Abstract
During the computations performed by the nervous system, its ‘wiring diagram’—the map of its neurons and synaptic connections—is dynamically modified and supplemented by multiple actions of neuromodulators that can be so complex that they can be thought of as constituting a biochemical network that combines with the neuronal network to perform the computation. Thus, the neuronal wiring diagram alone is not sufficient to specify, and permit us to understand, the computation that underlies behaviour. Here I review how such modulatory networks operate, the problems that their existence poses for the experimental study and conceptual understanding of the computations performed by the nervous system, and how these problems may perhaps be solved and the computations understood by considering the structural and functional ‘logic’ of the modulatory networks.
Keywords: neurotransmitter, neuromodulator, neuropeptide, biological system architecture, neural computation, computational modelling
1. Beyond the wiring diagram
Ideas about how the brain works have always looked for inspiration to the current technology of the era. After the pneumatic, hydraulic and mechanical analogies of earlier centuries, in the twentieth century came electrical and electronic analogies such as the telephone exchange and the computer (Churchland 1986; Kirkland 2002). Fundamental to these latter analogies is the concept of a network of interconnected components, and this remains the standard paradigm for thinking about the nervous system today. The nervous system is seen as composed of interconnected subsystems, which themselves are composed of lower-level subsystems, until at the lowest level there are components, such as individual neurons, that are (from the top-down perspective of the network) relatively simple and stereotyped, but there are very many of them and they are interconnected in a rich, structured way to make a very large, complex network. In such a network, given some understanding of the capabilities of the individual components, the key fact is how the components are connected together—the wiring diagram. The wiring diagram specifies and organizes the computation performed by the network that, in the case of the nervous system, emerges ultimately in the behaviour of the animal. Naturally, this paradigm has motivated an intensive effort to determine the wiring diagrams of nervous systems. We now have high-level wiring diagrams for many parts of the vertebrate, including human, brain (Sporns et al. 2004; Grillner & Graybiel 2006; Bullmore & Sporns 2009), wiring diagrams down to the level of individual neurons for some simple circuits, such as the crustacean stomatogastric ganglion (STG; Harris-Warrick et al. 1992a), and even the complete wiring diagram for one entire nervous system, that of the nematode worm Caenorhabditis elegans (White et al. 1986; http://www.wormatlas.org).
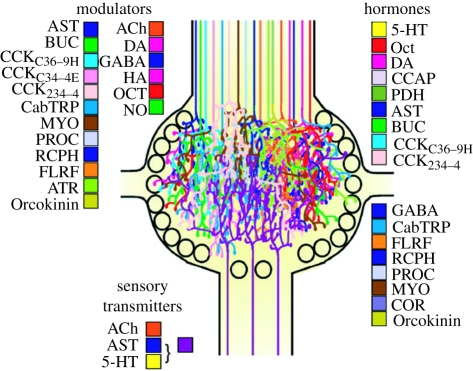
In this article, I will not argue that knowledge of the wiring diagram is not required to understand how a nervous system works. I will argue, however, that it is not sufficient—at least, if the wiring diagram is thought of, as it often is, as a relatively static, hard-wired set of neuronal connections. In this article, I will focus on one reason for this insufficiency: the fact that the neuronal wiring diagram is not static, but rather is dynamically modified by multiple actions of neuromodulators that indeed can be so complex that they can be thought of as constituting a biochemical network that combines with the neuronal network to perform the computation. Thus, the neuronal wiring diagram alone is not sufficient to specify, and permit us to understand, the computation that underlies behaviour. Ironically, it is often in the nervous systems whose neuronal wiring diagrams are best known and that therefore might be thought to be closest to being ‘solved’, such as in the STG, C. elegans, and Drosophila whose nervous system is being proposed as ripe for definitive ‘circuit-cracking’ (Olsen & Wilson 2008), where the greatest multiplicity of neuromodulators and their effects has been identified (Harris-Warrick et al. 1992b; Brownlee & Fairweather 1999; Orchard et al. 2001; Skiebe 2001; Nässel 2002; Taghert & Veenstra 2003; Husson et al. 2007; Ma et al. 2009; Stein 2009). Figure 1, for example, shows, in the now classic diagram of Marder and colleagues, the multiplicity of modulators—probably still a substantial underestimate of the true number (Fu et al. 2005; Ma et al. 2009)—identified in the STG. That the complexity of the modulatory networks appears greatest in these simple systems may, however, merely reflect the fact that these systems are the most completely characterized genetically, biochemically and physiologically: there are already many indications that similar modulatory complexity exists also in the vertebrate brain (Vizi & Lábos 1991; McCormick 1992; Herbert 1993; Hasselmo 1995; Hökfelt et al. 2000: Merighi 2002; Baraban & Tallent 2004; Salio et al. 2006; Abbracchio et al. 2008).
Figure 1.
Multiplicity of neuromodulators identified in the crustacean stomatogastric ganglion. Reproduced with permission from Marder & Bucher (2001).
In this article, I will review how such modulatory networks operate, the problems that their existence poses for the experimental study and conceptual understanding of the computations performed by the nervous system, and how these problems may perhaps be solved and the computations understood by considering the structural and functional ‘logic’ of the modulatory networks. I will be able to present a sketch only; space limitations force me to refer to reviews rather than the vast primary literature and to omit many details and even substantial subsidiary topics, but with the hope that, if the sketch is clear, the reader will readily see how to complete the picture.
2. Multiplicity of neuromodulators
What is a neuromodulator? Neurochemical signalling agents have traditionally been classified primarily by their chemical identity, distinguishing for example between the classical small-molecule neurotransmitters, biogenic amines and neuropeptides. From the systems-level point of view emphasized in this article, however, it is more relevant to distinguish primarily between transmitter and modulator functions. Whether a signalling agent functions as a transmitter or a modulator depends ultimately on the spatial and temporal scale on which the agent acts. If the agent acts so locally in both space and time that it merely communicates the current activity of one component of the neuronal network to another component—for example, the fact that a spike has just occurred in a presynaptic neuron to a postsynaptic neuron—then it is acting as a transmitter that merely implements one of the connections of the neuronal wiring diagram and so, for the purposes of this article, can be subsumed into that diagram. Indeed, such signalling has been termed ‘wiring transmission’ (Agnati et al. 1995; Zoli & Agnati 1996). A modulator, in contrast, acts globally on multiple components, over longer times, or usually both, so that its effects are not directly correlated with the current activity of any particular component or connection of the neuronal network: the modulator does not mediate that activity but rather modifies or modulates it. It is this spatial and temporal dissociation from the activity of the neuronal network that is responsible for many of the problems posed by neuromodulation as well as for its computational power.
There is of course considerable correspondence between the traditional chemical classes of signalling agents and this functional classification. In particular, the transmitter and modulator roles are played by certain classes of molecules that are adapted for these roles by the cell biology of their synthesis, storage and release. In the nervous system, the local transmitter role that subserves wiring transmission is played predominantly by classical small-molecule neurotransmitters such as ACh, GABA and glutamate, which signal locally because they are released from neurons by rapid spike-activated presynaptic mechanisms, and then rapidly open postsynaptic ligand-gated ion channels, at discrete, highly organized synapses that are ‘closed’ in that the transmitter remains confined within them (Zoli & Agnati 1996). The global modulator role, on the other hand, is played predominantly by biogenic amines such as dopamine, norepinephrine, serotonin and histamine, by adenosine and adenine nucleotides (Abbracchio et al. 2008; Gourine et al. 2009), by neuropeptides (discussed in more detail below), and probably by a number of other classes of molecules (Zoli et al. 1999; Barañano et al. 2001; Hofer & Lefkimmiatis 2007), the best established of which are gases such as nitric oxide (NO; Barañano et al. 2001; Guix et al. 2005). These agents signal globally because they are released, from both neurons and glia, more slowly at less organized, ‘open’ synapses or indeed non-synaptic sites from which they are then able to diffuse for long times over long distances through the intercellular space, or travel even longer distances as neurohormones through the circulatory system, in what has been termed ‘volume transmission’ (Agnati et al. 1995; Stjärne & Stjärne 1995; Zoli & Agnati 1996; Zoli et al. 1999). These agents then furthermore exert inherently slow actions through metabotropic receptors coupled to second-messenger cascades and other slow intracellular signalling processes. Perhaps the most extreme case of such global volume signalling is that of the gaseous NO, which passes through membranes and so can diffuse from the interior of one cell to directly modulate intracellular processes throughout the entire volume of surrounding tissue, largely irrespective of cellular boundaries.
