimated in Barnes et al. (2007), this probability is less than 0.05 only at small (less than 10–100) female effective population sizes. Different generation times had very little effect on the outcome of the simulations, except for a low generation time (10 years) and then only for a very small population size (Nef = 10; electronic supplementary material, table S2). From Damuth's equation, the carrying capacity of mammoths on Wrangel Island was estimated to 149–819 individuals. Assuming a 1 : 1 sex ratio and a 0.5 ratio of effective to actual number of females, this would correspond crudely to an effective female population size of 40–200.
Figure 3.
Simulated probability of obtaining five or more haplotypes during the isolation period on Wrangel Island assuming that the population was monomorphic in generation t = 0, and that all the variation observed was generated by mutation. Simulations were run for four different mutation rates per generation and seven female effective population sizes during a time span of 353 generations.
4. Discussion
(a). Genetic composition before and after the isolation of Wrangel Island
During the Late Pleistocene, sea levels were considerably lower than today and Wrangel Island constituted an elevated area of the mainland (Fairbanks 1989; Saarnisto & Karhu 2004). At this time, mammoths were still abundant in northern Eurasia (Kuzmin & Orlova 2004). Based on the low frequency of mammoth specimens on Wrangel Island that dates to the Late Pleistocene, Vartanyan et al. (2008) suggested that mammoths only visited the Wrangel area occasionally during this period. This was further supported by Arppe et al. (2009) who interpreted a difference in strontium isotope values in Late Pleistocene and Mid-Holocene mammoth bones as a shift in mammoth range in the area. The high genetic variation that we found in the samples from Wrangel Island before 12 000 yr BP in our study is in accordance with a genetic signature of a large population inhabiting the continuous tundra–steppe region on the mainland. Although the sample size is small, we also find no indication of a loss of haplotype diversity in connection with the last glacial maximum (electronic supplementary material, figure S1).
As melting ice sheets caused the sea levels to rise at the end of Pleistocene, Wrangel Island was gradually separated from the mainland. Around 9000 yr BP, the connection to the mainland had been submerged and the mammoths on Wrangel became isolated (Fairbanks 1989; Vartanyan et al. 2008). When we compared the mitochondrial DNA sequences from before and after the gap in the fossil record (12 000–9000 yr BP), we found a different genetic composition and a significantly lower genetic variation in the samples from after 9000 yr BP (table 2). This could be explained by a population extinction around 12 000 yr BP followed by a recolonization of Wrangel some 3000 years later. However, the dominant haplotype (W1/E1) on Wrangel Island after 9000 yr BP is also present before 12 000 yr BP (figure 1). An alternative explanation to the differences in genetic variation and haplotype frequencies could therefore be a demographic bottleneck that temporarily reduced the amount of genetic variation to a few or even a single haplotype (W1/E1). In conclusion, it seems clear that the observed gap in the fossil record (Vartanyan et al. 2008) represents a demographic event in the Wrangel Island mammoth population, rather than a being a sampling artefact, although we are at present unable to resolve whether this was owing to an extinction/recolonization or a bottleneck.
The haplotypes observed after 9000 yr BP are all closely related, and distributed in a star-like pattern in the haplotype network (figure 1). This type of pattern is often indicative of an expansion in population size that follows a bottleneck or a founder event (Slatkin & Hudson 1991). This is consistent with the results from our simulations, which indicate that the genetic variation observed in the population after the isolation of Wrangel Island could have arisen through mutation from a single haplotype.
(b). The extinction of mammoths on Wrangel Island
Small populations risk extinction through demographic and genetic stochasticity (Lande 1993; Frankham 2003, 2005), and such conditions could in the case of Wrangel Island be further aggravated by a reduction in carrying capacity related to the decreasing area connected to rising sea levels. However, we would in this case expect to find a gradual loss of genetic variation owing to genetic drift. By contrast, our results suggest that genetic variation was maintained at a relatively stable level, and even increased slightly (figure 2; electronic supplementary material, figure S1), up until the final extinction. The genetic data thus point towards a relatively large Holocene population on Wrangel Island, which is also congruent with the high density of mammoth fossils dating to this period (Vartanyan et al. 2008). This suggests that the final extinction was caused by a relatively sudden, rather than gradual, change in the mammoths' environment. One possible explanation for such a sudden change could be the arrival of humans on Wrangel Island. According to Jones et al. (2008), human extermination of a large mammal, such as the mammoth, should be readily visible in the archaeological record. In the case of Wrangel Island, such direct evidence is lacking. The only pre-historic human settlement has no record of mammoth hunting and is dated to 3360 ± 155 yr BP, and is thus at least 100 years younger than the last mammoth date (Dikov 1988). However, a gap of only a few hundred years is very small, and it is likely that the final extinction of mammoths happened later than the most recently dated mammoth specimen. Furthermore, the first arrival of humans probably pre-dates the age of the oldest human settlement on the island. It therefore does not seem unlikely that humans came into contact with the mammoths on Wrangel Island.
