Abstract
To date, the processing of wildlife location data has relied on a diversity of software and file formats. Data management and the following spatial and statistical analyses were undertaken in multiple steps, involving many time-consuming importing/exporting phases. Recent technological advancements in tracking systems have made large, continuous, high-frequency datasets of wildlife behavioural data available, such as those derived from the global positioning system (GPS) and other animal-attached sensor devices. These data can be further complemented by a wide range of other information about the animals' environment. Management of these large and diverse datasets for modelling animal behaviour and ecology can prove challenging, slowing down analysis and increasing the probability of mistakes in data handling. We address these issues by critically evaluating the requirements for good management of GPS data for wildlife biology. We highlight that dedicated data management tools and expertise are needed. We explore current research in wildlife data management. We suggest a general direction of development, based on a modular software architecture with a spatial database at its core, where interoperability, data model design and integration with remote-sensing data sources play an important role in successful GPS data handling.
Keywords: GPS tracking, biotelemetry, wildlife ecology, spatial database, data modelling, GIS
1. Introduction: gps data, new perspectives, new challenges
Global navigation satellite systems (GNSS) are constellations of orbiting satellites working in conjunction with a network of ground stations that provide geo-spatial positioning of a user's receiver with global coverage (Tomkiewicz et al. 2010). At the moment, the most used GNSS is the global positioning system (GPS) network. This technology represents a powerful tool for wildlife studies (Hebblewhite & Haydon 2010). GPS tracking systems can record huge amounts of highly accurate animal locations with minimal work by operators, thus allowing reduced sampling intervals, and increased accuracy and performance when compared with very high-frequency (VHF) radio-tracking systems (Rodgers 2001; Frair et al. 2004, 2010; Ropert-Coudert & Wilson 2005). Furthermore, data can be remotely transferred to operators (e.g. using GPS for mobile communications GSM network, or the Argos satellite system), making near-real-time monitoring of animals possible.
GPS tracking datasets can be used to address animal ecology questions (e.g. resource selection: Beyer et al. 2010, Fieberg et al. 2010; animal movement: Morales et al. 2010, Smouse et al. 2010; foraging behaviour: Owen-Smith et al. 2010; predation: Merrill et al. 2010) from a completely new perspective (i.e. closer to the animal point of view).
However, the availability of large datasets also poses a number of challenges, for example the need for appropriate analytical techniques to deal with spatially and temporally autocorrelated data (e.g. Boyce et al. 2010; Fieberg et al. 2010), or the development of modelling approaches to exploit the information embedded in continuous time series of animal locations (e.g. Kie et al. 2010; Smouse et al. 2010). On a more pragmatic and underlying level, GPS tracking routinely generates larger datasets than software tools commonly used by biologists in the recent past could handle (Rutz & Hays 2009).
Existing dedicated software tools for wildlife studies were mainly developed on the basis of VHF radiotracking data, which are characterized by small and discontinuous datasets, and focused on data analysis rather than data management (e.g. Ranges V: Kenward & Hodder 1996; HRE extension for ESRI ArcView: Rodgers & Carr 1998; Animal Movement extension to ESRI ArcView: Hooge & Eichenlaub 2000; Biotas: Ecological Software Solutions LLC 2004; Hawth's Tools extension to ESRI ArcGIS: Beyer 2004; HRT extension for ESRI ArcGIS: Rodgers et al. 2005). Spatial data, such as animal locations and home ranges, were traditionally stored locally in flat files, accessible to a single user at a time, and analysed by a number of independent applications without any common standards for interoperability. This approach can require data replication and export/import procedures that are time-consuming and potential sources of error. Moreover, data preprocessing has to be repeated for every scientific question to be addressed, resulting in task replication and wasted time. Instead, good scientific practice requires that data are securely, consistently and efficiently managed, to minimize errors, increase the reliability and reproducibility of inferences, and ensure data persistence (e.g. consistent use of data on multiple occasions and by several persons).
In this paper, we critically evaluate the requirements for good management of GPS wildlife tracking data. We highlight that dedicated data management tools and expertise are needed. We explore current research in wildlife data management and suggest a possible direction of development, based on a modular software architecture with a spatial database at its core. Specific concerns, including interoperability, data model design and integration with remote-sensing data sources are discussed.
