Abstract
Dictyostelium discoideum myosin II heavy chain kinase A (MHCK A) disrupts the assembly and cellular activity of bipolar filaments of myosin II by phosphorylating sites within its α-helical, coiled-coil tail. MHCK A is a member of the atypical α-kinase family of serine and threonine protein kinases and displays no sequence homology to typical eukaryotic protein kinases. We report the crystal structure of the α-kinase domain (A-CAT) of MHCK A. When crystallized in the presence of adenosine triphosphate (ATP), A-CAT contained adenosine monophosphate (AMP) at the active site. However, when crystallized in the presence of ATP and a peptide substrate, which does not appear in the structure, adenosine diphosphate (ADP) was found at the active site and an invariant aspartic acid residue (Asp766) at the active site was phosphorylated. The aspartylphosphate group was exposed to the solvent within an active-site pocket that might function as a docking site for substrates. Access to the aspartylphosphate was regulated by a conformational switch in a loop that bound to a magnesium ion (Mg2+), providing a mechanism that allows α-kinases to sense and respond to local changes in Mg2+.
INTRODUCTION
Myosin II is a hexameric protein composed of a pair of heavy chains and two pairs of light chains that plays a central role in cytokinesis, cell migration, and adhesion (1). The heavy chain of myosin II can be divided into three domains: a globular N-terminal motor domain that binds to actin and hydrolyzes adenosine triphosphate (ATP), a neck region that binds to the light chains, and a long C-terminal tail that forms an α-helical, coiled-coil dimer. The rod-like tails self-associate to assemble myosin II molecules into functional bipolar filaments that are required to generate a contractile force. In the social amoeba Dictyostelium discoideum, the formation of myosin II filaments is regulated by a group of closely related kinases, myosin heavy chain kinases (MHCKs) A through D (2–4). MHCK A, the first of these protein kinases to be purified and characterized, phosphorylates three threonine residues (Thr1823, Thr1833, and Thr2029) within the C-terminal portion of the coiled-coil tail of myosin II (5–7). Phosphorylation of these residues is sufficient to convert myosin II from a filamentous to a monomeric state (5, 8). When overexpressed in Dictyostelium cells, MHCK A drives the disassembly of myosin II filaments, which results in defects in cytokinesis and development that are comparable to those found in myosin II null cells (3, 9, 10).
The 130-kD MHCK A protein consists of an N-terminal coiled-coil domain, a central kinase domain, and a C-terminal WD-repeat domain (11). The coiled-coil domain mediates the oligomerization of MHCK A into trimers or tetramers (5, 9, 11) and is responsible for targeting MHCK A to actin-rich cellular protrusions, including the leading edge of migrating cells (12–16). The WD domain binds directly to filamentous myosin II and is required for the efficient phosphorylation of myosin II in vivo (9, 17); however, removal of the coiled-coil and WD domains does not affect the ability of the central kinase domain to phosphorylate peptide substrates (18). The kinase activity of MHCK A is strongly enhanced by autophosphorylation in vitro. MHCK A incorporates up to 10 mol of phosphate per mole of enzyme, although maximal activation is achieved after the incorporation of the first three moles of phosphate (19). Little is known concerning the signaling pathways that regulate MHCK A in the cell, but actin filaments, myosin II, and acidic phospholipids stimulate the rate of autophosphorylation of MHCK A in vitro (20, 21).
The kinase domain of MHCK A does not share any detectable sequence similarity with members of the superfamily of conventional, eukaryotic protein kinases (ePKs) (18). MHCK A represents one of the founding members of an atypical kinase family that has been termed the “α-kinases” on the basis of the proposal that they are adapted to recognize and phosphorylate residues located within α helices of target proteins (22, 23). Currently, however, there is little evidence to support this proposal. MHCKs A, B, and C exhibit an unusually strong preference for the phosphorylation of threonine residues in protein and peptide substrates, but this preference is not shared by all of the α-kinases (24–27).
Representatives of the α-kinase family are found in various protozoa, fungi, and animals but are absent from higher plants, insects, and yeast (24, 28). Only a few α-kinases exist in each organism, which suggests that they fulfill some highly specific function. Dictyostelium contains a total of six α-kinases. In addition to MHCKs A to D, these include alpha kinase 1 (AK1), which contains an Arf guanosine triphosphatase (GTPase)–activating protein (GAP) domain, and VwkA, which contains a von Willebrand factor A–like domain (29, 30). Mammals also have six α-kinases, the best-characterized of which are eukaryotic elongation factor 2 (eEF2) kinase and the ion channels TRPM6 (transient receptor potential melastatin-like 6) and TRPM7 (22). eEF2 kinase is a Ca2+-calmodulin–regulated enzyme that phosphorylates and inhibits the ribosomal translocation factor eEF-2 (31). TRPM6 and TRPM7 are closely related proteins that combine a Ca2+- and Mg2+-permeable channel with a C-terminal α-kinase domain (32). TRPM6 and TRPM7 phosphorylate sites in the tail of nonmuscle myosin II, which destabilizes the assembly of bipolar filaments, thus demonstrating a conserved role for α-kinases in the regulation of myosin II–mediated contractility from Dictyostelium to mammals (26, 33, 34).
The crystal structure of the α-kinase domain of mouse TRPM7 (TRPM7-CAT) reveals that the overall architecture of the α-kinases resembles that of ePKs (35). Moreover, functional counterparts for many of the invariant catalytic residues present within the active sites of ePKs can be identified within the active site of TRPM7-CAT. These results support the view that, despite a lack of primary sequence similarity, the α-kinases share a common, albeit distant, ancestor with the ePKs (35–37). Here, we describe the crystal structure of the α-kinase domain of Dictyostelium MHCK A (A-CAT). The results show that although A-CAT and TRPM7-CAT are divergent members of the α-kinase family, they share a well-conserved, common core kinase domain. Surprisingly, crystallization of A-CAT in the presence of ATP and a peptide substrate resulted in a structure containing a phosphorylated aspartic acid residue, Asp766, at the active site. Phosphorylated Asp766 (pAsp766) was exposed within an active-site pocket that could potentially be a docking site for substrates. Comparison of the structures of A-CAT and TRPM7-CAT showed that access to the active-site pocket was regulated by a C-terminal glycine-rich loop that bound to Mg2+ and switched between two distinct conformations. The structure of A-CAT provides insights into the catalytic and regulatory properties of the α-kinases and raises the possibility that, in contrast to the ePKs, α-kinases transfer the γ-phosphate of ATP to protein substrates through an aspartylphosphate intermediate.
RESULTS
The crystal structure of A-CAT
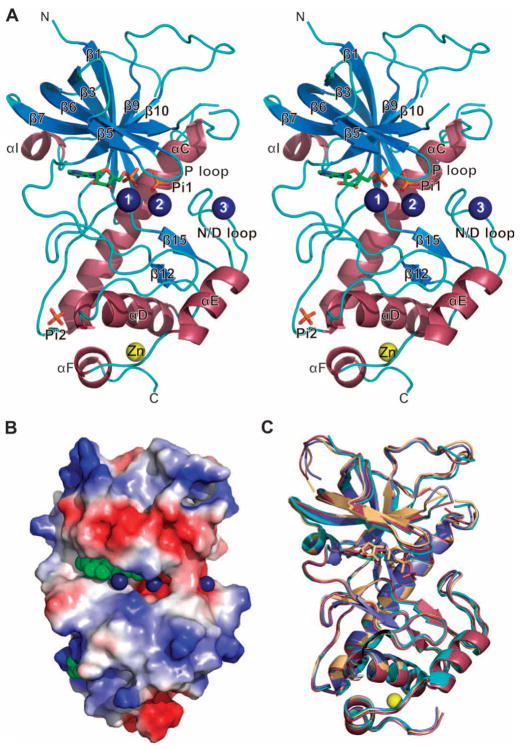
We solved the crystal structures of A-CAT in individual complexes with AMP, ADP, and β,γ-methyleneadenosine 5′-triphosphate (AMPPCP). The cocrystals of A-CAT–AMP and A-CAT–ADP were obtained after the crystallization of A-CAT in the presence of ATP. A crystal structure was also determined for an Asp766→Ala mutant of A-CAT (A-CAT-D766A) in a complex with ATP (Table 1). A-CAT consisted of N-terminal and C-terminal lobes that were connected and supported by a 21–amino acid residue central α helix (αC) that ran diagonally across the molecule from top to bottom (Fig. 1A). The N-terminal lobe consisted mainly of a highly twisted, seven-stranded, antiparallel β sheet (consisting of strands β1, β3, β5, β6, β7, β9, and β10), whereas the C-terminal lobe was built around three α helices (αD to αF) and contained one short stretch of antiparallel β strands (containing strands β12 and β15). The nucleotide was bound into a cleft located at the junction between the N- and C-terminal lobes. A surface representation shows the relatively open nature of the active-site cleft of A-CAT, which extends to the right of the nucleotide to form an active-site pocket (Fig. 1B).
Table 1.