The accumulating evidence has, however, also revealed considerable overlap in all these properties. In addition to acting through metabotropic receptors, serotonin, the adenine nucleotides, and even some neuropeptides (Lingueglia et al. 1995) open ligand-gated channels. ATP often has the hallmarks of a local transmitter, even being coreleased from the same synaptic vesicles as a classical transmitter such as ACh or GABA (Abbracchio et al. 2008; Zimmermann 2008). In many invertebrates, dopamine, histamine and serotonin function prominently as local transmitters (Gerschenfeld 1973). Conversely, there is abundant evidence that, upon release even at ‘closed’ synapses, the classical transmitters not only rapidly open ligand-gated channels but also exert parallel but slower modulatory actions through metabotropic receptors (see numerous examples in Katz 1999). Furthermore, ‘closed’ synapses can become ‘open’ through transmitter spillover at times of high release or reduced strength of the mechanisms that confine the transmitter within the synapse, so that the released transmitter acquires a very different spatio-temporal profile and can reach additional receptors that may mediate additional, modulatory actions (Agnati et al. 1995; Zoli & Agnati 1996). The overall picture emerges not of an absolute differentiation between transmitters and modulators but rather of a continuum between the transmitter and modulator functions, along which the signalling agents occupy various, often multiple, positions that can shift dynamically depending on the circumstances. Furthermore, as will be seen in the rest of this article, the transmitter and modulator functions are intimately intertwined at the points of contact between the neuronal and modulatory networks.
The neuropeptides (Strand 1999; Hökfelt et al. 2000; Skiebe 2001; Nässel 2002, 2009; Baraban & Tallent 2004; Landgraf & Neumann 2004; Salio et al. 2006) have a special place in this account because, in addition to being quintessential modulators in the sense just described, they are so diverse. Whereas there are only a few molecular species of the other modulators, there is a seemingly infinite variety of neuropeptide amino acid sequences (e.g. Kobayashi & Muneoka 1990; Schoofs et al. 1997; Brownlee & Fairweather 1999; Orchard et al. 2001; Taghert & Veenstra 2003; Boonen et al. 2009). The combinatorial nature of the neuropeptide structure and its linear encoding in the genome, then operated on by the evolutionary processes of coding-sequence and gene duplication followed by divergence under relaxed evolutionary pressure (Niall 1982; Strand 1999), has led to the encoding of multiple variant neuropeptide sequences in most neuropeptide genes, and multiple genes encoding neuropeptides that are closely as well as more distantly related in the genome. The complexity of the actual neuropeptidergic signalling is (fortunately) somewhat reduced by the fact that all of the neuropeptides synthesized by a neuron are usually copackaged into the same exocytotic vesicles and coreleased. Selective sorting of the neuropeptides for selective release from different processes of the same neuron is known, however (Sossin & Scheller 1991; Perone et al. 1997). Furthermore, different neurons release different complements of the neuropeptides, thanks to such mechanisms as alternative splicing of the neuropeptide mRNAs (Buck et al. 1987; Benjamin & Burke 1994; Perone et al. 1997) and, of course, the transcription of different subsets of the neuropeptide genes. It is now clear that very many neurons—in some nervous systems, seemingly all neurons—release some complement of neuropeptides as cotransmitters alongside their ‘primary’ classical transmitter (Kupfermann 1991; Maggi 1995; Lundberg 1996; Hökfelt et al. 2000; Merighi 2002). Strikingly illustrating the cell-biological basis of the transmitter–modulator distinction, the large dense-core vesicles that contain the neuropeptides are typically segregated, even within the same synaptic terminals, away from the small clear vesicles that contain the classical transmitter (Merighi 2002; Salio et al. 2006), to sites from which the neuropeptides are released more diffusely by smaller, slower elevations of intracellular Ca2+ (Verhage et al. 1994), so that the release of the neuropeptides and the classical transmitter can occur to some degree independently, for example in response to different firing patterns of the neuron. Altogether, it is the diversity of the neuropeptides that accounts for much of the modulator multiplicity in a system such as the crustacean STG (figure 1). The most recent study of a representative crustacean ‘neuropeptidome’ has identified 142 neuropeptides belonging to 17 different families (Ma et al. 2009).
3. Multiplicity of neuromodulatory actions
An immense amount of data has accumulated to show what the modulators can do. The standard experimental paradigm to gather such data is to exogenously apply a particular modulator and observe the perturbation of a particular measured variable in the system. It is then assumed that the modulator, if released endogenously within the system, is at least capable of exerting the same action. A fair summary of the data gathered over the past several decades with this approach is that modulators are capable of modulating essentially every variable, every process, at all levels of organization of the nervous system. Perturbation can be observed of the activities of the molecules and pathways of gene expression, intermediary metabolism and intracellular signal transduction in both neurons and glial cells, of essentially every type of membrane ion channel, transmission at many synapses, and the electrical behaviour of neurons and neuronal circuits of all kinds (numerous examples can be found in Lundberg 1996; Katz 1999; Marder & Thirumalai 2002; Dickinson 2006; Abbracchio et al. 2008). Moreover, the nervous system is very dense with these modulatory actions. Examining just a single synapse, for instance, we observe modulation of the propagation of action potentials into the presynaptic terminal, of multiple presynaptic ion channels, of the presynaptic Ca2+ concentration, of the molecular machinery of transmitter release and the actual release of the classical transmitter as well as any modulatory cotransmitters, then of multiple postsynaptic ion channels and the postsynaptic electrical properties that govern the generation of a postsynaptic action potential, and so, overall, of virtually every aspect of the transmission of information through the synapse (Vizi & Lábos 1991; Lundberg 1996; Marder & Thirumalai 2002).
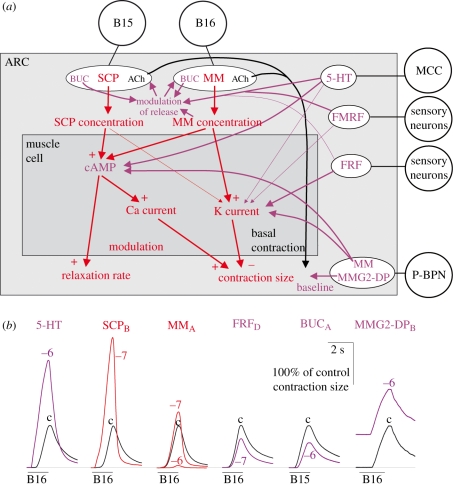
One experimental system in which the issues raised in this article have been particularly well investigated, and which, along with the crustacean STG, I shall discuss as an example, is the feeding system of the sea slug Aplysia (Kupfermann 1974; Weiss et al. 1993; Hooper et al. 1999). In this system, although much is now known also about the modulation of the central pattern generator (CPG) that drives the rhythmic feeding movements (e.g. Fujisawa et al. 1999; Furukawa et al. 2001; Sweedler et al. 2002; Proekt et al. 2005; Vilim et al. 2010), the best understood modulation is in the periphery of the neuromuscular plant that executes the movements. Figure 2a shows a schema of the modulation in a representative muscle, the accessory radula closer (ARC) muscle. Numerous modulators, in particular neuropeptides belonging to the small cardioactive peptide (SCP), myomodulin (MM), buccalin, FMRFamide and other families, are released as cotransmitters from the muscle's two motorneurons, B15 and B16, and from additional modulatory and even sensory neurons (for details and references see the figure legend). These modulators then act presynaptically to modulate the release of the motorneurons' primary, contraction-inducing transmitter, ACh, as well as of the modulatory cotransmitters—in some cases, therefore, of themselves through a feedback loop—and postsynaptically on the muscle cells to modulate their intracellular second-messenger systems, membrane ion channels and ultimately multiple parameters—baseline, latency, amplitude, relaxation rate—of the shape of the muscle's contractions (figure 2a,b). This example illustrates furthermore how the modulatory events in the nervous system join seamlessly with, and for the purposes of this article are no different from, those in the other tissues with which the nervous system interacts and indeed most other tissues of the body (see also Furness et al. 1992; Maggi 1995; Stjärne & Stjärne 1995; Lundberg 1996; Hooper et al. 1999; Gourine et al. 2009).
Figure 2.
Network of modulatory actions that shapes the contractions of the ARC muscle of Aplysia. (a) Schema of the ARC muscle (grey box), its two motorneurons B15 and B16 (circles at top) that release ACh to elicit the basal contractions of the muscle (black arrow) but also modulatory neuropeptides belonging to the SCP, MM and buccalin (BUC) families, other neurons (circles down right-hand side) that release additional modulators, and the network of the principal modulatory actions (coloured arrows). Thick arrows denote strong actions, thin arrows weaker actions. The subnetwork analysed in figure 3 is shown in red, the rest of the modulatory network in purple. Modified from Brezina et al. (2003a) and based in the first instance on the body of previous work summarized there, then brought up to date with the more recent findings of Orekhova et al. (2003), Proekt et al. (2005) and Vilim et al. (2010). (b) The diversity of ARC muscle contraction shapes produced by the various modulators. Representative contractions elicited in vitro by firing of either motor neuron B15 or B16 under control conditions (‘c’) and after exogenous application of 10−7 or 10−6 M (‘− 7’, ‘− 6’) of the modulators. Modified from Hooper et al. (1999), with the MMG2-DP traces taken from an experiment of Proekt et al. (2005). Abbreviations: MMG2-DP, myomodulin gene 2-derived peptides; FMRF, FMRFamide peptides; FRF, FRFamide peptides; 5-HT, serotonin; MCC, metacerebral cells; P-BPN, pedal–buccal projecting neurons.