This paper is focused on GPS-based location data, but is also valid for other wildlife monitoring data acquired with remote, automatic device-based techniques (Rutz & Hays 2009), known as biotelemetry (Cooke et al. 2004; Hooker et al. 2007; Tomkiewicz et al. 2010) or animal-attached remote sensing (Ropert-Coudert & Wilson 2005). Examples include datasets produced by activity sensors/accelerometers (e.g. Watanabe et al. 2005) that are frequently associated with GPS sensors (Coulombe et al. 2006; Tomkiewicz et al. 2010), depth sensors (often combined with other animal-borne loggers, e.g. Mitani et al. 2004; Wilson et al. 2008a,b), video-monitoring (Moll et al. 2007), GSM networks (Cronin & McConnell 2008) or wireless sensor network monitoring (e.g. Handcock et al. 2009; Pásztor et al. 2010). All these techniques can potentially produce near-real-time, high-frequency, large datasets that share the advantages and challenges of data storage and management with GPS-based location data.
2. Exploring the new information framework: a requirement analysis
GPS data characteristics and users' needs drive the conceptual definition of a suitable software architecture that can be developed with specific tools on different platforms. The main requirements and needs can be summarized as follows:
Data scalability. The main challenge is that GPS-based devices can record hundreds, or in some cases thousands, of locations per animal per day, and improvements in automated data collection is increasing the volume of data available from individual animals. Also, as the cost of this technology decreases, the number of monitored individuals will increase, which will increase the volume of data. Recent multi-sensor devices (Cooke et al. 2004) amplify the data volume by orders of magnitude and also complicate the data structure. To handle this large amount of data consistently, a persistent and very large data storage capability is needed.
Long-term storage for data reuse. Data must be consistently stored in the long term, independently from a specific application, to permit data reuse for different studies.
Periodic and automatic data acquisition. Many GPS data management issues are linked to the frequency of data recording and near-real-time access to data (Tomkiewicz et al. 2010). This requires automated procedures to receive, review and store data from GPS telemetry devices either continuously or at regular intervals.
Efficient data retrieval. Fast data search and retrieval tools are needed to support efficient data analysis and management.
Management of spatial information. GPS data are basically spatial data. They therefore require retrieval, manipulation and management tools specific to spatial domains.
Global spatial and time references. Studies with regional or global perspectives imply the development of specific tools to manage global time and spatial reference systems efficiently (i.e. to handle Coordinated Universal Time versus local time and different spatial reference systems in a common framework).
Heterogeneity of applications. The complex nature of movement ecology implies that GPS data should be visualized, explored and analysed by a wide range of specific task-oriented applications (e.g. for mapping, spatial statistics and reporting). This requires a software architecture that supports the integration of different software tools.
Easy implementation of new algorithms. Methods of analysis for wildlife location data have undergone extensive development, especially in recent years (e.g. Worton 1989; Morales et al. 2004; Moorcroft et al. 2006; Calenge et al. 2009; Smouse et al. 2010). Such rapid development of analytical techniques should be supported by a software environment where new algorithms can be easily implemented and customized.
Integration of different data sources. The availability of spatial data derived from remote sensing, environmental and socio-economic databases, and other animal-related data (e.g. capture details, life-history traits), provide many opportunities to expand the information embedded in GPS-based locations if these spatial and non-spatial datasets can be correctly managed and efficiently integrated into a comprehensive data structure.
Multi-user support. Within research groups, public institutions and environmental organizations, several users might need to access data simultaneously, both locally and remotely, with different access privileges (e.g. Wong et al. 2007). This multi-user environment calls for concurrency-control mechanisms and security functionalities.
Data sharing. The need for multi-disciplinary answers to global environmental problems makes data sharing among research groups a high priority (e.g. Movebank, http://www.movebank.org; Global Biodiversity Information Facility, http://www.gbif.org; INSPIRE, http://inspire.jrc.ec.europa.eu). This requires adherence to standard data formats, definition of metadata and methods for data storage and management that, in turn, guarantee interoperability (Yeung & Hall 2007).