Data collection and refinement statistics for each of the complexes.
| A-CAT–AMP | A-CAT–AMPPCP | A-CAT–ADP | A-CAT-D766A–ATP | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P21212 | P21212 | P21212 | P41 |
| Cell dimensions | ||||
| a, b, c (Å) | 76.2, 83.7, 44.3 | 82.5, 109.4, 79.4 | 82.8, 110.1, 78.7 | 81.2, 81.2, 187.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50–2.3 (2.30–2.38)* | 25–2.1 (2.1–2.18) | 30–2.0 (2.0–2.07) | 30–2.2 (2.2–2.28) |
| Rmerge | 0.06 (0.19) | 0.08 (0.40) | 0.09 (0.48) | 0.058 (0.13) |
| I/σI | 46.5 (9.8) | 37.9 (5.1) | 32 (4.8) | 18.6 (6.7) |
| Completeness (%) | 96.6 (78.7) | 99.9 (99.7) | 99.7 (99.9) | 92.1 (80.8) |
| Redundancy | 12.3 (7.4) | 9.8 (7.9) | 7.2 (6.1) | 8.4 (8.2) |
| Refinement | ||||
| Resolution (Å) | 28.3–1.6 | 23.9–2.1 | 19.7–2.0 | 30–2.2 |
| No. of reflections | 34,910 | 40,221 | 46,634 | 50,350 |
| Rwork/Rfree | 0.19/0.22 | 0.21/0.26 | 0.21/0.26 | 0.20/0.25 |
| No. of atoms | ||||
| Protein | 1964 | 4021 | 3992 | 7990 |
| Nucleotide/Mg2+/Zn2+/H2PO4 | 23/3/1/10 | 62/–/2/10 | 54/–/2/10 | 124/4/4/20 |
| Water | 305 | 369 | 391 | 599 |
| B-factors | ||||
| Protein | 12.97 | 29.80 | 31.83 | 23.36 |
| Nucleotide/Mg2+/Zn2+/H2PO4 | 16.5/14.2/8.9/14.9 | 47.5/–/24.2/33.4 | 45.1/–/26.8/40.9 | 23.5/24.1/17.1/21.2 |
| Water | 26.34 | 37.11 | 42.91 | 28.65 |
| RMSDs | ||||
| Bond lengths (Å) | 0.006 | 0.010 | 0.012 | 0.006 |
| Bond angles (°) | 1.135 | 1.427 | 1.776 | 1.395 |
Values in parentheses are for the highest-resolution shell.
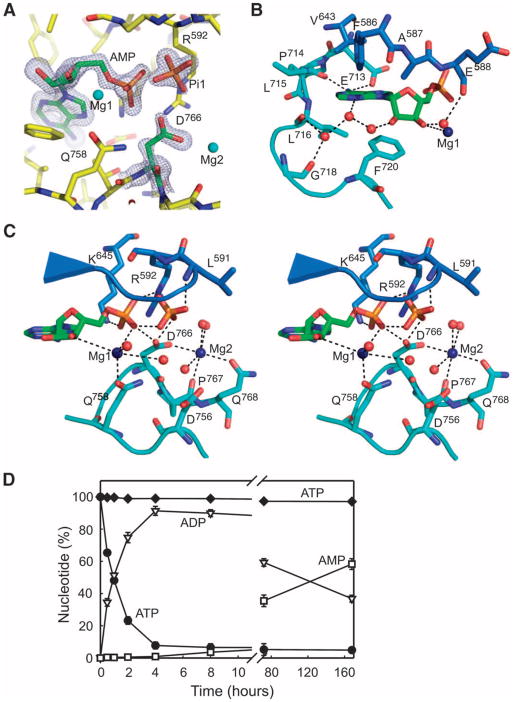
Fig. 1.
Structure of A-CAT. (A) A stereo view of the A-CAT–AMP complex with the β strands (blue) and α helices (strawberry) numbered according to the sequence alignment with TRPM7-CAT (see Fig. 4B). Mg2+ ions are rendered as dark blue numbered spheres and the zinc atom as a yellow sphere. AMP, Pi1, and Pi2 are shown as sticks with the C, O, and P atoms colored green, red, and orange, respectively. N and C termini are indicated. (B) Distribution of electrostatic potential shown on the molecular surface of A-CAT. AMP, Pi1, and Pi2 are rendered as green spheres and the Mg2+ ions as dark blue spheres. (C) Superimposed structures of the A-CAT–AMPPCP (light orange), A-CAT–AMP (cyan), A-CAT–ADP (blue), and A-CAT-D766A–ATP complexes (raspberry). Nucleotides in each complex are shown as sticks of the same color, and the zinc atom is depicted as a yellow sphere. All diagrams of the structure of A-CAT were generated with PyMOL (http://www.pymol.org).
The A-CAT–AMP complex contained an inorganic phosphate molecule (Pi1) in the active site and three Mg2+ ions (Fig. 1A). The first Mg2+ (Mg1) was bound to AMP, the second (Mg2) was located in the active-site pocket, and the third (Mg3) was coordinated by a loop that we have named the N/D loop (see below). The binding sites for both Mg2 and Mg3 exhibited an overall negative charge. In the A-CAT–ADP and A-CAT–AMPPCP complexes, Pi1 was missing and the three Mg2+ ions were replaced by water molecules (for the interaction distances for ions and water molecules, see table S1). The absence of Mg2+ ions from the A-CAT–ADP complex was unexpected, because these crystals were grown in the presence of 0.2 M MgSO4. The presence or absence of Mg2+ and the nature of the bound nucleotide had little effect on the overall structure of A-CAT (Fig. 1C). The root mean square deviation (RMSD), a measure of the average distances between the Cα atoms of the two superimposed structures, was 0.60 Å for the A-CAT–AMP and A-CAT–ADP complex structures, 0.61 Å for the A-CAT–AMP and A-CAT–AMPPCP complex structures, and 0.21 Å for the A-CAT–ADP and A-CAT–AMPPCP complex structures. A metal ion, which was identified as zinc by analysis of A-CAT with inductively coupled plasma optical emission spectroscopy, was coordinated by His742 (from αD), His794 and Cys796 (in the linker between αE and αF), and Cys800 (from αF). Mutation of Cys796 or Cys800 to alanine drastically reduced the amount of A-CAT that could be purified and its kinase activity, indicating that the zinc-binding module was crucial for the stability and the activity of the kinase domain (Fig. 2A). A phosphate molecule (Pi2) was bound between Lys684 in the loop following αC and Arg734 and Ser735 at the start of αD (Fig. 1A).
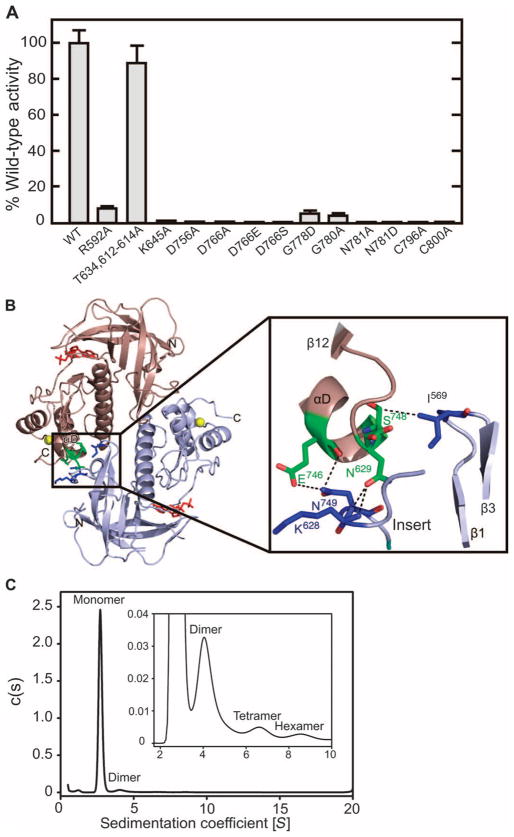
Fig. 2.
Mutational analysis and intermolecular interactions. (A) Wild-type (WT) A-CAT and the indicated site-directed A-CAT mutant proteins were assayed for kinase activity in experiments with myelin basic protein as the substrate as described in Materials and Methods. The activity of each mutant protein is expressed as a percentage of that of WT A-CAT. Error bars represent the SD, and the data are representative of five experiments. (B) The ribbon diagram shows molecule 1 (salmon) and molecule 2 (light blue) in the asymmetric unit of the A-CAT–AMPPCP complex (left). Nucleotides are shown as red sticks and the zinc atoms as yellow spheres. The region enclosed by the box shows residues in the αD to β12 turn in molecule 1 (green) that interact with residues in molecule 2 (dark blue). An expanded view of this region is shown in the box on the right. Dashed lines indicate the interactions between the residues. (C) Sedimentation velocity analysis of A-CAT. The main graph shows the full species distribution, whereas in the inset, the vertical scale is magnified by a factor of 50 to better show the minor components. c(s), continuous sedimentation coefficient distribution. A monomer peak at 2.7S, a dimer peak at 4.1S, a tetramer peak at 6.6S, and a hexamer peak at 8.6S were resolved, which constituted 91.2, 3.4, 1.4, and 0.7% of the total amount of protein, respectively. The data are representative of three separate experiments.
In the case of the crystallized A-CAT–AMP complex, one asymmetric unit contained one molecule of the complex, whereas for the structures of the A-CAT–ADP and the A-CAT–AMPPCP complexes, one asymmetric unit contained two molecules of the appropriate complex. In the A-CAT–ADP and A-CAT–AMPPCP complexes, both molecules within each structural unit associated in a head-to-tail and back-to-back arrangement, which placed the active sites at opposite ends to each other (Fig. 2B, left panel). The intermolecular interactions were not extensive and primarily involved residues in the loop that connected αD to β12 (Fig. 2B, right panel). Non-polar contacts were also made between Ile569 in the turn that connected strands β1 and β3 of one molecule with Trp673 and Lys676 in αC of the other molecule. Sedimentation velocity studies in the analytical ultracentrifuge demonstrated that A-CAT was primarily a monomer in solution (Fig. 2C); however, small amounts of dimeric, tetrameric, and hexameric A-CAT were also found, consistent with the presence of weak intermolecular interactions between A-CATs. These intermolecular interactions may be important in the context of MHCK A, because multiple α-kinase domains are held in close proximity to each other by the N-terminal coiled-coil domain.
Comparison of the structures of A-CAT and TRPM7-CAT
Dictyostelium MHCK A and mouse TRPM7 have no structural features in common except for the presence of an α-kinase domain (Fig. 3A). A-CAT and TRPM7-CAT share only 24% sequence identity (Fig. 3B) and represent evolutionarily divergent members of the α-kinase family. [We also performed a structure-guided sequence alignment of the α-kinases (fig. S1) and prepared a phylogenetic tree (fig. S2).] Nevertheless, superposition of the structures of A-CAT and TRPM7-CAT revealed the presence of a conserved, core α-kinase domain that began with the β1 stand and continued to the end of αF (Fig. 4A). The RMSD for the Cα atoms between the two core kinase domains was 1.53 Å. Because the secondary structural elements are so well-conserved, the names assigned for the secondary elements of TRPM7-CAT were adopted for A-CAT, wherever possible (Figs. 1A and 3B) (35).