The alternative to human predation as a cause for the extinction could include the emergence of a novel disease (MacPhee & Marx 1997) or a previously undetected short-term change in the climate. Thus, in this study, we have found support for an extinction scenario involving a rapid decrease in population size, however, the detailed mechanism for why the last remaining mammoth population suddenly vanished remains unresolved.
Acknowledgements
This work was supported by grants from Oscar and Lili Lamm's foundation and Helge Axelsson Johnson's foundation. L.D. acknowledges funding from the EU Marie Curie scheme (FP6 041545). N.R. thanks Francois Balloux for making the source code for Easypop available. Thanks also to three anonymous reviewers for comments on an earlier draft of the manuscript.
References
- Alcover J. A., Sans A., Palmer M.1998The extent of extinctions of mammals on islands. J. Biogeogr. 25, 913–918 (doi:10.1046/j.1365-2699.1998.00246.x) [Google Scholar]
- Arppe L., Karhu J. A., Vartanyan S. L.2009Bioapatite 87Sr/86Sr of the last woolly mammoths: implications for the isolation of Wrangel Island. Geology 37, 347–350 (doi:10.1130/G25467A.1) [Google Scholar]
- Balloux F.2001Easypop (version 1.7): a computer program for population genetics simulations. J. Hered. 92, 301–302 (doi:10.1093/jhered/92.3.301) [DOI] [PubMed] [Google Scholar]
- Barnes I., Shapiro B., Lister A., Kuznetsova T., Sher A., Guthrie D., Thomas M. G.2007Genetic structure and extinction of the woolly mammoth Mammuthus primigenius. Curr. Biol. 17, 1–4 (doi:10.1016/j.cub.2007.05.035) [DOI] [PubMed] [Google Scholar]
- Barnosky A. D., Koch P. L., Feranec R. S., Wing S. L., Shabel A. B.2004Assessing the causes of Late Pleistocene extinctions on the continents. Science 306, 70–75 (doi:10.1126/science.1101476) [DOI] [PubMed] [Google Scholar]
- Breyne J. P., S. T., Wolochowicz M.1737A letter from John Phil. Breyne, M. D. F. R. S. to Sir Hans Sloane, Bart. Pres. R. S. with observations, and a description of some mammoth's bones dug up in Siberia, proving them to have belonged to elephants. Phil. Trans. (1683–1775) 40, 124–138 (doi:10.1098/rstl.1737.0026) [Google Scholar]
- Clement M., Posada D., Crandall K.2000TCS: a computer program to estimate gene genealogies. Mol. Ecol. 9, 1657–1660 (doi:10.1046/j.1365-294x.2000.01020.x) [DOI] [PubMed] [Google Scholar]
- Damuth J.1981Population density and body size in mammals. Nature 290, 699–700 (doi:10.1038/290699a0) [Google Scholar]
- Debruyne R., et al. 2008Out of America: ancient DNA evidence for a new world origin of Late Quaternary woolly mammoths. Curr. Biol. 18, 1–6 (doi:10.1016/j.cub.2008.07.061) [DOI] [PubMed] [Google Scholar]
- Dikov N. N.1988The earliest sea mammal hunters of Wrangel Island. Arctic Anthropol. 25, 80–93 [Google Scholar]
- Excoffier L., Laval G., Schneider S.2005Arlequin v. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 1, 47–50 [PMC free article] [PubMed] [Google Scholar]
- Fairbanks R. G.1989A 17 000-year glacio-eustatic sea level record: influence of glacial melting rates on the Younger Dryas event and deep-ocean circulation. Nature 342, 637–642 (doi:10.1038/342637a0) [Google Scholar]
- Frankham R.2003Genetics and conservation biology. Compt. Rend. Biol. 326, 22–29 (doi:10.1016/S1631-0691(03)00023-4) [DOI] [PubMed] [Google Scholar]
- Frankham R.2005Genetics and extinction. Biol. Conserv. 126, 131–140 (doi:10.1016/j.biocon.2005.05.002) [Google Scholar]
- Gilbert M. T. P., et al. 2008Intraspecific phylogenetic analysis of Siberian woolly mammoths using complete mitochondrial genomes. Proc. Natl Acad. Sci. USA 105, 8327–8332 (doi:10.1073/pnas.0802315105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie R. D.2004Radiocarbon evidence of mid-Holocene mammoths stranded on an Alaskan Bering Sea island. Nature 429, 746–749 (doi:10.1038/nature02612) [DOI] [PubMed] [Google Scholar]