Data dissemination. Dissemination of data among the scientific community and general public is important to promote management decisions (e.g. by public administrators), raise funds and make people aware of conservation and more general environmental issues (e.g. WILDSPACE project, http://wildspace.ec.gc.ca, Wong et al. 2003; Seaturtle project, http://www.seaturtle.org; Tagging of Pacific Predators, http://www.topp.org; OBIS Seamap, http://seamap.env.duke.edu; MooseResearch.SE, http://www.moose-research.se). Moreover, the general public can become a source of information (Dettki et al. 2004). All these instances require the integration of specific tools to make data accessible (e.g. Data Web interface and Web-GIS tools).
Cost-effectiveness. Cost-effectiveness of software tools is an important accessibility factor for institutions with limited financial resources that can be applied to production and analysis of data, instead of data handling.
These requirements must all be satisfied to take full advantage of the information that wildlife tracking devices can provide, and to avoid the risk of drowning in data. The software used in the past cannot provide for all these needs with the increasing volume and complexity of data. There is thus an urgent need for new software architectures.
3. Towards a solution: spatial database management systems
Advanced information systems currently developed to manage wildlife tracking data include: Information System for Analysis and Management of Ungulate Data (ISAMUD) and European Roe Deer project (EURODEER; E. Mach Foundation, Trento, Italy; Cagnacci & Urbano 2008; http://sites.google.com/site/eurodeerproject); Wireless Remote Animal Monitoring (WRAM; Swedish University of Agricultural Sciences, Umeå, Sweden; http://www-wram.slu.se); MoveBank (New York State Museum, New York State, USA and Max Planck Institute for Ornithology, Radolfzell, Germany; http://www.movebank.org); CSIRO integrated database system (Hartog et al. 2009); HeardMap (Australian Antarctic Division; Frydman & Gales 2007); GPS Plus Database Manager (Vectronic Aerospace GmbH, Berlin, Germany; http://www.vectronic-aerospace.com); and BioMap for GPS (Lotek Wireless Inc, Newmarket, Canada; http://www.lotek.com). Similar approaches have been adopted for data from sensors other than GPS (e.g. The Penguiness book, Ropert-Coudert et al. 2006). All these software solutions share a common feature, namely the use of a relational or object-relational database management system (DBMS), with dedicated spatial tools. DBMSs are efficient tools for storage, fast retrieval and manipulation of data (e.g. Elmasri & Navathe 2003). From a strictly technical point of view, advantages of a DBMS for wildlife behavioural studies include:
— storage capacity: virtually unlimited data storage capacity if compared with the potential volume of data from wildlife GPS tracking (requirement 1);
— backup and recovery: efficient tools to manage backup and recovery processes (requirement 2);
— data integrity: controls can be added to force data inserts, changes and deletions to respect specific rules (requirement 2);
— data consistency: full support of transactions, which are units of work performed in a database to ensure complete data operations and correct data management (requirements 2, 9);
— automation of processes: DBMSs can be empowered by defining internal functions and triggers, thus a wide range of routinely complex workflows can be automatically and efficiently performed inside the database itself (requirement 3);
— data retrieval performance: querying hundreds of thousands of GPS-based locations can be very time consuming—DBMSs can speed up this process using database indexes (requirement 4);
— time data type management: relevant properties linked to temporal data types (e.g. time stamp with Coordinated Universal Time, time interval) are supported (e.g. time difference; requirement 6);
— reduced data redundancy: using a database as a central data repository avoids data replication, preventing error derived from asynchrony between different versions (requirements 7, 10, 11);
— client/server architecture: an advanced DBMS provides data as a central service, to which a number of different applications can be connected and used as database front-end, including spatial, statistical, and internet tools (requirements 7, 10, 12);
— advanced exploratory data analysis: databases support data mining techniques for automatic knowledge discovery of potentially useful information embedded in large spatial datasets (Miller & Han 2001; requirement 8);
— database data models: in a database context, data models can link and integrate different data sources defining complex relationships (requirement 9; see §§5, 6);
— multi-user environment: once centralized in a DBMS, data can be accessed by multiple users at the same time, keeping control on the coherence between operations performed by them (requirement 10);
— data security: a wide range of data access controls can be implemented, where each user is constrained to the use of specific sets of operations on defined subsets of data (requirement 10);
— standards: consolidated industry standards for databases facilitate interoperability with client applications and data sharing among different research groups (requirement 11; see §4).