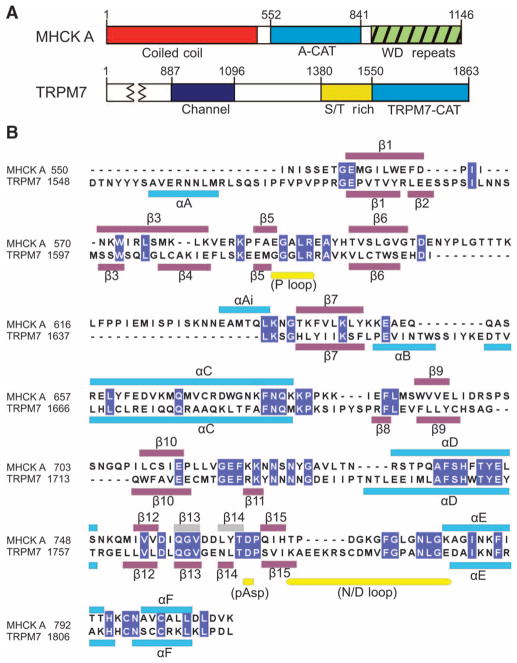
Fig. 3.
Alignment of the sequences of the α-kinase domains of MHCK A and TRPM7. (A) The diagrams show the domain structures of Dictyostelium MHCK A and mouse TRPM7. S/T, serine-threonine. (B) The α-kinase domains of Dictyostelium MHCK A and mouse TRPM7 (UniprotKB accession codes Q54EX1 and Q923J1, respectively) were aligned on the basis of similarities in their sequence and structure. The positions of the β strands (purple boxes) and α helices (blue boxes) in the structures of the A-CAT–AMPPCP complex and the TRPM7-CAT–AMPPNP complex are shown above and below the alignment, respectively. β strands 13 and 14 (gray boxes) appear in some, but not all, of the structures of A-CAT. Yellow boxes below the alignment indicate the P loop, A-CAT insert, phosphorylated aspartate residue (pAsp), and the N/D loop. Identical residues in A-CAT–AMPPCP and TRPM7-CAT–AMPPNP are shown in white text on a blue background. Abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
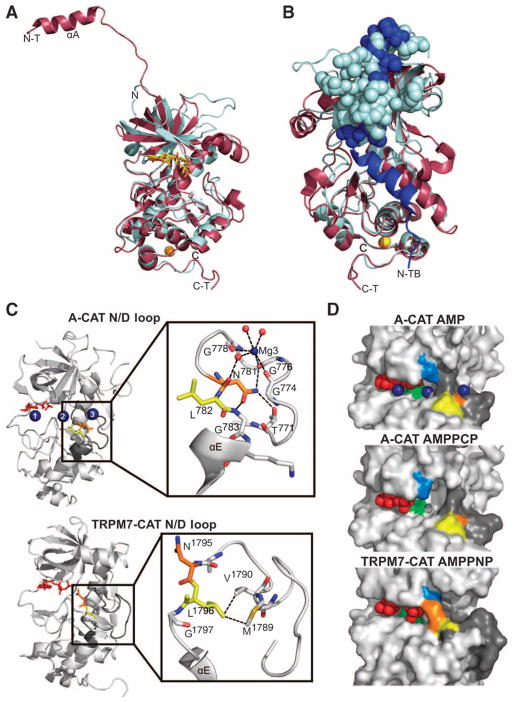
Fig. 4.
Comparison of the structures of A-CAT and TRPM7-CAT. (A) Front view of the superimposed structures of A-CAT (cyan) and TRPM7-CAT (raspberry) (PDB code 1IA9). In both structures, the nucleotide and zinc atoms are colored yellow and orange, respectively. N and C refer to the N terminus and C terminus, respectively, of A-CAT, whereas N-T and C-T refer to the N terminus and C terminus, respectively, of TRPM7-CAT. (B) Rear view of the superimposed structures showing the N-terminal extension (residues 1549 to 1577) from the second subunit (chain B) of TRPM7-CAT in the dimer in dark blue. For clarity, the N-terminal extension of the first TRPM7-CAT molecule (chain A) is omitted. Amino acid residues 1567 to 1577 of chain B and residues 604 to 624 of A-CAT (the insert) are rendered as spheres. N-TB indicates the N terminus of chain B and C-T denotes the C terminus of chain A. (C) The location of the N/D loop (dark gray) in the context of the entire structure of A-CAT (top panel) and TRPM7-CAT (bottom panel) is shown on the left, with the nucleotide shown as red sticks, the Mg2+ ions as dark blue spheres, the invariant Asn781 and Asn1795 residues colored in orange, and the conserved Leu782 and Leu1796 residues in yellow. An expanded view of the N/D loop is shown on the right, with water molecules illustrated as red spheres. Interactions made by Mg3, Asn781, and Leu1796 are shown as dashed lines. The interaction distances for Mg3 are given in table S1. (D) Surface representations of A-CAT and TRPM7-CAT highlighting the active-site pocket. The color scheme is the same as that used in (C). Leu591 in the P loop of A-CAT (Leu1621 in TRPM7-CAT) is colored marine blue and the catalytic Asp766 of A-CAT (Asp1775 in TRPM7-CAT) is colored green. Asn781 in the center of the N/D loop is far from the active site of A-CAT, whereas Asn1795, which is flipped out of the N/D loop, blocks access to the active-site pocket of TRPM7-CAT.
TRPM7-CAT contained an extended segment, located N-terminal to the common core, that was absent in A-CAT (Fig. 4A). The N-terminal segment, which contained αA and an unstructured sequence, is an integral part of TRPM7-CAT and is required for the assembly of dimers and for kinase activity (35, 38). In A-CAT, the absence of the N-terminal segment may be compensated for by a 29–amino acid residue sequence inserted after Glu606. This insert wrapped around the back of the N-terminal lobe of A-CAT to interact with a region that, in TRPM7-CAT, constituted a critical part of the dimerization interface (Fig. 4B). One function of the insert may therefore be to stabilize the monomeric state of A-CAT. Mass spectrometric analysis identified four sites of autophosphorylation within the insert: Thr634 and the poorly ordered residues Thr612, Thr613, and Thr614. The Thr634→Ala mutation (T634A) reduced the extent of autophosphorylation of A-CAT from 1.8 to 1.1 mol of phosphate per mole of enzyme. Mutation of Thr612, Thr613, and Thr614 to alanine in the T634A mutant reduced the extent of autophosphorylation to ~1.0 mol of phosphate per mole of enzyme, showing that Thr634 was the major autophosphorylation site in the insert. Mutation of all four threonine residues in the insert to alanine did not substantially alter the kinase activity of A-CAT (Fig. 2A). Ser553 and Thr825, both of which lie outside of the core kinase domain, were identified as the other sites at which autophosphorylation of A-CAT occurred. Further studies will be required to determine whether any of these sites is involved in the autophosphorylation-dependent activation of MHCK A (19).
The N/D loop
The sequence connecting the β15 strand to αE is highly variable within the α-kinase family, although it is often glycine-rich and is characterized by an invariant asparagine (in 59% of α-kinases) or aspartic acid (in 40% of α-kinases) residue (table S2). In light of the conserved nature of this residue (Asn781 in A-CAT and Asn1795 in TRPM7-CAT), we have named the β15-to-αE connecting sequence the N/D loop. In the A-CAT–AMP complex, the N/D loop folded into a cloverleaf-shaped structure that bound to Mg3 (Fig. 4C, top panel). The loop adopted a very similar conformation in the complexes of A-CAT–ADP and A-CAT–AMPPCP, but bound to a water molecule instead of Mg2+ ion. The Mg2+ ion was coordinated in a distorted, octahedral arrangement by three water molecules and by the backbone carbonyl groups of Gly774, Gly776, and Gly778. Of these three glycine residues, only Gly778, the first residue in the GxGN/DLG motif, is conserved in most α-kinases (table S2). The side chain of the invariant Asn781 was concealed within the center of the N/D loop, where it formed hydrogen bonds with the side-chain hydroxyl group of Thr771, the main-chain carbonyl group of Gly776, and the main-chain amide group of Gly783. The side chain of Leu782 was directed outward, away from the N/D loop. The base of the N/D loop was closed off, with the Cα atom of Gly783 only 2.8 Å from the main-chain carbonyl group of Thr771 at the start of the loop. Although the N/D loop of TRPM7-CAT contains five more residues than does that of A-CAT, the leucine and glycine residues that follow the invariant Asn1795 are conserved (Fig. 3B). In the TRPM7-CAT structure, the side chain of the invariant Asn1795 was directed outward, away from the N/D loop, and the side chain of Leu1796 was buried in the center of the loop (Fig. 4C, lower panel). The base of the loop was widened, with the Cα atom of Gly1797 positioned 12.5 Å from the main-chain carbonyl group of Ala1781 at the start of the loop. The large shift in the position of Gly1797 in TRPM7-CAT relative to that of Gly783 in A-CAT caused the N terminus of αE to partially unwind (Fig. 4C, compare upper and lower panels).
The N/D loop formed a wall at the right-hand side of the active site to create a solvent-accessible pocket that contained Mg2 in the A-CAT–AMP complex (Fig. 4D, upper panel). The top part of the active-site pocket was occluded by the side chain of Leu591 in the P loop, which extended outward to make contact with the N/D loop. In the A-CAT–AMPPCP complex, the top part of the active-site pocket was exposed as a result of a displacement of 3 to 4 Å in the relative positions of Leu591 and the N/D loop (Fig. 4D, middle panel). It can be concluded that the size and shape of the active-site pocket is extremely sensitive to small alterations in the conformations of the P loop and the N/D loop. Such movements could be induced by the binding of Mg2 to the active-site pocket and that of Mg3 to the N/D loop. The segment linking the β7 strand to αC, which lacked electron density in all of the A-CAT structures, was located just above the active-site pocket. The flexibility of this segment is likely to facilitate the movements of Leu591 and the N/D loop. In all of the A-CAT structures, the side chain of the invariant Asn781 residue, which was buried in the center of the N/D loop, was far from the active-site pocket, as was the side chain of Leu782 (Fig. 4D, upper and middle panels). The situation was markedly different in TRPM7-CAT (Fig. 4D, lower panel). In this structure, the side chain of Asn1795, which was flipped out of the N/D loop, made contact with the side chain of Leu1621 in the P loop to obstruct the entrance to the active-site pocket. Together, these results suggest that the N/D loop not only dictates the size and shape of the active-site pocket but may also act as a regulatory switch to control access to it.