- Hall T. A.1999BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 41, 95–98 [Google Scholar]
- Hofreiter M., Serre D., Poinar H. N., Kuch M., Pääbo S.2001Ancient DNA. Nat. Rev. Genet. 2, 353–359 (doi:10.1038/35072071) [DOI] [PubMed] [Google Scholar]
- Jones T. L., Porcasi J. F., Erlandson J. M., Dallas H., Jr, Wake T. A., Schwaderer R.2008The protracted Holocene extinction of California's flightless sea duck (Chendytes lawi) and its implications for the Pleistocene overkill hypothesis. Proc. Natl Acad. Sci. USA 105, 4105–4108 (doi:10.1073/pnas.0711140105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J., et al. 2005Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature 439, 724–727 (doi:10.1038/nature04432) [DOI] [PubMed] [Google Scholar]
- Kuzmin Y. V., Orlova L. A.2004Radiocarbon chronology and environment of woolly mammoth (Mammuthus primigenius Blum.) in northern Asia: results and perspectives. Earth Sci. Rev. 68, 133–169 (doi:10.1016/j.earscirev.2004.04.002) [Google Scholar]
- Lande R.1993Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am. Nat. 142, 911–927 (doi:10.1086/285580) [DOI] [PubMed] [Google Scholar]
- Lozhkin A. V., Anderson P. M., Vartanyan S. L., Brown T. A., Belaya B. V., Kotov A. N.2001Late Quaternary paleoenvironments and modern pollen data from Wrangel Island (Northern Chukotka). Quat. Sci. Rev. 20, 217–233 (doi:10.1016/S0277-3791(00)00121-9) [Google Scholar]
- MacPhee R. D. E., Marx P. A.1997The 40 000-year plague: humans, hyperdisease and first-contact extinctions. In Natural change and human impact in Madagascar (eds Goodman S., Patterson D.), pp. 169–217 Washington, DC: Smithsonian Institution Press [Google Scholar]
- Raymond M., Rousset F.1995Genepop (version 1.2): population genetics software for exact tests and ecumenicism. J. Hered. 86, 248–249 [Google Scholar]
- Rogaev E. I., Moliaka Y. K., Malyarchuk B. A., Kondrashov F. A., Derenko M. V., Chumakov I., Grigorenko A. P.2006Complete mitochondrial genome and phylogeny of Pleistocene mammoth Mammuthus primigenius. PLoS Biol. 4, e73 (doi:10.1371/journal.pbio.0040073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarnisto M., Karhu J. A.2004The last mammoths: palaeoenvironment of the Holocene mammoth on Wrangel Island. Quat. Perspec. 14, 126–129 [Google Scholar]
- Slatkin M., Hudson R. R.1991Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129, 555–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumar R.1989The Asian elephant: ecology and management, p. 179 Cambridge, UK: Cambridge University Press [Google Scholar]
- Vartanyan S. L.2007Wrangel Island in the end of Quaternary period: geology and paleogeography, p. 144 Saint Petersburg, Russia: Ivan Limbakh Publishing House; (in Russian) [Google Scholar]
- Vartanyan S. L., Garutt V. E., Sher A. V.1993Holocene dwarf mammoths from Wrangel Island in the Siberian Arctic. Nature 362, 336–339 (doi:10.1038/362337a0) [DOI] [PubMed] [Google Scholar]
- Vartanyan S. L., Arslanov K. A., Karhu J. A., Possnert G., Sulerzhitsky L. D.2008Collection of radiocarbon dates on the mammoths (Mammuthus primigenius) and other genera of Wrangel Island, northeast Siberia, Russia. Quat. Res. 70, 51–59 (doi:10.1016/j.yqres.2008.03.005) [Google Scholar]
- Yang D. Y., Eng B., Waye J. S., Dudar J. C., Saunders S. R.1998Technical note: improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 4, 539–543 (doi:10.1002/(SICI)1096-8644(199804)105:4>539::AID-AJPA10>3.0.CO;2-1) [DOI] [PubMed] [Google Scholar]
- Yesner D., Veltre D., Crossen K., Graham R.20055700-year-old mammoth remains from Qagnax Cave, Pribilof Islands, Alaska. In World of Elephants. Short Papers and Abstracts of the 2nd Int. Congress, Mammoth Site Scientific Papers 4, pp. 200–204 Hot Springs, SD: Mammoth Site of Hot Springs Publication [Google Scholar]