4. The neuromodulatory network
Figure 2a exhibits a very characteristic feature of modulatory pathways: their extensive divergence and convergence (Brezina & Weiss 1997). Each modulator exerts multiple actions, and the same action is exerted by multiple modulators, and similarly at all levels of the modulatory system so that it forms a single, richly interconnected network, often with multiple feedback loops. As a result, a perturbation at any point in the network—the release of a single modulator, for example—will ramify throughout the network to perturb many of its components.
The modulatory network therefore has a structure, with its own ‘wiring diagram’, that is no less complex than that of the neuronal network in the same region of nervous tissue. In important respects, it is more complex. Whereas the discrete nodes and arrows of the network diagram constitute a natural representation of the discrete neurons and synaptic connections of the actual neuronal network, they constitute only a very abstract representation of the actual events in the modulatory network, which are inescapably continuous in three-dimensional space as well as time. More realistically, we should picture cocktails of multiple modulators released from many neurons, diffusing through the extracellular space over different distances to create spatio-temporal concentration profiles that are highly dependent on the dynamics of the release and the particular geometry of the tissue, and then exerting their many actions on a multiplicity of time-scales, all this dynamically changing with different patterns of neuronal firing and the state of the nervous system (Furness et al. 1992; Stjärne & Stjärne 1995; Zoli & Agnati 1996; Zoli et al. 1999; Gourine et al. 2009; Nässel 2009).
The neuronal and modulatory networks, moreover, are intimately coupled through their myriad interactions so that information flows repeatedly from one network to the other and back again. Thus, the neuronal firing that constitutes the activity of the neuronal network typically leads to the release of the neurons' modulatory cotransmitters—these are therefore intrinsic modulators in the terminology of Katz & Frost (1996)—that modulate, in turn, that firing. At synapses, even phenomena such as classical synaptic plasticity (Fisher et al. 1997; Zucker & Regehr 2002) that normally are interpreted as inherent to the release of the classical transmitter itself, and so part of the intrinsic dynamics of the neuronal network, may actually be due in some cases to slow modulatory actions of coreleased neuropeptides (Fox & Lloyd 2001; Koh & Weiss 2005). In the crustacean STG, modulatory neurons projecting from other ganglia (coloured lines leading to the STG in figure 1) release modulators that initiate and maintain the rhythmic motor patterns that are produced by the neuronal STG network, and those patterns, in turn, impose a rhythmicity also on the modulator release from the projection neuron terminals (Nusbaum et al. 2001; Nusbaum 2002). Altogether, it is through this intimate coupling of the neuronal and modulatory networks that the application of a modulator can affect seemingly any component not only of the modulatory but also the neuronal network, as is observed.
5. Problems posed by neuromodulation
The existence of the modulatory network and its coupling to the neuronal network poses a number of problems for our attempts to investigate and understand how the nervous system performs its computations, indeed what computations it performs.
First, we have very imperfect knowledge of what modulators are contained in the cocktail of modulators that acts at any point in the nervous system. With the neuropeptides in particular, when each neuron can express its own individual complement of multiple neuropeptides, to map this expression comprehensively is a heroic task. Even in such a well-explored system as the Aplysia feeding system, entire new neuropeptide families continue to be discovered (Fujisawa et al. 1999; Furukawa et al. 2001; Sweedler et al. 2002; Proekt et al. 2005; Vilim et al. 2010). The issue is complicated by the fact that, like all biological systems, the modulatory system is a contingently evolved system that in some respects is bound to be far from optimized, and, as already mentioned, the neuropeptides in particular pay little penalty for multiplication and structural diversification. Thus, it is not surprising that some neuropeptides that are expressed and released into the cocktail appear to be redundant, or more properly degenerate (Edelman & Gally 2001), variants (e.g. Brezina et al. 1995; Hewes et al. 1998) or even appear inactive (Bowers 1994). It is difficult to be sure that they really are so, of course, when the modulatory actions, too, are only imperfectly understood.
The modulatory actions are imperfectly understood because not only the modulators but also the rest of the modulatory network is known only in part, with many additional network nodes remaining unidentified and many potential connections untested. However, it is also because the standard experimental paradigm, mentioned above, that uses exogenous modulator application to identify the modulatory actions is grossly inadequate. Typically, only one modulator, at a series of concentrations, is applied at a time. Mixtures of even just two or three modulators—when the real physiological cocktail may well contain dozens!—have been systematically tested only in a handful of studies (Wood 1995; Brezina et al. 1996; Dickinson et al. 1997; Djokaj et al. 2001; Mesce et al. 2001; Svensson et al. 2001). Mixtures of more than three modulators have not been studied because, even when the identities of more modulators are known, their physiologically relevant concentrations are not known, and systematic testing of all possible combinations of concentrations presents an experimentally insuperable combinatorial problem. However, those few studies that have tested two modulators have invariably found not only strong concentration-dependence of their actions but strong nonlinearities in their interactions, so that the actions of either modulator in the presence of the other were often very different from its actions alone (see further below). This implies that adding another modulator to the studied mixture, or perhaps in the future discovering a novel modulator in the system, may change all of the results obtained so far using this paradigm. The finding that a modulator can exert a certain action in such experiments does not mean that it does exert that action in the real system.
Yet a further problem with the standard paradigm is that the exogenous modulator application, even at the real average concentration, does not reproduce the complex real spatio-temporal concentration profile that can be critical for the proper expression of the modulator actions (Brezina et al. 1997). Rhythmic oscillations of concentration, for example, are required in the crustacean STG (Nusbaum 2002) and for the actions of a number of vertebrate neuropeptide hormones (Strand 1999). Similarly in the spatial domain, the precise distribution of the modulator in the tissue can have profound functional and computational consequences (Stjärne & Stjärne 1995; Nässel 2009), as has been modelled for NO signalling in different geometries of NO sources and targets (Ott et al. 2007) and reported, once more, in the STG where the sculpting of concentration profiles by extracellular peptidases so as to restrict the released modulatory neuropeptides to only one or another part of the ganglion appears to play a significant functional role (Nusbaum 2002; Wood & Nusbaum 2002; Stein 2009).
Finally, by modifying the electrical properties of the neurons in the neuronal network and their synaptic connections, neuromodulators can profoundly change the functional—as opposed to the anatomical—connectivity of the neuronal network (Dickinson 2006). The first and still the best example is again provided by the crustacean STG, where the different modulators can variously reconfigure the STG neurons into different functional assemblies that generate different motor patterns (Dickinson & Moulins 1992; Nusbaum et al. 2001; Scholz et al. 2001; Skiebe 2001; Stein 2009). Thus, until the modulation is understood, even the basic neuronal wiring diagram, apparently so hard-wired and well defined, becomes uncertain.
6. Computation performed by the neuromodulatory network
Why is the neuromodulatory system so complex? A more tractable subsidiary question that has often been asked (Segal 1983; Bloom 1984; Kupfermann 1991; Marder et al. 1995; Brezina & Weiss 1997) is, why are there so many neuromodulators? The answer has usually been sought by assuming that the structure of the modulatory system has a functional logic that aims, fundamentally, to provide a separate, unique addressability and controllability of each of the systems' ‘outputs’—the various modulator actions, or sometimes the neurons of the coupled neuronal system—by the ‘input’ modulators. Since there are many outputs, and, in contrast to the anatomically labelled lines of the neuronal wiring diagram, the modulators are all mixed together in the modulator cocktail, there must then be at least as many input modulators as output modulator actions (Brezina & Weiss 1997). Such a system could in principle consist of chemically labelled ‘signalling wires’ (Weng et al. 1999) of one-to-one mapping of modulators to actions; in such a system, we could speak of the function of each modulator. As already discussed, however, all real modulatory systems are divergent–convergent networks in which each modulator exerts many actions, and each action is exerted by many modulators. Unique addressability and controllability can still be achieved, nevertheless, in a combinatorial manner, no longer by single modulators but by the cocktail of all of the modulators controlling simultaneously the ensemble of all of the actions. It is then no longer meaningful to attribute any specific function to any modulator alone (Brezina & Weiss 1997; Olypher & Calabrese 2007; Boonen et al. 2009).