Mutations within the N/D loop had a profound effect on the activity of A-CAT. Replacement of Gly778 with aspartic acid or of Gly780 with alanine reduced the kinase activities of the mutant proteins by a factor of ~20 relative to that of wild-type A-CAT (Fig. 2A). Replacement of the invariant Asn781 residue with alanine or aspartic acid abolished the kinase activities of the resulting mutant proteins (Fig. 2A). The introduction of these mutations might destabilize the N/D loop and prevent formation of the active-site pocket or perhaps lock the loop into an inactive conformation that blocks access to the active-site pocket.
Hydrolysis of ATP to AMP by A-CAT
Attempts to crystallize A-CAT in the absence of any nucleotide were unsuccessful, which suggested that a nucleotide in the central cleft is required to stabilize the conformation of A-CAT. When A-CAT was crystallized in the presence of ATP, a molecule of AMP and a separate molecule of phosphate (Pi1) were found at the active site (Fig. 5A). The adenine base and ribose moiety of AMP fit into a hydrophobic pocket composed of residues from both the N-terminal and C-terminal lobes of A-CAT (Fig. 5B). The exocyclic amino group, N6, was engaged in a salt bridge with the invariant Glu713 and formed hydrogen bonds with the main-chain carbonyl group of Pro714. The adenine N-3 atom formed hydrogen bonds through a network of ordered water molecules to the main-chain carbonyl groups of Leu716 and Gly718 and to the 2′-hydroxyl group of ribose. The ribose moiety adopted an unusual, almost planar configuration in the A-CAT–AMP complex; however, in the A-CAT–ADP and A-CAT–AMPPCP complexes, the ribose adopted a more common C3′-endo (3E) pucker (see below).
Fig. 5.
Active-site interactions with AMP, Pi1, Mg1, and Mg2. (A) The 2Fo − Fc electron density map, contoured at the 2σ level (gray mesh), of AMP, Pi1, and Asp766 in the active site of A-CAT. Mg1 and Mg2 are shown as cyan spheres. (B) Summary of the interactions made by the adenine base and ribose moiety of AMP (green) with residues from the N-terminal (blue) and C-terminal (cyan) lobes of A-CAT. Ordered water molecules are shown as red spheres and Mg1 as a dark blue sphere. Interactions are indicated by dashed lines. (C) A stereo view of the active site of A-CAT showing interactions made by the α-phosphate of AMP, Pi1, Mg1, and Mg2. The color scheme is the same as that used in (B). Interactions are indicated by dashed lines, and the distances involved are provided in table S1. (D) Time course of the hydrolysis of [α-32P]ATP by A-CAT (black circles) and A-CAT-D766A (black diamonds). A-CAT, but not A-CAT-D766A, hydrolyzed ATP to ADP (open triangles) and AMP (open squares). Error bars show the SD for three independent experiments, carried out as described in Materials and Methods.
The 3′-hydroxyl group of ribose coordinated with Mg1, the α-phosphate of AMP, the side chain of Gln758, and two water molecules (Fig. 5C). An inspection of the electron density maps did not reveal other peaks that could function as the sixth ligand coordinating Mg1. The side chains of two invariant basic residues, Arg592 and Lys645, extended from the N-terminal lobe to form ion pairs with the α-phosphate of AMP. Mutation of Arg592 or Lys645 to alanine reduced the kinase activities of the mutant proteins to 8% and 1.5% of that of wild-type A-CAT, respectively (Fig. 2A). The invariant Asp766 residue was located at the center rear of the active site where it interacted with Pi, the α-phosphate of AMP, and Mg2 in the active-site pocket. Replacement of Asp766 with alanine, glutamic acid, or serine eliminated the kinase activities of the resulting mutant proteins (Fig. 2A). Pi1 formed hydrogen bonds to Asp766, the α-phosphate of AMP, and the main-chain amide groups of Leu591 and Arg592 in the P loop. Apparently, these interactions were strong enough to prevent Pi1 from leaving the active site. Mg2 was coordinated in an octahedral arrangement by the side chains of Asp766 and Gln768, the main-chain carbonyl group of Pro767, and three water molecules.
The ability of A-CAT to catalyze the removal of the β-phosphoryl group of ATP was unexpected and so was further investigated. Experiments with [γ-32P]ATP showed that, in the absence of a peptide substrate, A-CAT hydrolyzed ATP to ADP with a Michaelis constant (Km) of 60 μM and a kcat of 0.03 s−1. By way of comparison, A-CAT exhibits a Km for ATP of 50 μM and a kcat value of 2.7 s−1 when myelin basic protein is its substrate (18). Experiments with [α-32P]ATP showed that A-CAT hydrolyzed ADP to AMP, albeit at a rate slower by a factor of ~100 than that of its ATPase activity (Fig. 5D). Hydrolysis of ATP was eliminated by mutation of Asp766 to alanine, which showed that the reaction was dependent on a functional active site in A-CAT (Fig. 5D). Although the ADPase activity of A-CAT was slow and therefore unlikely to be of physiological relevance, it might explain the appearance of AMP in the A-CAT active site after the 3- to 4-week period of crystal formation.
Phosphorylation of Asp766 in the A-CAT–ADP complex
Previous work identified peptides with the sequences YAYDTRYRR and AAYKTKKKK (39) as good substrates for A-CAT (24, 27). We co-crystallized A-CAT in the presence of ATP and a 10-fold molar excess of each peptide in an attempt to obtain a peptide-bound structure. Structures obtained after crystallization under these conditions did not contain a peptide in the active site; however, the electron density maps showed ADP instead of AMP at the active site and the presence of a strong and unambiguous additional density that extended from the density of the invariant Asp766 residue (Fig. 6A). This additional density ideally houses a phosphate molecule in a covalent linkage (1.5 Å) with Oδ2 of Asp766. On the basis of this result, we inferred that the γ-phosphoryl group of ATP was transferred to Asp766 during the crystallization process. If pAsp766 represents an intermediate in the catalytic mechanism, then A-CAT would be predicted to catalyze the partial reaction (A-CAT + ATP ↔ A-CAT-P + ADP) in the absence of peptide substrate (40). Isotopic studies with ATP and [8-14C]ADP showed that A-CAT catalyzed the exchange of ATP for ADP at a rate of 0.0015 s−1 (fig. S3). No exchange activity was detected for two kinase-defective mutants of A-CAT (A-CAT-D766S and A-CAT-D765A), showing that the exchange reaction was indeed catalyzed by A-CAT (fig. S3).
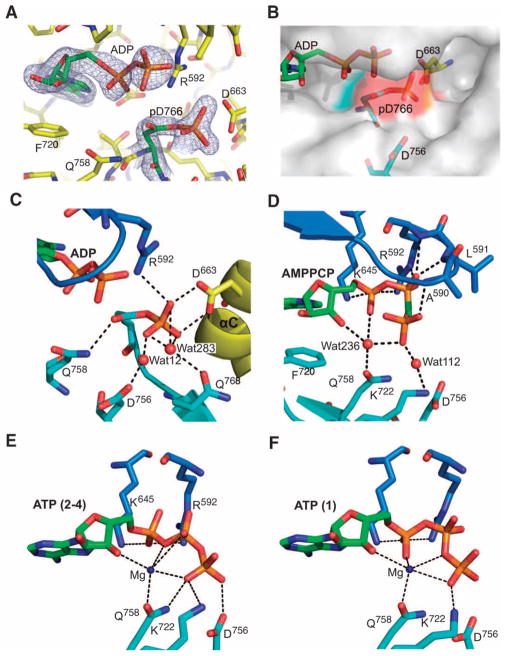
Fig. 6.
Active-site interactions with pAsp766, ATP, and AMPPCP. (A) The 2Fo − Fc electron density map of ADP and pAsp766 (pD766) contoured at 2σ (gray mesh). Residues are shown as yellow sticks, with ADP and pAsp766 shown as green sticks. (B) A surface representation of the structure of A-CAT–ADP showing the location of pAsp766 near to the entrance of the active-site pocket. The surface area contributed by the aspartylphosphate residue is colored in red. (C to F) The interactions made by (C) pAsp766, (D) the AMPPCP phosphoryl groups, (E) the ATP phosphoryl groups in molecules 2 to 4, and (F) the ATP phosphoryl groups in molecule 1 of the four-molecule asymmetric unit of A-CAT-D766A. The color scheme used in (C) to (F) is the same as that used in Fig. 5B. Interactions are indicated by dashed lines.
The pAsp766 residue was exposed to solvent at the entrance to the active-site pocket, which suggested that it had the potential to transfer the phosphate group to a substrate docked within the pocket (Fig. 6B). The aspartylphosphate made an ion pair interaction with Arg592 and formed hydrogen bonds to two water molecules and to the side chain of Asp663, which protruded into the back of the active-site pocket from αC (Fig. 6C). This arrangement suggested that the carboxyl group of Asp663 was protonated and acted as both a donor and an acceptor of hydrogen bonds.
Complexes of A-CAT with AMPPCP or ATP
To obtain a structure of A-CAT in a complex with a nucleotide triphosphate, A-CAT was crystallized in the presence of the nonhydrolyzable ATP analog AMPPCP. The phosphoryl groups of AMPPCP were bent into a U-shape configuration, which enabled the γ-phosphate to form hydrogen bonds through a water molecule (Wat245) to the 3′-hydroxyl group of ribose (Fig. 6D). Wat245, which also formed hydrogen bonds to Gln758 and the α-phosphate of AMP, took the place occupied by Mg1 in the structure of the A-CAT–AMP complex. The γ-phosphate pointed out into the solvent and made only a single direct interaction with A-CAT: a hydrogen bond to the main-chain amide of Ala590 in the P loop (Fig. 6D). The γ-phosphate also made a water-mediated hydrogen bond to the side chain of Lys722. The β-phosphoryl group was more firmly held in place through ion-pair and hydrogen bond interactions with Leu591 and Arg592 in the P loop.