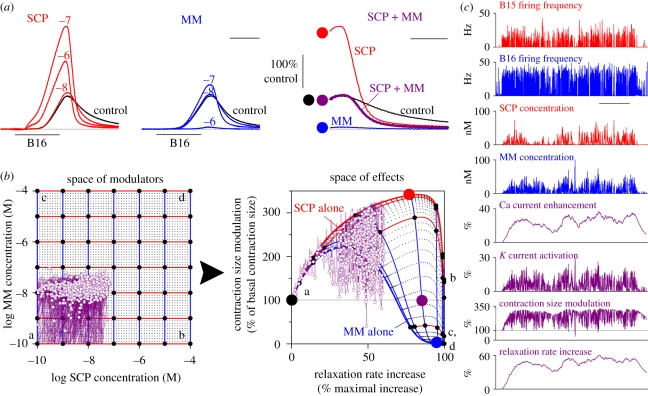
Figure 3 summarizes an analysis along these lines of the core of the peripheral modulatory network in the Aplysia feeding system, specifically of the subnetwork shown in red in figure 2, of just two of the modulators, SCP and MM, released from the motorneurons B15 and B16, respectively, then each modulating both the amplitude and the relaxation rate of the ARC muscle's contractions. Figure 3b shows the mapping, generated by a realistic mathematical model of the subnetwork, of the space of all possible combinations of the concentrations of SCP and MM to the space of all possible magnitudes of the two effects. Note how in combination, although not either one alone, SCP and MM uniquely address and control a large region of the effect space. Note also the nonlinearity of the mapping and how the addition of SCP changes the effect of MM and vice versa (Brezina et al. 1996). As already noted, of course, this implies that the entire mapping might be changed by the addition of yet a third modulator, several candidates for which certainly exist in the system (figure 2a). Figure 3a shows experimental confirmation of the mathematical analysis that indeed combinations of the effects on contraction amplitude and relaxation rate can be achieved by the combinations of SCP and MM that cannot be achieved by SCP or MM alone. Generalizing to higher dimensions, it is tempting to speculate that the additional modulators in the system offer the possibility of simultaneously controlling even more of the features of the contractions (figure 2b).
Figure 3.
Analysis of the actions of modulator combinations in the Aplysia ARC muscle system. This partial analysis considers the combinatorial action of just two modulators, SCP released from motor neuron B15 and MM released from motor neuron B16, through the subnetwork of actions shown in red in figure 2a, on just two parameters, the size and the relaxation rate, of the ARC muscle contractions. (a) Representative ARC muscle contraction shapes, recorded as in figure 2b, produced by various exogenously applied steady concentrations of SCP alone (red), MM alone (blue) and both SCP and MM applied together (purple; the peak and relaxation phase of just one contraction is shown; scale bar 1 s, 500 ms). (b) Mapping from the space of all combinations of SCP and MM concentrations (over the range from 10−10 to 10−4 M) to the space of all combinations of the effects on contraction size and relaxation rate. The grid of small dots in the two spaces shows the steady-state mapping generated by the computational model of Brezina et al. (1996, 2003a). The larger black dots and blue and red curves show how the steady-state mapping is traversed as the SCP concentration varies in the presence of constant MM concentration (red curves) or the converse (blue curves); the letters ‘a’–‘d’ identify the mapping of the four corners of the modulator space for orientation. The large black, red, blue and purple dots mark the approximate locations of the contraction shapes in (a), right. The purple overlay then shows the dynamical trajectory through the modulator and effect spaces of the entire meal modelled in (c), in the modulator space plotting against each other the SCP and MM concentration waveforms shown in (c) and in the effect space the contraction size and relaxation rate waveforms, plotting either the entire continuous waveforms (thin purple curve) or just their values at the end of the retraction phase of each feeding cycle (small purple circles). (c) Modelled waveforms of the principal variables during an entire meal eaten by a real Aplysia in vivo, comprising 749 feeding cycles over approximately 2.5 h, during which the firing frequencies of the motorneurons B15 and B16 (top two waveforms) were recorded with chronically implanted electrodes by Horn et al. (2004) and were then used by Brezina et al. (2005) to drive the computational model of Brezina et al. (2003a). Scale bar, 30 min. (a,b) modified from Brezina et al. (2003a) and (c) from Brezina et al. (2005).
Similar combinatorial thinking, although overlaid by many details, informs also the work on the crustacean STG, where the different modulatory neuropeptides released from different projection neurons both diverge and converge to act on partially overlapping complements of ion currents, synaptic connections and STG neurons so as to sculpt a number of multidimensional motor patterns (Nusbaum et al. 2001; Marder & Thirumalai 2002; Stein 2009).
The multidimensional combinatorial mechanism just sketched out automatically operates in modulatory networks simply in consequence of their network structure. However, the structure appears unlikely to have evolved solely to implement this rather logically severe mechanism. Indeed, recent work in systems biology strongly suggests an alternative set of primary design criteria. It is striking to what extent the complex network structure of the modulatory system resembles the structures that have now been well mapped out for intracellular biochemical networks of various kinds—protein–protein interaction networks, G-protein, second-messenger and other signal-transduction networks, and genetic networks (Hille 1992; Bray 1995; Weng et al. 1999; Jordan et al. 2000; Alon 2007)—into which the modulatory network indeed transforms at the interface of the cell membrane (Boonen et al. 2009). Similar questions concerning the functional logic of divergence, convergence, pleiotropy and specificity have been asked and answered rather similarly in these networks (Ross 1989; Gudermann et al. 1996; Jordan et al. 2000). It is therefore tempting to extend to the modulatory network also the more general design principles that have been postulated for these networks.
These principles are of two kinds. First, the nonlinearities that are generated by the properties of the nodes and connections at various sites within the network perform various local computations. In modulatory networks, more than sufficient scope for very complex nonlinearities, and so complex computations, appears to exist, for example, in the interactions between multiple modulators at the synapses where they modulate each other's release (Vizi & Lábos 1991; Stjärne & Stjärne 1995; Lundberg 1996; Salio et al. 2006)—sometimes even differentially the release of just a specific subset of the total modulator complement (DeLong et al. 2009)—and in their modulation, through intracellular signal-transduction pathways, of each other's receptors and so actions (Sebastião & Ribeiro 2000). Rooted in such interactions are then higher-order functional phenomena such as metamodulation (Katz & Edwards 1999; Stein 2009), the gating of the actions of one modulator by another (Dickinson et al. 1997; Svensson et al. 2001), complex dependence of the expression of the modulator actions on the state and activity of the underlying neuronal network (Ito & Schuman 2008; Sakurai & Katz 2009), and ultimately, it has been proposed, even such top-level functionalities of the vertebrate nervous system as the representations of memory, salience, reward, expectation and uncertainty (Hasselmo 1995; Yu & Dayan 2005; Cragg 2006). Very complex computations are inherent also in the processing of the released modulators—in particular, the purines and the neuropeptides—by the extracellular processing enzymes that transform the cocktail of active modulators into another cocktail with different activities, with dynamics that themselves can be modulated (Dale & Gilday 1996; Konkoy & Davis 1996; Dale 2002; Nusbaum 2002; Huxtable et al. 2009). In more abstract terms, the computations can be modelled as mathematical operations (Kupfermann 1991) or logical operations such as associativity (Dale 2002), coincidence detection (Bourne & Nicoll 1993; Offermanns & Schultz 1994) and decision-making (Helikar et al. 2008). Altogether, the modulatory network thus offers a powerful example of the idea that computation can be performed by biochemical networks just as well as it can be performed by traditional neuronal networks (see Bray 1995, 2009; Katz & Clemens 2001; Koch 1999, ch. 20).
In most nervous systems, as I have already argued, it is actually not the modulatory network alone but rather an intimate combination of the modulatory and neuronal networks that performs the computations. There are, however, some systems where the computation is truly mostly modulatory. This may be the case in the enteric ‘brain’ of the vertebrate gut (Furness et al. 1992; Strand 1999), the vertebrate hypothalamic system (Leng & Ludwig 2006), and other systems where the dynamics of release, diffusion, and feedback action of the released modulators can give rise to such dynamic phenomena as oscillations and travelling waves of modulatory signalling (Přibyl et al. 2003).
In this respect, the Aplysia feeding system again presents an instructive case study, since, in the neuromuscular periphery, essentially all of the system's complexity reflects the operation of the modulatory network. Specifically, an extended analysis (Brezina et al. 2003a,b, 2005) has revealed how particular dynamics of the modulatory processes can help endow the modulatory network with a truly independent computational role. Figure 3c, for example, plots the principal variables of the mathematical model of the system when the model was driven by firing patterns of the motorneurons B15 and B16 that were previously recorded with chronically implanted electrodes during an approximately 2.5 h meal consumed by a real animal. The purple trajectories in figure 3b show the corresponding motions through the modulator and effect spaces. The key point to note is that whereas some of the modulatory variables have fast dynamics, others—in particular, the modulation of the calcium current and of the relaxation rate—are much slower than the characteristic time-scale of the motorneuronal firing input. These slow dynamics act to decouple the modulatory network from the instantaneous motorneuronal input, endowing the network with its own autonomous memory of past activity that filters the current motorneuronal input in a history-dependent manner. At the same time, there is reason to believe that the combination of the fast and slow dynamics of the different variables endows the network with a multidimensional robustness whereby the modulated muscle contractions and movement are, at least on average, always optimal even as they vary greatly in response to a highly variable feeding environment (Brezina et al. 2005).
Such conclusions lead directly to the second design principle that the example of other networks also suggests for modulatory networks: that, quite apart from the specific identities of any particular nodes and connections and their local computations, the complex network structure as a whole is an end in itself. It provides multiple compensatory pathways of information and activity flow and feedback loops that allow autoregulation and stability of outputs (Beckskei & Serrano 2000), indeed usually multiple outputs from the same multifunctional network (Bhalla & Iyengar 1999; Jordan et al. 2000). Concomitantly, the structure provides what is arguably the most important global attribute of a biological system selected for by evolution (Kitano 2004), robustness with respect to parameter variation and stochastic noise as well as resistance to and graceful degradation upon damage (Barkai & Leibler 1997; Alon et al. 1999; Gonze et al. 2002; Kitano 2004; Li et al. 2004).
If we attribute such features to the modulatory networks as well, their study becomes a branch of systems biology and can be pursued by its methods (Boonen et al. 2009). We may, for instance, expect to find in modulatory networks such features of the intracellular biochemical networks as modularity (Qi & Ge 2006), perhaps involving the repetition of simple structural motifs that perform elementary computational operations (Bourne & Nicoll 1993; Milo et al. 2002; Ma'ayan et al. 2005; Prill et al. 2005; Brandman & Meyer 2008). Given the complexity of modulatory networks, the methods for their study are increasingly likely to involve mathematical modelling (Hasselmo 1995; Brezina et al. 1996, 2003a,b; Fellous & Linster 1998), analytical methods such as multidimensional sensitivity analysis, multivariable regression, and other methods that examine the global structure of the network (Olypher & Calabrese 2007; Weaver & Wearne 2008), and the application of engineering methods and principles to ‘reverse-engineer’ the biological system to understand simultaneously its computational mechanisms and the computational problems to which those mechanisms provide the solution (Hartwell et al. 1999; Csete & Doyle 2002; El-Samad et al. 2005; Tomlin & Axelrod 2005; Reeves & Fraser 2009).
Acknowledgements
The author's work described in this paper was funded by US National Institutes of Health grant NS41497.
Footnotes
One contribution of 8 to a Theme Issue ‘Neuronal network analyses: progress, problems, and uncertainties’.
References
- Abbracchio M. P., Burnstock G., Verkhratsky A., Zimmermann H.2008Purinergic signalling in the nervous system: an overview. Trends Neurosci. 32, 19–29 (doi:10.1016/j.tins.2008.10.001) [DOI] [PubMed] [Google Scholar]
- Agnati L. F., Zoli M., Strömberg I., Fuxe K.1995Intercellular communication in the brain: wiring versus volume transmission. Neuroscience 69, 711–726 (doi:10.1016/0306-4522(95)00308-6) [DOI] [PubMed] [Google Scholar]
- Alon U.2007An introduction to systems biology: design principles of biological circuits Boca Raton, FL: Chapman & Hall/CRC [Google Scholar]
- Alon U., Surette M. G., Barkai N., Leibler S.1999Robustness in bacterial chemotaxis. Nature 397, 168–171 (doi:10.1038/16483) [DOI] [PubMed] [Google Scholar]
- Baraban S. C., Tallent M. K.2004Interneuronal neuropeptides—endogenous regulators of neuronal excitability. Trends Neurosci. 27, 135–142 (doi:10.1016/j.tins.2004.01.008) [DOI] [PubMed] [Google Scholar]
- Barañano D. E., Ferris C. D., Snyder S. H.2001Atypical neural messengers. Trends Neurosci. 24, 99–106 (doi:10.1016/S0166-2236(00)01716-1) [DOI] [PubMed] [Google Scholar]
- Barkai N., Leibler S.1997Robustness in simple biochemical networks. Nature 387, 913–917 (doi:10.1038/43199) [DOI] [PubMed] [Google Scholar]
- Beckskei A., Serrano L.2000Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (doi:10.1038/35014651) [DOI] [PubMed] [Google Scholar]
- Benjamin P. R., Burke J. F.1994Alternative mRNA splicing of the FMRFamide gene and its role in neuropeptidergic signalling in a defined neural network. BioEssays 16, 335–342 (doi:10.1002/bies.950160508) [DOI] [PubMed] [Google Scholar]
- Bhalla U. S., Iyengar R.1999Emergent properties of networks of biological signaling pathways. Science 283, 381–387 (doi:10.1126/science.283.5400.381) [DOI] [PubMed] [Google Scholar]
- Bloom F. E.1984The functional significance of neurotransmitter diversity. Am. J. Physiol. (Cell Physiol.) 246, C184–C194 [DOI] [PubMed] [Google Scholar]
- Boonen K., Creemers J. W., Schoofs L.2009Bioactive peptides, networks and systems biology. BioEssays 31, 300–314 (doi:10.1002/bies.200800055) [DOI] [PubMed] [Google Scholar]
- Bourne H. R., Nicoll R.1993Molecular machines integrate coincident synaptic signals. Cell 72/Neuron 10, 65–75 [DOI] [PubMed] [Google Scholar]
- Bowers C. W.1994Superfluous neurotransmitters? Trends Neurosci. 17, 315–320 (doi:10.1016/0166-2236(94)90168-6) [DOI] [PubMed] [Google Scholar]
- Brandman O., Meyer T.2008Feedback loops shape cellular signals in space and time. Science 322, 390–395 (doi:10.1126/science.1160617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray D.1995Protein molecules as computational elements in living cells. Nature 376, 307–312 (doi:10.1038/376307a0) [DOI] [PubMed] [Google Scholar]
- Bray D.2009Wetware: a computer in every living cell. New Haven, CT: Yale University Press [Google Scholar]
- Brezina V., Weiss K. R.1997Analyzing the functional consequences of transmitter complexity. Trends Neurosci. 20, 538–543 (doi:10.1016/S0166-2236(97)01120-X) [DOI] [PubMed] [Google Scholar]
- Brezina V., Bank B., Cropper E. C., Rosen S., Vilim F. S., Kupfermann I., Weiss K. R.1995Nine members of the myomodulin family of peptide cotransmitters at the B16-ARC neuromuscular junction of Aplysia. J. Neurophysiol. 74, 54–72 [DOI] [PubMed] [Google Scholar]
- Brezina V., Orekhova I. V., Weiss K. R.1996Functional uncoupling of linked neurotransmitter effects by combinatorial convergence. Science 273, 806–810 (doi:10.1126/science.273.5276.806) [DOI] [PubMed] [Google Scholar]
- Brezina V., Orekhova I. V., Weiss K. R.1997Control of time-dependent biological processes by temporally patterned input. Proc. Natl Acad. Sci. USA 94, 10 444–10 449 (doi:10.1073/pnas.94.19.10444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brezina V., Orekhova I. V., Weiss K. R.2003aNeuromuscular modulation in Aplysia. I. Dynamic model. J. Neurophysiol. 90, 2592–2612 (doi:10.1152/jn.01091.2002) [DOI] [PubMed] [Google Scholar]
- Brezina V., Orekhova I. V., Weiss K. R.2003bNeuromuscular modulation in Aplysia. II. Modulation of the neuromuscular transform in behavior. J. Neurophysiol. 90, 2613–2628 (doi:10.1152/jn.01093.2002) [DOI] [PubMed] [Google Scholar]
- Brezina V., Horn C. C., Weiss K. R.2005Modeling neuromuscular modulation in Aplysia. III. Interaction of central motor commands and peripheral modulatory state for optimal behavior. J. Neurophysiol. 93, 1523–1556 (doi:10.1152/jn.00475.2004) [DOI] [PubMed] [Google Scholar]
- Brownlee D. J. A., Fairweather I.1999Exploring the neurotransmitter labyrinth in nematodes. Trends Neurosci. 22, 16–24 (doi:10.1016/S0166-2236(98)01281-8) [DOI] [PubMed] [Google Scholar]
- Buck L. B., Bigelow J. M., Axel R.1987Alternative splicing in individual Aplysia neurons generates neuropeptide diversity. Cell 51, 127–133 (doi:10.1016/0092-8674(87)90017-1) [DOI] [PubMed] [Google Scholar]
- Bullmore E., Sporns O.2009Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198 (doi:10.1038/nrn2575) [DOI] [PubMed] [Google Scholar]
- Churchland P. S.1986Neurophilosophy: toward a unified science of the mind/brain Cambridge, MA: MIT Press [Google Scholar]
- Cragg S. J.2006Meaningful silences: how dopamine listens to the ACh pause. Trends Neurosci. 29, 125–131 (doi:10.1016/j.tins.2006.01.003) [DOI] [PubMed] [Google Scholar]