A crystal structure was also obtained of the inactive A-CAT-D766A mutant protein in a complex with ATP. Comparison of the structures of A-CAT-D766A and A-CAT AMPPCP yielded a Cα atom RMSD of 0.50 Å, showing that the Asp766→Ala mutation produced little, if any, change in the conformation of A-CAT (Fig. 1C). Four molecules of A-CAT-D766A were present in the asymmetric unit, with molecules 1 and 2 and molecules 3 and 4 exhibiting intermolecular interactions equivalent to those shown for A-CAT–ADP and A-CAT–AMPPCP (Fig. 2B). Additional hydrophobic contacts between His596 in molecule 2 and Val717 and Lys641 in molecule 4 linked the two bimolecular units together. The ATP in the active site of A-CAT-D766A bound to a Mg2+ ion that is equivalent to Mg1 in the A-CAT–AMP complex (Fig. 6, E and F). The Mg2+ ion formed a bridge between the γ-phosphate of ATP and the 3′-hydroxyl group of ribose and Gln758. The other two coordinating ligands for the Mg2+ ion were provided by different phosphoryl groups of ATPs in molecule 1 and molecules 2 to 4 of the four-molecule, asymmetric unit of A-CAT-D766A, indicating a degree of conformational flexibility in the phosphate chain of ATP (Fig. 6, E and F). As with Mg1 in the A-CAT–AMP complex, a sixth coordinating ligand for the ATP-bound Mg2+ ion could not be detected. The phosphoryl groups of ATP sat lower in the active site than did those of AMPPCP, which weakened interactions with the P loop and enabled direct interactions between the γ-phosphoryl group and residues in the C-terminal lobe, including the side chains of Lys722, Gln758, and Asp756.
DISCUSSION
A-CAT, the α-kinase domain of Dictyostelium MHCK A, shares a similar overall secondary structure and architecture with TRPM7-CAT, the α-kinase domain of mouse TRPM7, even though these proteins are divergent members of the α-kinase family (35). Superposition of the active sites of A-CAT and TRPM7-CAT showed that their key catalytic residues exhibited a near-perfect spatial match (fig. S4). The sequences of all known members of the α-kinase family can be aligned with a high degree of confidence to the secondary structural elements present in the core domain common to A-CAT and TRPM7-CAT, which suggests that this core domain is well conserved in all of the α-kinases (fig. S1). Many α-kinase domains contain large inserts, but these are always restricted to the loops and turns that connect the conserved elements of secondary structure.
The A-CAT–ADP complex, obtained by crystallization of A-CAT in the presence of ATP and a peptide substrate, provided convincing evidence of the presence of a phosphate covalently attached to the invariant Asp766 residue at the active site. It should be noted that three independent x-ray data sets obtained in the presence of two different peptide substrates unambiguously demonstrated the existence of the pAsp766. The stability of the aspartylphosphate in the crystal structure is unusual but not unprecedented. Although aspartylphosphate has a high free energy of hydrolysis, it is chemically stable at physiological pH with half-lives of about 20 hours at 25°C and 14 days at 4°C (41, 42). An aspartylphosphate intermediate has been observed at active sites in the crystal structures of at least two members of the haloacid dehalogenase (HAD) family of phosphotransferases: Lactococcus lactis β-phosphoglucomutase and the phosphoacceptor domain of the Bacillus stearothermophilus response regulator Spo0A (43, 44). In these cases, the persistence of the aspartylphosphate was ascribed to the absence of a proximal residue capable of acting as a general base catalyst, the lack of a Mg2+ cofactor, or the exclusion of water from the active site. The situation is more complicated with A-CAT, because Asp766 was phosphorylated only when a peptide substrate was included in the crystallization process. The peptide substrate was not found in the active site, and so it is not clear how it promoted the formation of the aspartylphosphate or how it might block the hydrolysis of, or the transfer of a phosphate from, the aspartylphosphate. Indeed, it cannot be stated with certainty that the peptide substrate, rather than some other factor, was responsible for the stability of pAsp766 in the crystal.
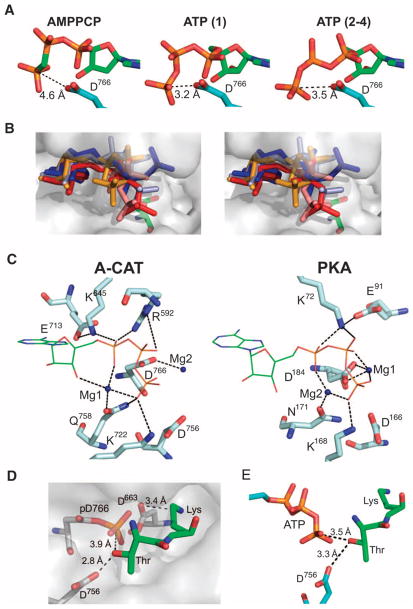
Mutational analysis showed that Asp766 was essential for both the ATPase and the kinase activities of A-CAT (Fig. 2A). Similarly, TRPM6 and TRPM7 are rendered phosphotransferase-deficient by mutation of the residues equivalent to Asp766 (Asp1933 and Asp1775, respectively) (25, 45, 46). We speculate that the side-chain carboxyl group of Asp766 makes a direct nucleophilic attack on the γ-phosphate of ATP. Support for this reaction is provided by experiments showing that A-CAT catalyzes an exchange reaction between [14C]ADP and ATP and that the exchange activity coincides with kinase activity (40). The rate of the ATP-ADP exchange reaction was slower by a factor of ~20 than the rate of ATP hydrolysis, which may be explained if the rate of the dissociation of ADP from the intermediate complex is slow. Alternatively, the slow rate of the exchange reaction may indicate that pAsp766 is formed in a nonproductive side reaction. Studies of members of the HAD superfamily, which includes phosphatases, phosphomutases, bacterial response regulators, and P-type adenosine triphosphatases (ATPases), have established that aspartic acid acts as the nucleophile in various phosphotransferase reactions (47, 48). In the P-type ATPases, such as sarcoplasmic reticulum Ca2+-ATPase (SERCA), an aspartic acid residue makes a nucleophilic attack on the γ-phosphate of ATP. The structure of Ca2+-bound SERCA shows that Asp351 (the phosphorylation site) is 3.2 Å from, and directly aligned with, the γ-phosphate of AMPPCP, in agreement with an associative, nucleophilic reaction mechanism (SN2) of phosphoryl transfer (49). In the A-CAT–AMPPCP complex, the distance from the Oδ2 of Asp766 to the γ-phosphate of ATP was 4.6 Å (Fig. 7A). When an aspartic acid residue was modeled in place of Ala766 in the A-CAT-D766A–ATP complex, a distance of 3.2 to 3.5 Å was obtained (Fig. 7A). However, in the A-CAT structures, the γ-phosphate pointed away from Asp766 and would have to pivot inward to achieve a geometry that is compatible with an SN2 substitution reaction (Fig. 7A).
Fig. 7.
ATP and peptide substrate binding. (A) The positions of the γ-phosphoryl groups of ATP and AMPPCP relative to the side chain of Asp766. ATP adopts different conformations in molecule 1 and molecules 2 to 4 of the asymmetric unit of A-CAT-D766A. Aspartic acid was modeled in place of Ala766 in the structure of A-CAT-D766A. (B) The stereo view shows the superimposed structures of AMPPCP in A-CAT (orange), ATP in molecule 1 (salmon), ATP in molecules 2 to 4 of A-CAT-D766A (red), and AMPPNP in chain A (light blue) and chain B (dark blue) of TRPM7-CAT. The surface representation is of the A-CAT–AMPPCP complex with Asp766 rendered as green sticks. (C) Comparison of the active sites of A-CAT (left panel) and PKA (right panel) showing key catalytic residues, ATP (thin green sticks), and Mg2+ ions (dark blue spheres). The high- and low-affinity Mg2+ ions bound to PKA are labeled Mg1 and Mg2, respectively. (D) A Thr-Lys dipeptide (green sticks) was docked into the active-site pocket of the A-CAT–ADP complex (surface shown in gray). (E) The Thr-Lys dipeptide, in the same position and configuration as that shown in (D), was docked onto the ACAT-D766A–ATP complex.
A comparison of the structures of the A-CAT–AMPPCP, A-CAT-D766A–ATP, and TRPM7-CAT–AMPPNP complexes showed that the γ-phosphate of the nucleotide exhibited a striking degree of mobility within the active site (Fig. 7B). Although some of the conformational differences in the phosphate chain may be due to substitution of the β,γ-bridging oxygen atom with an NH or CH2 group (50), the results nonetheless showed that the active site of the α-kinase could accommodate the γ-phosphate in multiple positions. The structure of TRPM7-CAT also revealed the presence of two different side-chain rotamers for Asp1775 (equivalent to Asp766 in A-CAT) (fig. S4). Although this high degree of conformational freedom may enable the γ-phosphate of ATP and the side chain of Asp766 to achieve the correct geometry required for phosphotransfer to occur, the possibility that Asp766 is phosphorylated by a more complicated, indirect route cannot be ruled out. For example, the γ-phosphate of ATP may first be transferred to the hydroxyl group of threonine on the peptide substrate and subsequently transferred by a phosphatase-type reaction to Asp766. Further studies, including the crystallization of A-CAT in a complex with a high-affinity peptide substrate or inhibitor, or with ADP and ALF4− to mimic the transition state of phosphoryl transfer, may help to resolve the question of exactly how the phosphorylation of Asp766 occurs.