- Csete M. E., Doyle J. C.2002Reverse engineering of biological complexity. Science 295, 1664–1669 (doi:10.1126/science.1069981) [DOI] [PubMed] [Google Scholar]
- Dale N.2002Resetting intrinsic purinergic modulation of neural activity: an associative mechanism? J. Neurosci. 22, 10 461–10 469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale N., Gilday D.1996Regulation of rhythmic movements by purinergic neurotransmitters in frog embryos. Nature 383, 259–263 (doi:10.1038/383259a0) [DOI] [PubMed] [Google Scholar]
- DeLong N. D., Beenhakker M. P., Nusbaum M. P.2009Presynaptic inhibition selectively weakens peptidergic cotransmission in a small motor system. J. Neurophysiol. 102, 3492–3504 (doi:10.1152/jn.00833.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P. S.2006Neuromodulation of central pattern generators in invertebrates and vertebrates. Curr. Opin. Neurobiol. 16, 604–614 (doi:10.1016/j.conb.2006.10.007) [DOI] [PubMed] [Google Scholar]
- Dickinson P. S., Moulins M.1992Interactions and combinations between different networks in the stomatogastric nervous system. In Dynamic biological networks: the stomatogastric nervous system (eds Harris-Warrick R. M., Marder E., Selverston A. I., Moulins M.), pp. 139–160 Cambridge, MA: MIT Press [Google Scholar]
- Dickinson P. S., Fairfield W. P., Hetling J. R., Hauptman J.1997Neurotransmitter interactions in the stomatogastric system of the spiny lobster: one peptide alters the response of a central pattern generator to a second peptide. J. Neurophysiol. 77, 599–610 [DOI] [PubMed] [Google Scholar]
- Djokaj S., Cooper R. L., Rathmayer W.2001Presynaptic effects of octopamine, serotonin, and cocktails of the two modulators on neuromuscular transmission in crustaceans. J. Comp. Physiol. A 187, 145–154 (doi:10.1007/s003590100187) [DOI] [PubMed] [Google Scholar]
- Edelman G. M., Gally J. A.2001Degeneracy and complexity in biological systems. Proc. Natl Acad. Sci. USA 98, 13 763–13 768 (doi:10.1073/pnas.231499798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Samad H., Kurata H., Doyle J. C., Gross C. A., Khammash M.2005Surviving heat shock: control strategies for robustness and performance. Proc. Natl Acad. Sci. USA 102, 2736–2741 (doi:10.1073/pnas.0403510102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellous J.-M., Linster C.1998Computational models of neuromodulation. Neural Comput. 10, 771–805 (doi:10.1162/089976698300017476) [DOI] [PubMed] [Google Scholar]
- Fisher S. A., Fischer T. M., Carew T. J.1997Multiple overlapping processes underlying short-term synaptic enhancement. Trends Neurosci. 20, 170–177 (doi:10.1016/S0166-2236(96)01001-6) [DOI] [PubMed] [Google Scholar]
- Fox L. E., Lloyd P. E.2001Evidence that post-tetanic potentiation is mediated by neuropeptide release in Aplysia. J. Neurophysiol. 86, 2845–2855 [DOI] [PubMed] [Google Scholar]
- Fu Q., Kutz K. K., Schmidt J. J., Hsu Y.-W. A., Messinger D. I., Cain S. D., de la Iglesia H. O., Christie A. E., Li L.2005Hormone complement of the Cancer productus sinus gland and pericardial organ: an anatomical and mass spectrometric investigation. J. Comp. Neurol. 493, 607–626 (doi:10.1002/cne.20773) [DOI] [PubMed] [Google Scholar]
- Fujisawa Y., et al. 1999The Aplysia Mytilus inhibitory peptide-related peptides: identification, cloning, processing, distribution, and action. J. Neurosci. 19, 9618–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furness J. B., Bornstein J. C., Murphy R., Pompolo S.1992Role of peptides in transmission in the enteric nervous system. Trends Neurosci. 15, 66–71 (doi:10.1016/0166-2236(92)90029-8) [DOI] [PubMed] [Google Scholar]
- Furukawa Y., et al. 2001The enterins: a novel family of neuropeptides isolated from the enteric nervous system and CNS of Aplysia. J. Neurosci. 21, 8247–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerschenfeld H. M.1973Chemical transmission in invertebrate central nervous systems and neuromuscular junctions. Physiol. Rev. 53, 1–119 [DOI] [PubMed] [Google Scholar]
- Gonze D., Halloy J., Goldbeter A.2002Robustness of circadian rhythms with respect to molecular noise. Proc. Natl Acad. Sci. USA 99, 673–678 (doi:10.1073/pnas.022628299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourine A. V., Wood J. D., Burnstock G.2009Purinergic signalling in autonomic control. Trends Neurosci. 32, 241–248 (doi:10.1016/j.tins.2009.03.002) [DOI] [PubMed] [Google Scholar]
- Grillner S., Graybiel A. M.2006Microcircuits: the interface between neurons and global brain function. Cambridge, MA: MIT Press [Google Scholar]
- Gudermann T., Kalkbrenner F., Schultz G.1996Diversity and selectivity of receptor–G protein interaction. Annu. Rev. Pharmacol. Toxicol. 36, 429–459 [DOI] [PubMed] [Google Scholar]
- Guix F. X., Uribesalgo I., Coma M., Munoz F. J.2005The physiology and pathophysiology of nitric oxide in the brain. Prog. Neurobiol. 76, 126–152 (doi:10.1016/j.pneurobio.2005.06.001) [DOI] [PubMed] [Google Scholar]
- Harris-Warrick R. M., Marder E., Selverston A. I., Moulins M.1992aDynamic biological networks: the stomatogastric nervous system. Cambridge, MA: MIT Press [Google Scholar]
- Harris-Warrick R. M., Nagy F., Nusbaum M. P.1992bNeuromodulation of stomatogastric networks by identified neurons and transmitters. In Dynamic biological networks: the stomatogastric nervous system (eds Harris-Warrick R. M., Marder E., Selverston A. I., Moulins M.), pp. 87–137 Cambridge, MA: MIT Press [Google Scholar]
- Hartwell L. H., Hopfield J. J., Leibler S., Murray A. W.1999From molecular to modular cell biology. Nature 402, C47–C52 (doi:10.1038/35011540) [DOI] [PubMed] [Google Scholar]
- Hasselmo M. E.1995Neuromodulation and cortical function: modeling the physiological basis of behavior. Behav. Brain Res. 67, 1–27 (doi:10.1016/0166-4328(94)00113-T) [DOI] [PubMed] [Google Scholar]
- Helikar T., Konvalina J., Heidel J., Rogers J. A.2008Emergent decision-making in biological signal transduction networks. Proc. Natl Acad. Sci. USA 105, 1913–1918 (doi:10.1073/pnas.0705088105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert J.1993Peptides in the limbic system: neurochemical codes for co-ordinated adaptive responses to behavioural and physiological demand. Prog. Neurobiol. 41, 723–791 (doi:10.1016/0301-0082(93)90033-O) [DOI] [PubMed] [Google Scholar]
- Hewes R. S., Snowdeal E. C., Taghert P. H.1998Functional redundancy of FMRFamide-related peptides at the Drosophila larval neuromuscular junction. J. Neurosci. 18, 7138–7151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B.1992G protein-coupled mechanisms and nervous signaling. Neuron 9, 187–195 (doi:10.1016/0896-6273(92)90158-A) [DOI] [PubMed] [Google Scholar]
- Hofer A. M., Lefkimmiatis K.2007Extracellular calcium and cAMP: second messengers as ‘third messengers’? Physiology 22, 320–327 (doi:10.1152/physiol.00019.2007) [DOI] [PubMed] [Google Scholar]
- Hökfelt T., Broberger C., Xu Z.-Q. D., Sergeyev V., Ubink R., Diez M.2000Neuropeptides—an overview. Neuropharmacology 39, 1337–1356 (doi:10.1016/S0028-3908(00)00010-1) [DOI] [PubMed] [Google Scholar]
- Hooper S. L., Brezina V., Cropper E. C., Weiss K. R.1999Flexibility of muscle control by modulation of muscle properties. In Beyond neurotransmission: neuromodulation and its importance for information processing (ed. Katz P. S.), pp. 241–274 Oxford, UK: Oxford University Press [Google Scholar]
- Horn C. C., Zhurov Y., Orekhova I. V., Proekt A., Kupfermann I., Weiss K. R., Brezina V.2004Cycle-to-cycle variability of neuromuscular activity in Aplysia feeding behavior. J. Neurophysiol. 92, 157–180 (doi:10.1152/jn.01190.2003) [DOI] [PubMed] [Google Scholar]
- Husson S. J., Mertens I., Janssen T., Lindemans M., Schoofs L.2007Neuropeptide signaling in the nematode Caenorhabditis elegans. Prog. Neurobiol. 82, 33–55 (doi:10.1016/j.pneurobio.2007.01.006) [DOI] [PubMed] [Google Scholar]
- Huxtable A. G., Zwicker J. D., Poon B. Y., Pagliardini S., Vrouwe S. Q., Greer J. J., Funk G. D.2009Tripartite purinergic modulation of central respiratory networks during perinatal development: the influence of ATP, ectonucleotidases, and ATP metabolites. J. Neurosci. 29, 14 713–14 725 (doi:10.1523/JNEUROSCI.2660-09.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H. T., Schuman E. M.2008Frequency-dependent signal transmission and modulation by neuromodulators. Front. Neurosci. 2, 138–144 (doi:10.3389/neuro.01.027.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan J. D., Landau E. M., Iyengar R.2000Signaling networks: the origins of cellular multitasking. Cell 103, 193–200 (doi:10.1016/S0092-8674(00)00112-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz P. S.1999Beyond neurotransmission: neuromodulation and its importance for information processing. Oxford, UK: Oxford University Press [Google Scholar]
- Katz P. S., Clemens S.2001Biochemical networks in nervous systems: expanding neuronal information capacity beyond voltage signals. Trends Neurosci. 24, 18–25 (doi:10.1016/S0166-2236(00)01686-6) [DOI] [PubMed] [Google Scholar]
- Katz P. S., Edwards D. H.1999Metamodulation: the control and modulation of neuromodulation. In Beyond neurotransmission: neuromodulation and its importance for information processing (ed. Katz P. S.), pp. 349–381 Oxford, UK: Oxford University Press [Google Scholar]
- Katz P. S., Frost W. N.1996Intrinsic neuromodulation: altering neuronal circuits from within. Trends Neurosci. 19, 54–61 (doi:10.1016/0166-2236(96)89621-4) [DOI] [PubMed] [Google Scholar]
- Kirkland K. L.2002High-tech brains. Persp. Biol. Med. 45, 212–223 (doi:10.1353/pbm.2002.0033) [DOI] [PubMed] [Google Scholar]
- Kitano H.2004Biological robustness. Nat. Rev. Genet. 5, 826–837 (doi:10.1038/nrg1471) [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Muneoka Y.1990Structure and action of molluscan neuropeptides. Zool. Sci. 7, 801–814 [DOI] [PubMed] [Google Scholar]
- Koch C.1999Biophysics of computation: information processing in single neurons. New York, NY: Oxford University Press [Google Scholar]
- Koh H.-Y., Weiss K. R.2005Peptidergic contribution to posttetanic potentiation at a central synapse of Aplysia. J. Neurophysiol. 94, 1281–1286 (doi:10.1152/jn.00073.2005) [DOI] [PubMed] [Google Scholar]
- Konkoy C. S., Davis T. P.1996Ectoenzymes as sites of peptide regulation. Trends Pharmacol. Sci. 17, 288–294 (doi:10.1016/0165-6147(96)10036-5) [DOI] [PubMed] [Google Scholar]
- Kupfermann I.1974Feeding behavior in Aplysia: a simple system for the study of motivation. Behav. Biol. 10, 1–26 (doi:10.1016/S0091-6773(74)91644-7) [DOI] [PubMed] [Google Scholar]
- Kupfermann I.1991Functional studies of cotransmission. Physiol. Rev. 71, 683–732 [DOI] [PubMed] [Google Scholar]
- Landgraf R., Neumann I. D.2004Vasopressin and oxytocin release within the brain: a dynamic concept of multiple and variable modes of neuropeptide communication. Front. Endocrinol. 25, 150–176 (doi:10.1016/j.yfrne.2004.05.001) [DOI] [PubMed] [Google Scholar]
- Leng G., Ludwig M.2006Information processing in the hypothalamus: peptides and analogue computation. J. Neuroendocrinol. 18, 379–392 (doi:10.1111/j.1365-2826.2006.01428.x) [DOI] [PubMed] [Google Scholar]
- Li F., Long T., Lu Y., Ouyang Q., Tang C.2004The yeast cell-cycle network is robustly designed. Proc. Natl Acad. Sci. USA 101, 4781–4786 (doi:10.1073/pnas.0305937101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingueglia E., Champigny G., Lazdunski M., Barbry P.1995Cloning of the amiloride-sensitive FMRFamide-gated sodium channel. Nature 378, 730–733 (doi:10.1038/378730a0) [DOI] [PubMed] [Google Scholar]
- Lundberg J. M.1996Pharmacology of cotransmission in the autonomic nervous system: integrative aspects of amines, neuropeptides, adenosine triphosphate, amino acids and nitric oxide. Pharmacol. Rev. 48, 113–178 [PubMed] [Google Scholar]
- Ma M., Wang J., Chen R., Li L.2009Expanding the crustacean neuropeptidome using a multifaceted mass spectrometric approach. J. Proteome Res. 8, 2426–2437 (doi:10.1021/pr801047v) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A., et al. 2005Formation of regulatory patterns during signal propagation in a mammalian cellular network. Science 309, 1078–1083 (doi:10.1126/science.1108876) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi C. A.1995Tachykinins and calcitonin gene-related peptide (CGRP) as co-transmitters released from peripheral endings of sensory nerves. Prog. Neurobiol. 45, 1–98 (doi:10.1016/0301-0082(94)E0017-B) [DOI] [PubMed] [Google Scholar]
- Marder E., Bucher D.2001Central pattern generators and the control of rhythmic movements. Curr. Biol. 11, R986–R996 (doi:10.1016/S0960-9822(01)00581-4) [DOI] [PubMed] [Google Scholar]
- Marder E., Thirumalai V.2002Cellular, synaptic and network effects of neuromodulation. Neural Netw. 15, 479–493 (doi:10.1016/S0893-6080(02)00043-6) [DOI] [PubMed] [Google Scholar]
- Marder E., Christie A. E., Kilman V. L.1995Functional organization of cotransmission systems: lessons from small nervous systems. Invert. Neurosci. 1, 105–112 (doi:10.1007/BF02331908) [DOI] [PubMed] [Google Scholar]
- McCormick D. A.1992Neurotransmitter actions in the thalamus and cerebral cortex and their role in neuromodulation of thalamocortical activity. Prog. Neurobiol. 39, 337–388 (doi:10.1016/0301-0082(92)90012-4) [DOI] [PubMed] [Google Scholar]
- Merighi A.2002Costorage and coexistence of neuropeptides in the mammalian CNS. Prog. Neurobiol. 66, 161–190 (doi:10.1016/S0301-0082(01)00031-4) [DOI] [PubMed] [Google Scholar]
- Mesce K. A., Crisp K. M., Gilchrist L. S.2001Mixtures of octopamine and serotonin have nonadditive effects on the CNS of the medicinal leech. J. Neurophysiol. 85, 2039–2046 [DOI] [PubMed] [Google Scholar]
- Milo R., Shen-Orr S., Itzkovitz S., Kashtan N., Chklovskii D., Alon U.2002Network motifs: simple building blocks of complex networks. Science 298, 824–827 (doi:10.1126/science.298.5594.824) [DOI] [PubMed] [Google Scholar]
- Nässel D. R.2002Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog. Neurobiol. 68, 1–84 (doi:10.1016/S0301-0082(02)00057-6) [DOI] [PubMed] [Google Scholar]
- Nässel D. R.2009Neuropeptide signaling near and far: how localized and timed is the action of neuropeptides in brain circuits? Invert. Neurosci. (doi:10.1007/s10158-009-0090-1) [DOI] [PubMed] [Google Scholar]
- Niall H. D.1982The evolution of peptide hormones. Annu. Rev. Physiol. 44, 615–624 (doi:10.1146/annurev.ph.44.030182.003151) [DOI] [PubMed] [Google Scholar]
- Nusbaum M. P.2002Regulating peptidergic modulation of rhythmically active neural circuits. Brain Behav. Evol. 60, 378–387 (doi:10.1159/000067791) [DOI] [PubMed] [Google Scholar]
- Nusbaum M. P., Blitz D. M., Swensen A. M., Wood D., Marder E.2001The roles of co-transmission in neural network modulation. Trends Neurosci. 24, 146–154 (doi:10.1016/S0166-2236(00)01723-9) [DOI] [PubMed] [Google Scholar]
- Offermanns S., Schultz G.1994Complex information processing by the transmembrane signaling system involving G proteins. Naunyn-Schmied Arch. Pharmacol. 350, 329–338 [DOI] [PubMed] [Google Scholar]
- Olsen S. R., Wilson R. I.2008Cracking neural circuits in a tiny brain: new approaches for understanding the neural circuitry of Drosophila. Trends Neurosci. 31, 512–520 (doi:10.1016/j.tins.2008.07.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olypher A. V., Calabrese R. L.2007Using constraints on neuronal activity to reveal compensatory changes in neuronal parameters. J. Neurophysiol. 98, 3749–3758 (doi:10.1152/jn.00842.2007) [DOI] [PubMed] [Google Scholar]
- Orchard I., Lange A. B., Bendena W. G.2001FMRFamide-related peptides: a multifunctional family of structurally related neuropeptides in insects. Adv. Insect. Physiol. 28, 268–329 [Google Scholar]