Asp766 was located in the active site of A-CAT in a position similar to that of Asp184 in the active site of the cyclic adenosine monophosphate (cAMP)–dependent protein kinase (PKA) (Fig. 7C). In PKA, Asp184 coordinates the activating Mg2+ ion that bridges the β- and γ-phosphates of ATP (51, 52). In contrast, the Mg2+ ion in the A-CAT-D766A–ATP complex was coordinated by the γ-phosphate and ribose 3′-hydroxyl group of ATP and the side chain of Gln758 (Fig. 7C). The single Mg2+ ion present in the ACAT–AMP and TRPM7-CAT–ADP complexes was chelated in a similar manner (Fig. 4C) (35). Thus, there is convincing evidence that nucleotides bound to the active sites of α-kinases contain a single Mg2+ ion that is located similarly to the low-affinity Mg2+ ion that is coordinated by Asn171 in PKA (Fig. 7C). It is notable that the α- and β-phosphoryl groups of ATP interacted with two basic residues (Arg592 and Lys645) in the active site of A-CAT but with only a single basic residue (Lys72) in PKA (Fig. 7C). The negative charge on ATP may therefore be more effectively neutralized in the α-kinases than in the ePKs, eliminating the need for a second ATP-bound metal ion and freeing Asp766 to act as a nucleophile in a phosphotransferase reaction. Mutation of Lys645 or Arg592 in A-CAT, or of the equivalent residues (Arg1622 and Lys1646) in TRPM7, severely inhibited kinase activity, showing that both invariant basic residues played an important role in catalysis (Fig. 2A) (25).
A key question concerns the function of the pAsp766 residue. In the P-type ATPases and bacterial response regulators, the phosphorylation of aspartic acid is coupled to a global conformational change (49, 53). This was not the case for A-CAT. The phosphorylation of Asp766 produced no substantial alteration in the overall structure of A-CAT or in the disposition of active-site residues (Fig. 1C). Once transferred to Asp766, the phosphoryl group was exposed to solvent at the edge of the active-site pocket (Fig. 6B). Although no structural information is available to indicate how α-kinases recognize protein substrates, the active-site pocket provides an obvious docking site for an amino acid side chain. Because A-CAT strongly prefers to phosphorylate a threonine residue that is followed in sequence by a lysine or arginine residue (24, 27), a Thr-Lys dipeptide was modeled into the active-site pocket (Fig. 7D; for distances to neighboring residues, see fig. S5). The active-site pocket was large enough to accommodate the lysine side chain, which formed an electrostatic interaction with Asp663 at the back of the pocket. This suggests that Asp663 may play a role in determining substrate specificity. Sequence alignments showed that 74% of α-kinases have an acidic residue at the position that corresponds to Asp663, which suggests that many α-kinases may recognize a basic residue near the site of phosphorylation (table S2).
The side-chain hydroxyl group of threonine in the Thr-Lys dipeptide might be positioned so that it forms hydrogen bonds to the invariant Asp756 residue. We propose that Asp756 functions as the catalytic base that accepts the proton from the attacking hydroxyl group of the threonine, which was within 3.9 Å of the pAsp766, although an outward rotation of the aspartylphosphate would be required to achieve an optimal orientation for an in-line, nucleophilic attack. When modeled into the structure of A-CAT-D766A, the threonine hydroxyl group was 3.5 Å from the γ-phosphate of ATP, although, again, the geometry was not suitable for a straight-line, nucleophilic attack on the γ-phosphate (Fig. 7E). The structural model prevents us from distinguishing between a catalytic mechanism that involves a direct, in-line transfer of the γ-phosphoryl of ATP to the peptide substrate and one that requires the formation of a pAsp766 intermediate. To differentiate between these two mechanisms, we need to rigorously establish the stereochemical course of the A-CAT–catalyzed phosphoryl-transfer reaction (54). If the formation of pAsp766 is not required for catalysis, it may serve to regulate the kinase or ATPase activities of A-CAT or to modulate interactions between A-CAT and its substrates.
A-CAT and TRPM7-CAT both require millimolar concentrations of Mg2+ or Mn2+, in excess of that required to bind to ATP, to exhibit maximal catalytic activity (18, 46, 55). The A-CAT–AMP complex contained two Mg2+ ions (Mg2 and Mg3), which are likely to play a role in the regulation of kinase activity (Fig. 1A). Mg2 was located in the active-site pocket and so might aid in binding to peptide or protein substrates. In addition, Mg2 was directly coordinated by Asp766 and so might help to position the side chain of Asp766 during phosphotransfer, play a role in stabilizing negative charges that develop during the transition state, or both. Mg3 was bound to the N/D loop, which played a critical role in forming the active-site pocket that enclosed the aspartylphosphate. Comparison of the structures of A-CAT and TRPM7-CAT showed that the N/D loop switched between two different configurations: an active conformation in which the side chain of the invariant asparagine residue was buried in the center of the loop, and an inactive conformation in which the side chain moved out of the loop to block the entrance to the active-site pocket (Fig. 4, C and D). The strict conservation of an asparagine or aspartic acid residue at this position in the N/D loop may reflect constraints on the size of the side chain (to enable it to fit inside the N/D loop) and the requirement to form hydrogen bonds with other residues in the loop to stabilize its active conformation. A-CAT did not tolerate mutation of Asn781 to aspartic acid, indicating that the N/D loops of individual α-kinases may be specialized to accommodate one or another of these residues. The mutation of individual glycine residues in the N/D loop to alanine or aspartic acid severely reduced the catalytic activities of A-CAT, TRPM6, and TRPM7 (Fig. 2A) (25, 45, 46). It is possible that these mutations sterically hinder formation of the active “asparagine-in” conformation of the N/D loop. It can be speculated that the configuration of the N/D loop in solution is controlled by the concentrations of divalent metal ions. If the active asparagine-in conformation of the N/D loop is stabilized by the binding of Mg2+ or Mn2+ ions, this would provide an elegant mechanism to couple the activity of A-CAT to the local concentration of cytoplasmic divalent cations. It will be important to obtain direct evidence for a divalent metal ion–induced conformational switch in the N/D loop with techniques such as fluorescence spectroscopy and nuclear magnetic resonance (NMR). It will also be interesting to determine whether the N/D loop of TRPM7-CAT functions as a regulatory metal ion–binding site and if the divergent sequences of the N/D loops of A-CAT and TRPM7-CAT correlate with differences in their affinities and selectivities for divalent cations.
Many questions remain concerning the functional and regulatory properties of the α-kinases. The data presented here show that an essential aspartic acid residue in the active site of an α-kinase is phosphorylated by ATP, but whether the aspartylphosphate represents an obligatory reaction intermediate or serves some alternate function remains to be determined. Evidence was also presented to show that a specialized loop, termed the N/D loop, may be responsible for regulating the activity of α-kinases in response to changes in the concentration of divalent cations. Together, the features described here may help to explain why a small number of α-kinases have been conserved throughout evolution to regulate key cellular processes.
MATERIALS AND METHODS
Protein expression and purification
We constructed a pET28a vector that encoded A-CAT (residues 552 to 841 of MHCK A) with an N-terminal hexahistidine tag and a tobacco etch virus (TEV) protease site. DNA constructs encoding site-directed mutants of A-CAT were generated with the QuikChange mutagenesis system (Stratagene). Wild-type and mutant forms of A-CAT were expressed in Escherichia coli strain BL21(DE3) and purified as described previously (27). All steps were carried out at 4°C. Briefly, cells were lysed by sonication in 20 mM tris-HCl (pH 8.0), 500 mM NaCl, and 5 mM imidazole, containing one Complete Mini EDTA-free protease inhibitor cocktail tablet (Roche Diagnostics) per 50 ml of buffer, and lysates were centrifuged at 75,000 g for 1 hour. The resulting supernatant was applied to a column of His-Bind resin (Novagen). The material that was eluted with 20 mM tris-HCl (pH 8.0), 250 mM NaCl, and 200 mM imidazole and was collected, dialyzed against 20 mM tris-HCl (pH 7.5), 50 mM NaCl, and 2% glycerol, and passed sequentially over columns of Whatman DE-53 cellulose and SP Sepharose Fast Flow (GE Biosciences). Protein to be used for crystallization trials was concentrated to 8 mg/ml with a 10,000 molecular weight cutoff Ultrafree-4 Centrifugal Filtration Unit (Millipore). The His tag was cleaved from the protein by treatment overnight at 4°C with AcTEV protease (Invitrogen), and the protease was removed by chromatography over a His-Bind column. Attempts to obtain a selenomethionine-substituted form of A-CAT by expression in BL41(DE3) and B834(DE3) cell lines were unsuccessful.
Crystallization procedures
The following samples were prepared: A-CAT with 1.5 mM ATP and 2 mM MgCl2; A-CAT with 1.5 mM AMPPCP and 2 mM MgCl2; A-CAT with 1.5 mM ATP, 2 mM MgCl2, and a 10:1 molar ratio of peptide substrate (either YAYDTRYRR or AAYKTKKKK); and A-CAT-D766A with 1.5 mM ATP and 2 mM MgCl2 (24, 27). A 1-μl aliquot of each sample was mixed with 1 μl of reservoir solution consisting of either 0.1 M tris-HCl (pH 8.5), 0.2 M NaH2PO4, 18% (w/v) polyethylene glycol (PEG) 8000 or 0.1 M sodium cacodylate (pH 6.0), 0.2 M MgSO4, 20% (w/v) PEG 3350. Crystallization was performed by the hanging-drop vapor-diffusion method at 4°C. Crystals appeared after 2 weeks and grew to their maximum size in 4 weeks. Crystals were flash-frozen in a stream of liquid nitrogen after being dipped in a cryoprotectant solution that consisted of the mother liquor component and 25% (w/v) ethylene glycol.