- Orekhova I. V., Alexeeva V., Church P. J., Weiss K. R., Brezina V.2003Multiple presynaptic and postsynaptic sites of inhibitory modulation by myomodulin at ARC neuromuscular junctions of Aplysia. J. Neurophysiol. 89, 1488–1502 (doi:10.1152/jn.00140.2002) [DOI] [PubMed] [Google Scholar]
- Ott S. R., Philippides A., Elphick M. R., O'Shea M.2007Enhanced fidelity of diffusive nitric oxide signalling by the spatial segregation of source and target neurones in the memory centre of an insect brain. Eur. J. Neurosci. 25, 181–190 [DOI] [PubMed] [Google Scholar]
- Perone M. J., Windeatt S., Castro M. G.1997Intracellular trafficking of prohormones and proneuropeptides: cell type-specific sorting and targeting. Exp. Physiol. 82, 609–628 [DOI] [PubMed] [Google Scholar]
- Přibyl M., Muratov C. B., Shvartsman S. Y.2003Long-range signal transmission in autocrine relays. Biophys. J. 84, 883–896 (doi:10.1016/S0006-3495(03)74906-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prill R. J., Iglesias P. A., Levchenko A.2005Dynamic properties of network motifs contribute to biological network organization. PLoS Biol. 3, 1881–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proekt A., et al. 2005Identification of a new neuropeptide precursor reveals a novel source of extrinsic modulation in the feeding system of Aplysia. J. Neurosci. 25, 9637–9648 (doi:10.1523/JNEUROSCI.2932-05.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Ge H.2006Modularity and dynamics of cellular networks. PLoS Comput. Biol. 2, 1502–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves G. T., Fraser S. E.2009Biological systems from an engineer's point of view. PLoS Biol. 7, e1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E. M.1989Signal sorting and amplification through G protein-coupled receptors. Neuron 3, 141–152 (doi:10.1016/0896-6273(89)90027-5) [DOI] [PubMed] [Google Scholar]
- Sakurai A., Katz P. S.2009State-, timing-, and pattern-dependent neuromodulation of synaptic strength by a serotonergic neuron. J. Neurosci. 29, 268–279 (doi:10.1523/JNEUROSCI.4456-08.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salio C., Lossi L., Ferrini F., Merighi A.2006Neuropeptides as synaptic transmitters. Cell Tissue Rev. 326, 583–598 (doi:10.1007/s00441-006-0268-3) [DOI] [PubMed] [Google Scholar]
- Scholz N. L., de Vente J., Truman J. W., Graubard K.2001Neural network partitioning by NO and cGMP. J. Neurosci. 21, 1610–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs L., Veelaert D., Vanden Broeck J., De Loof A.1997Peptides in the locusts, Locusta migratoria and Schistocerca gregaria. Peptides 18, 145–156 (doi:10.1016/S0196-9781(96)00236-7) [DOI] [PubMed] [Google Scholar]
- Sebastião A. M., Ribeiro J. A.2000Fine-tuning neuromodulation by adenosine. Trends Pharmacol. Sci. 21, 341–346 (doi:10.1016/S0165-6147(00)01517-0) [DOI] [PubMed] [Google Scholar]
- Segal M. M.1983Specification of synaptic action. Trends Neurosci. 6, 118–121 (doi:10.1016/0166-2236(83)90061-9) [Google Scholar]
- Skiebe P.2001Neuropeptides are ubiquitous chemical mediators: using the stomatogastric nervous system as a model system. J. Exp. Biol. 204, 2035–2048 [DOI] [PubMed] [Google Scholar]
- Sossin W. S., Scheller R. H.1991Biosynthesis and sorting of neuropeptides. Curr. Opin. Neurobiol. 1, 79–83 (doi:10.1016/0959-4388(91)90013-W) [DOI] [PubMed] [Google Scholar]
- Sporns O., Chialvo D. R., Kaiser M., Hilgetag C. C.2004Organization, development and function of complex brain networks. Trends Cogn. Sci. 8, 418–425 (doi:10.1016/j.tics.2004.07.008) [DOI] [PubMed] [Google Scholar]
- Stein W.2009Modulation of stomatogastric rhythms. J. Comp. Physiol. A 195, 989–1009 (doi:10.1007/s00359-009-0483-y) [DOI] [PubMed] [Google Scholar]
- Stjärne L., Stjärne E.1995Geometry, kinetics and plasticity of release and clearance of ATP and noradrenaline as sympathetic cotransmitters: roles for the neurogenic contraction. Prog. Neurobiol. 47, 45–94 (doi:10.1016/0301-0082(95)00018-Q) [DOI] [PubMed] [Google Scholar]
- Strand F. L.1999Neuropeptides: regulators of physiological processes. Cambridge, MA: MIT Press [Google Scholar]
- Svensson E., Grillner S., Parker D.2001Gating and braking of short- and long-term modulatory effects by interactions between colocalized neuromodulators. J. Neurosci. 21, 5984–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweedler J. V., Li L., Rubakhin S. S., Alexeeva V., Dembrow N. C., Dowling O., Jing J., Weiss K. R., Vilim F. S.2002Identification and characterization of the feeding circuit-activating peptides, a novel neuropeptide family of Aplysia. J. Neurosci. 22, 7797–7808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert P. H., Veenstra J. A.2003Drosophila neuropeptide signaling. Adv. Genet. 49, 1–65 (doi:10.1016/S0065-2660(03)01001-0) [DOI] [PubMed] [Google Scholar]
- Tomlin C. J., Axelrod J. D.2005Understanding biology by reverse engineering the control. Proc. Natl Acad. Sci. USA 102, 4219–4220 (doi:10.1073/pnas.0500276102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage M., Ghijsen W. E., Lopes da Silva F. H.1994Presynaptic plasticity: the regulation of Ca2+-dependent transmitter release. Prog. Neurobiol. 42, 539–574 (doi:10.1016/0301-0082(94)90050-7) [DOI] [PubMed] [Google Scholar]
- Vilim F. S., et al. 2010Distinct mechanisms produce functionally complementary actions of neuropeptides that are structurally related but derived from different precursors. J. Neurosci. 30, 131–147 (doi:10.1523/JNEUROSCI.3282-09.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizi E. S., Lábos E.1991Non-synaptic interactions at presynaptic level. Prog. Neurobiol. 37, 145–163 (doi:10.1016/0301-0082(91)90025-V) [DOI] [PubMed] [Google Scholar]
- Weaver C. M., Wearne S. L.2008Neuronal firing sensitivity to morphologic and active membrane parameters. PLoS Comput. Biol. 4, 130–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K. R., Brezina V., Cropper E. C., Heierhorst J., Hooper S. L., Probst W. C., Rosen S. C., Vilim F. S., Kupfermann I.1993Physiology and biochemistry of peptidergic cotransmission in Aplysia. J. Physiol. (Paris) 87, 141–151 (doi:10.1016/0928-4257(93)90025-O) [DOI] [PubMed] [Google Scholar]
- Weng G., Bhalla U. S., Iyengar R.1999Complexity in biological signaling systems. Science 284, 92–96 (doi:10.1126/science.284.5411.92) [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S.1986The structure of the nervous system of the nematode C. elegans. Phil. Trans. R. Soc. Lond. B 314, 1–340 (doi:10.1098/rstb.1986.0056) [DOI] [PubMed] [Google Scholar]
- Wood D. E.1995Neuromodulation of rhythmic motor patterns in the blue crab Callinectes sapidus by amines and the peptide proctolin. J. Comp. Physiol. A 177, 335–349 [DOI] [PubMed] [Google Scholar]
- Wood D. E., Nusbaum M. P.2002Extracellular peptidase activity tunes motor pattern modulation. J. Neurosci. 22, 4185–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A. J., Dayan P.2005Uncertainty, neuromodulation, and attention. Neuron 46, 681–692 (doi:10.1016/j.neuron.2005.04.026) [DOI] [PubMed] [Google Scholar]
- Zimmermann H.2008ATP and acetylcholine, equal brethren. Neurochem. Int. 52, 634–648 (doi:10.1016/j.neuint.2007.09.004) [DOI] [PubMed] [Google Scholar]
- Zoli M., Agnati L. F.1996Wiring and volume transmission in the central nervous system: the concept of closed and open synapses. Prog. Neurobiol. 49, 363–380 [DOI] [PubMed] [Google Scholar]
- Zoli M., Jansson A., Syková E., Agnati L. F., Fuxe K.1999Volume transmission in the CNS and its relevance for neuropsychopharmacology. Trends Pharmacol. Sci. 20, 142–150 (doi:10.1016/S0165-6147(99)01343-7) [DOI] [PubMed] [Google Scholar]
- Zucker R. S., Regehr W. G.2002Short-term synaptic plasticity. Annu. Rev. Physiol. 64, 355–405 (doi:10.1146/annurev.physiol.64.092501.114547) [DOI] [PubMed] [Google Scholar]