Data collection and structure determination
A sulfur single-wavelength anomalous dispersion (S-SAD) 2.3 Å data set was collected for a cocrystal of A-CAT and AMP with a Rigaku chromium Kα x-ray source operating at a wavelength of 2.29 Å and an R-AXIS IV detector. A nonanomalous 1.6 Å data set from the same crystal and a 2.1 Å A-CAT-AMPPCP data set, a 2.0 Å A-CAT-ADP data set, and a 2.2 Å A-CAT-D766A-ATP data set were collected by the F1 beam line at the Cornell High Energy Synchrotron Source (CHESS). The data sets were processed and scaled with DENZO and SCALEPACK or with the HKL2000 suite program (56). The initial phase was calculated for S-SAD by the program SHELX (57) followed by use of the autobuild function implemented in AUTOSHARP (58). From 290 residues, 223 were built with the correct side chains. Partial refinement of the model by CNS with simulated annealing (59) enabled the manual building of additional protein residues with XTALVIEW (60). At this stage, inspection of the Fo − Fc electron difference map facilitated the unambiguous placement of AMP, Pi, Mg2+ ions, and zinc atom. The complete model was further refined by CNS and restrained refinement in REFMAC5 (61). The nonanomalous data sets were solved by molecular replacement with the initially solved A-CAT structure, devoid of AMP and ions, as the search model with PHASER software (62). After several cycles of inspection of the 2Fo − Fc map and the Fo − Fc electron difference map, all of the data exhibited instantly recognizable nucleotides and ions. CNS and REFMAC5 were used to build and refine the final models. In the final model of A-CAT–AMP, residues Ile552 to Glu555, Thr612 to Thr614, Glu650, Ala651, and those residues C-terminal to Val807 were not included because of their poorly defined electron densities. Missing residues not included in the other models are given in table S3.
Phosphorylation and ATP hydrolysis assays
Wild-type and mutant A-CAT proteins were assayed at 22°C in kinase buffer [20 mM 2-{[2-hydroxy-1,1-bis(hydroxymethyl)methyl]amino}ethanesulfonic acid (TES) (pH 7.0), 2 mM MgCl2, 1 mM dithiothreitol, 0.25 mM ATP] incorporating [γ-32P]ATP (NEN PerkinElmer) at a specific activity of 100 to 500 cpm per picomole of ATP. Kinase assay reactions contained A-CAT (50 μg/ml) and myelin basic protein (1 mg/ml). Autophosphorylation assay reactions contained A-CAT (70 μg/ml) that had been dephosphorylated before use by treatment for 2 hours at room temperature with calf intestinal alkaline phosphatases immobilized on agarose beads (Sigma-Aldrich) (18). Aliquots of 20 μl were removed from the reactions after 1, 2, 3, 4, 5, 15, and 30 min and spotted onto squares of P81 phosphocellulose paper (Whatman). Under these assay conditions, the activity of wild-type A-CAT was 0.05 s−1. The squares were washed in 1% phosphoric acid and counted in ScintiVerse Universal LS Cocktail (Fisher Scientific) by a Beckman LS 9000 scintillation counter. Incorporation of 32P was quantified by spotting the samples onto Whatman filter followed by washing the filters in cold 5% trichloroacetic acid. ATP hydrolysis was measured by following the release of 32Pi from [γ-32P]ATP as described previously (63). Hydrolysis of ATP to ADP and AMP was measured in kinase buffer with [α-32P]ATP at a specific activity of 15 to 30 cpm per picomole ATP. Aliquots of 300 μl were removed from the reactions and flash-frozen in a dry ice–ethanol bath. For analysis, aliquots were rapidly thawed by addition of an equal volume of an unlabeled mixture of 10 mM ATP, 10 mM ADP, and 10 mM AMP and loaded onto a 1-ml HiTrap Q HP column (GE Healthcare). AMP, ADP and ATP were eluted with a stepwise gradient of 50, 115, and 300 mM NaCl. Fractions were collected and aliquots were counted as described above to measure the amounts of ATP, ADP, and AMP in each sample.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed with a Beckman Optima XL-1 instrument equipped with an AN 60-Ti rotor and interference and schlieren optics. A-CAT (1.0 mg/ml) was analyzed in 5 mM tris-HCl (pH 7.4), 50 mM NaCl with a rotor speed of 60,000 rpm. Between 350 and 400 scans by interference optics were taken for each experiment. Data were analyzed with the SEDFIT program with a partial specific volume of 0.73 cm3/g and a solution density of 0.9992 g/cm3.
Supplementary Material
Acknowledgments
We thank C. Yang at Rigaku Corporation for collection of the x-ray S-SAD data set, Y.-M. She at the Department of Chemistry, Queen’s University, for carrying out mass spectrometry experiments, and K. Munro of the Queen’s University Protein Function Discovery Facility, for help with the sedimentation velocity experiments. Portions of this research were carried out at the Cornell High Energy Synchrotron Source.
Funding: This research was funded by grants from the Canadian Institutes of Health Research (MOP8603) and Heart and Stroke Foundation of Ontario (T6054) to G.P.C. and Natural Sciences and Engineering Research Council of Canada grant 203705 to Z.J. Z.J. is a Canada Research Chair in Structural Biology.
Footnotes
Author contributions: Q.Y., S.W.C., Y.Y., and G.P.C. designed research; Q.Y., S.W.C., and Y.Y. performed research; Q.Y., S.W.C., Y.Y., Z.J., and G.P.C. analyzed data; G.P.C. wrote the paper with editorial input from Q.Y., S.W.C., Y.Y., and Z.J.
Competing interests: The authors declare no conflicts of interest.
Accession numbers: Coordinates have been deposited in the Protein Data Bank under ID codes 3LKM (for the A-CAT–AMP complex), 3LMH (for the A-CAT–ADP complex), 3LLA (for the A-CAT–AMPPCP complex), and 3LMI (for the A-CAT–A-CAT-D766A ATP complex).
www.sciencesignaling.org/cgi/content/full/3/111/ra17/DC1
Fig. S1. Alignment of the sequences of multiple α-kinase domains.
Fig. S2. Evolutionary relationships of the α-kinases.
Fig. S3. The ATP-ADP exchange reaction catalyzed by A-CAT.
Fig. S4. Comparison of the active sites of A-CAT and TRPM7-CAT.
Fig. S5. Docking of a Thr-Lys dipeptide into the active-site pocket of the A-CAT–ADP complex.
Table S1. Interaction distances for the relevant ions and water molecules in the various A-CAT complexes.
Table S2. Conservation of functionally important residues in the α-kinase domain.
Table S3. Summary of the missing residues in the structures of A-CAT and A-CAT-D766A.
Table S4. α-Kinase domains included in the multiple sequence alignment.
REFERENCES AND NOTES
- 1.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De la Roche MA, Smith JL, Betapudi V, Egelhoff TT, Côté GP. Signaling pathways regulating Dictyostelium myosin II. J Muscle Res Cell Motil. 2002;23:703–718. doi: 10.1023/a:1024467426244. [DOI] [PubMed] [Google Scholar]
- 3.Yumura S, Yoshida M, Betapudi V, Licate LS, Iwadate Y, Nagasaki A, Uyeda TQ, Egelhoff TT. Multiple myosin II heavy chain kinases: Roles in filament assembly control and proper cytokinesis in Dictyostelium. Mol Biol Cell. 2005;16:4256–4266. doi: 10.1091/mbc.E05-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosgraaf L, van Haastert PJ. The regulation of myosin II in Dictyostelium. Eur J Cell Biol. 2006;85:969–979. doi: 10.1016/j.ejcb.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Côté GP, Bukiejko U. Purification and characterization of a myosin heavy chain kinase from Dictyostelium discoideum. J Biol Chem. 1987;262:1065–1072. [PubMed] [Google Scholar]
- 6.Vaillancourt JP, Lyons C, Côté GP. Identification of two phosphorylated threonines in the tail region of Dictyostelium myosin II. J Biol Chem. 1988;263:10082–10087. [PubMed] [Google Scholar]
- 7.Lück-Vielmetter D, Schleicher M, Grabatin B, Wippler J, Gerisch G. Replacement of threonine residues by serine and alanine in a phosphorylatable heavy chain fragment of Dictyostelium myosin II. FEBS Lett. 1990;269:239–243. doi: 10.1016/0014-5793(90)81163-i. [DOI] [PubMed] [Google Scholar]
- 8.Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- 9.Kolman MF, Egelhoff TT. Dictyostelium myosin heavy chain kinase A subdomains. Coiled-coil and WD repeat roles in oligomerization and substrate targeting. J Biol Chem. 1997;272:16904–16910. doi: 10.1074/jbc.272.27.16904. [DOI] [PubMed] [Google Scholar]
- 10.Kolman MF, Futey LM, Egelhoff TT. Dictyostelium myosin heavy chain kinase A regulates myosin localization during growth and development. J Cell Biol. 1996;132:101–109. doi: 10.1083/jcb.132.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futey LM, Medley QG, Côté GP, Egelhoff TT. Structural analysis of myosin heavy chain kinase A from Dictyostelium. Evidence for a highly divergent protein kinase domain, an amino-terminal coiled-coil domain, and a domain homologous to the β-subunit of heterotrimeric G proteins. J Biol Chem. 1995;270:523–529. doi: 10.1074/jbc.270.2.523. [DOI] [PubMed] [Google Scholar]
- 12.Russ M, Croft D, Ali O, Martinez R, Steimle PA. Myosin heavy-chain kinase A from Dictyostelium possesses a novel actin-binding domain that cross-links actin filaments. Biochem J. 2006;395:373–383. doi: 10.1042/BJ20051376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang W, Licate L, Warrick H, Spudich J, Egelhoff T. Differential localization in cells of myosin II heavy chain kinases during cytokinesis and polarized migration. BMC Cell Biol. 2002;3:19. doi: 10.1186/1471-2121-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagasaki A, Itoh G, Yumura S, Uyeda TQ. Novel myosin heavy chain kinase involved in disassembly of myosin II filaments and efficient cleavage in mitotic Dictyostelium cells. Mol Biol Cell. 2002;13:4333–4342. doi: 10.1091/mbc.E02-04-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steimle PA, Licate L, Côté GP, Egelhoff TT. Lamellipodial localization of Dictyostelium myosin heavy chain kinase A is mediated via F-actin binding by the coiled-coil domain. FEBS Lett. 2002;516:58–62. doi: 10.1016/s0014-5793(02)02494-8. [DOI] [PubMed] [Google Scholar]
- 16.Steimle PA, Yumura S, Côté GP, Medley QG, Polyakov MV, Leppert B, Egelhoff TT. Recruitment of a myosin heavy chain kinase to actin-rich protrusions in Dictyostelium. Curr Biol. 2001;11:708–713. doi: 10.1016/s0960-9822(01)00182-8. [DOI] [PubMed] [Google Scholar]
- 17.Steimle PA, Naismith T, Licate L, Egelhoff TT. WD repeat domains target Dictyostelium myosin heavy chain kinases by binding directly to myosin filaments. J Biol Chem. 2001;276:6853–6860. doi: 10.1074/jbc.M008992200. [DOI] [PubMed] [Google Scholar]
- 18.Côté GP, Luo X, Murphy MB, Egelhoff TT. Mapping of the novel protein kinase catalytic domain of Dictyostelium myosin II heavy chain kinase A. J Biol Chem. 1997;272:6846–6849. doi: 10.1074/jbc.272.11.6846. [DOI] [PubMed] [Google Scholar]
- 19.Medley QG, Gariépy J, Côté GP. Dictyostelium myosin II heavy-chain kinase A is activated by autophosphorylation: Studies with Dictyostelium myosin II and synthetic peptides. Biochemistry. 1990;29:8992–8997. doi: 10.1021/bi00490a016. [DOI] [PubMed] [Google Scholar]
- 20.Egelhoff TT, Croft D, Steimle PA. Actin activation of myosin heavy chain kinase A in Dictyostelium: A biochemical mechanism for the spatial regulation of myosin II filament disassembly. J Biol Chem. 2005;280:2879–2887. doi: 10.1074/jbc.M410803200. [DOI] [PubMed] [Google Scholar]
- 21.Medley QG, Bagshaw WL, Truong T, Côté GP. Dictyostelium myosin II heavy-chain kinase A is activated by heparin, DNA and acidic phospholipids and inhibited by polylysine, polyarginine and histones. Biochim Biophys Acta. 1992;1175:7–12. doi: 10.1016/0167-4889(92)90003-t. [DOI] [PubMed] [Google Scholar]
- 22.Ryazanov AG. Elongation factor-2 kinase and its newly discovered relatives. FEBS Lett. 2002;514:26–29. doi: 10.1016/s0014-5793(02)02299-8. [DOI] [PubMed] [Google Scholar]
- 23.Ryazanov AG, Pavur KS, Dorovkov MV. Alpha-kinases: A new class of protein kinases with a novel catalytic domain. Curr Biol. 1999;9:R43–R45. doi: 10.1016/s0960-9822(99)80006-2. [DOI] [PubMed] [Google Scholar]
- 24.Crawley SW, Côté GP. Determinants for substrate phosphorylation by Dictyostelium myosin II heavy chain kinases A and B and eukaryotic elongation factor-2 kinase. Biochim Biophys Acta. 2008;1784:908–915. doi: 10.1016/j.bbapap.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Matsushita M, Kozak JA, Shimizu Y, McLachlin DT, Yamaguchi H, Wei FY, Tomizawa K, Matsui H, Chait BT, Cahalan MD, Nairn AC. Channel function is dissociated from the intrinsic kinase activity and autophosphorylation of TRPM7/ChaK1. J Biol Chem. 2005;280:20793–20803. doi: 10.1074/jbc.M413671200. [DOI] [PubMed] [Google Scholar]
- 26.Clark K, Middelbeek J, Morrice NA, Figdor CG, Lasonder E, van Leeuwen FN. Massive autophosphorylation of the Ser/Thr-rich domain controls protein kinase activity of TRPM6 and TRPM7. PLoS One. 2008;3:e1876. doi: 10.1371/journal.pone.0001876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo X, Crawley SW, Steimle PA, Egelhoff TT, Côté GP. Specific phosphorylation of threonine by the Dictyostelium myosin II heavy chain kinase family. J Biol Chem. 2001;276:17836–17843. doi: 10.1074/jbc.M009366200. [DOI] [PubMed] [Google Scholar]
- 28.Drennan D, Ryazanov AG. Alpha-kinases: Analysis of the family and comparison with conventional protein kinases. Prog Biophys Mol Biol. 2004;85:1–32. doi: 10.1016/S0079-6107(03)00060-9. [DOI] [PubMed] [Google Scholar]
- 29.Betapudi V, Mason C, Licate L, Egelhoff TT. Identification and characterization of a novel α-kinase with a von Willebrand factor A-like motif localized to the contractile vacuole and Golgi complex in Dictyostelium discoideum. Mol Biol Cell. 2005;16:2248–2262. doi: 10.1091/mbc.E04-07-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg JM, Manning G, Liu A, Fey P, Pilcher KE, Xu Y, Smith JL. The Dictyostelium kinome—Analysis of the protein kinases from a simple model organism. PLoS Genet. 2006;2:e38. doi: 10.1371/journal.pgen.0020038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Proud CG. Signalling to translation: How signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- 32.Fleig A, Penner R. The TRPM ion channel subfamily: Molecular, biophysical and functional features. Trends Pharmacol Sci. 2004;25:633–639. doi: 10.1016/j.tips.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Clark K, Middelbeek J, Dorovkov MV, Figdor CG, Ryazanov AG, Lasonder E, van Leeuwen FN. The α-kinases TRPM6 and TRPM7, but not eEF-2 kinase, phosphorylate the assembly domain of myosin IIA, IIB and IIC. FEBS Lett. 2008;582:2993–2997. doi: 10.1016/j.febslet.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 34.Clark K, Langeslag M, van Leeuwen B, Ran L, Ryazanov AG, Figdor CG, Moolenaar WH, Jalink K, van Leeuwen FN. TRPM7, a novel regulator of actomyosin contractility and cell adhesion. EMBO J. 2006;25:290–301. doi: 10.1038/sj.emboj.7600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamaguchi H, Matsushita M, Nairn AC, Kuriyan J. Crystal structure of the atypical protein kinase domain of a TRP channel with phosphotransferase activity. Mol Cell. 2001;7:1047–1057. doi: 10.1016/s1097-2765(01)00256-8. [DOI] [PubMed] [Google Scholar]
- 36.Scheeff ED, Bourne PE. Structural evolution of the protein kinase-like superfamily. PLoS Comput Biol. 2005;1:e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G. Structural and functional diversity of the microbial kinome. PLoS Biol. 2007;5:e17. doi: 10.1371/journal.pbio.0050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crawley SW, Côté GP. Identification of dimer interactions required for the catalytic activity of the TRPM7 α-kinase domain. Biochem J. 2009;420:115–122. doi: 10.1042/BJ20081405. [DOI] [PubMed] [Google Scholar]
- 39.Abbreviations for the amino acids are as follows: A, Ala; D, Asp; K, Lys; R, Arg; T, Thr; and Y, Tyr.
- 40.Knowles JR. Enzyme-catalyzed phosphoryl transfer reactions. Annu Rev Biochem. 1980;49:877–919. doi: 10.1146/annurev.bi.49.070180.004305. [DOI] [PubMed] [Google Scholar]
- 41.Stock JB, Stock AM, Mottonen JM. Signal transduction in bacteria. Nature. 1990;344:395–400. doi: 10.1038/344395a0. [DOI] [PubMed] [Google Scholar]
- 42.Goudreau PN, Lee PJ, Stock AM. Stabilization of the phospho-aspartyl residue in a two-component signal transduction system in Thermotoga maritima. Biochemistry. 1998;37:14575–14584. doi: 10.1021/bi980869i. [DOI] [PubMed] [Google Scholar]
- 43.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. Caught in the act: The structure of phosphorylated β-phosphoglucomutase from Lactococcus lactis. Biochemistry. 2002;41:8351–8359. doi: 10.1021/bi0202373. [DOI] [PubMed] [Google Scholar]
- 44.Lewis RJ, Brannigan JA, Muchova K, Barak I, Wilkinson AJ. Phosphorylated aspartate in the structure of a response regulator protein. J Mol Biol. 1999;294:9–15. doi: 10.1006/jmbi.1999.3261. [DOI] [PubMed] [Google Scholar]
- 45.Thebault S, Cao G, Venselaar H, Xi Q, Bindels RJ, Hoenderop JG. Role of the α-kinase domain in TRPM6 channel and regulation by intracellular ATP. J Biol Chem. 2008;283:19999–20007. doi: 10.1074/jbc.M800167200. [DOI] [PubMed] [Google Scholar]
- 46.Schmitz C, Perraud AL, Johnson CO, Inabe K, Smith MK, Penner R, Kurosaki T, Fleig A, Scharenberg AM. Regulation of vertebrate cellular Mg2+ homeostasis by TRPM7. Cell. 2003;114:191–200. doi: 10.1016/s0092-8674(03)00556-7. [DOI] [PubMed] [Google Scholar]
- 47.Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 48.Lahiri SD, Zhang G, Dunaway-Mariano D, Allen KN. The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science. 2003;299:2067–2071. doi: 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- 49.Sørensen TL, Moller JV, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 50.Kovári J, Barabas O, Varga B, Bekesi A, Tolgyesi F, Fidy J, Nagy J, Vertessy BG. Methylene substitution at the α-β bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site. Proteins. 2008;71:308–319. doi: 10.1002/prot.21757. [DOI] [PubMed] [Google Scholar]
- 51.Zheng J, Knighton DR, Ten Eyck LF, Karlsson R, Xuong N-H, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 52.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 53.Cho HS, Pelton JG, Yan D, Kustu S, Wemmer DE. Phosphoaspartates in bacterial signal transduction. Curr Opin Struct Biol. 2001;11:679–684. doi: 10.1016/s0959-440x(01)00271-8. [DOI] [PubMed] [Google Scholar]
- 54.Ho MF, Bramson HN, Hansen DE, Knowles JR, Kaiser ET. Stereochemical course of the phospho group transfer catalyzed by cAMP-dependent protein kinase. J Am Chem Soc. 1988;110:2680–2681. [Google Scholar]
- 55.Ryazanova LV, Dorovkov MV, Ansari A, Ryazanov AG. Characterization of the protein kinase activity of TRPM7/ChaK1, a protein kinase fused to the transient receptor potential ion channel. J Biol Chem. 2004;279:3708–3716. doi: 10.1074/jbc.M308820200. [DOI] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 58.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 59.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 60.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 62.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 63.Pollard TD, Korn ED. Acanthamoeba myosin. I. Isolation from Acanthamoeba castellanii of an enzyme similar to muscle myosin. J Biol Chem. 1973;248:4682–4690. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.