In addition to these features, most DBMSs also can support the spatial dimension of GPS tracking data. Spatial tools are increasingly integrated within databases that now accommodate native spatial data types (e.g. points, lines, polygons). These spatial DBMSs are designed to store, query and manipulate spatial data, including spatial reference systems (requirements 5, 6). Spatial databases integrate the geometric data types of spatial objects with standard data types that store the object's associated attributes (requirement 9). Some spatial databases also include support for storing and managing raster data (requirement 9). In a spatial database, spatial data types can be manipulated by a spatial extension of the structured query language (SQL; Shekhar & Chawla 2003; OGC 2006; Yeung & Hall 2007), where complex geospatial queries can be generated and optimized with specific spatial indexes. Today, practically all major relational databases offer native spatial information capabilities and functions in their products, including IBM DB2 (Spatial Extender), SQL Server (SQL Server 2008 Spatial), Oracle (Oracle Spatial), Informix (Spatial DataBlade), and the open source PostgreSQL (PostGIS), MySQL (Spatial Extension) and SQLite (Spatialite and SQLiteGeo), while ESRI ArcSDE is a middleware application that can spatially enable a set of DBMSs.
Spatial databases can be integrated with GIS software that can be connected to the server database as client applications. In fact, traditional GIS software is focused on specific analysis and data visualization, providing a rich set of spatial operations, but few are optimized for managing large vector datasets and complex data structures. Spatial databases, in turn, allow simple spatial operations that can be efficiently undertaken on a large set of elements (Shekhar & Chawla 2003), like GPS datasets. Thus, in an ideal information system, simple but massive operations are preprocessed within the spatial database, while more advanced spatial analyses rely on the GIS and spatial statistics packages connected to it (requirement 8).
Finally, the need of a cost-effective system architecture (requirement 13) can be achieved using, partially or totally, open source software (James 2003). Most relevant open source tools are available for wildlife GPS applications (Tufto & Cavallini 2005; Hall & Leahy 2008), addressing all the requirements listed in §2, including strong adherence to standards. Notable open source software tools available in the most used operation systems include: spatial databases (e.g. PostgreSQL, MySQL, SQLite), spatial libraries (e.g. PROJ4, GDAL/OGR, GEOS, Geotools, Geoserver), desktop GIS (e.g. GRASS GIS, Quantum GIS, uDIG, gvSIG, ILWIS, SAGA), Web-GIS packages (e.g. UMN Mapserver, OpenLayers, MapFish) and statistical tools (R and its several specific packages). R (R Development Core Team 2009, http://www.r-project.org), in particular, as an open advanced software environment for statistical computing and graphics, supports the quick implementation of new statistics and spatial analysis algorithms (requirement 8). Popular examples are the home range and trajectory analysis tools in the Adehabitat package (Calenge et al. 2009). R functions also can be loaded inside the database itself as native procedures (Conway 2008).
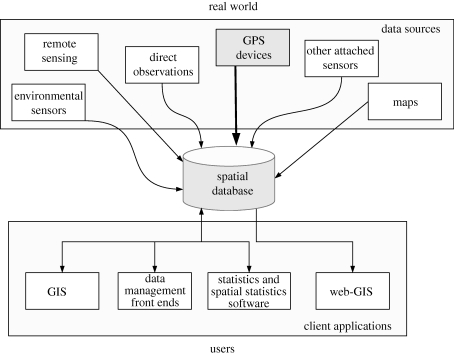
A general schema of a client/server architecture based on a spatial database able to accommodate all the relevant data sources is illustrated in figure 1.
Figure 1.
Schema of a possible client/server software system. Information from several data sources, including core GPS data, are integrated in the central spatial database and here accessed, locally or remotely, by client applications for manipulation, visualization and analysis. Outputs are stored back in the database.
An example of a modular software platform developed to handle GPS telemetry data and built on a spatial database is offered by ISAMUD, described by Cagnacci & Urbano (2008). This is based on an open source spatial database (PostgreSQL and PostGIS) and includes, as client applications, a statistics and spatial statistics package (R), GIS software (QGIS; GRASS), Web-GIS (MapServer; Ka-Map), a database management tool (PGAdmin) and a user-friendly interface for data entry, querying and reporting (Microsoft Access).
4. Interoperability and standards
To answer the requirements 7, 10 and 11 (heterogeneity of software client applications, multi-user environment, data sharing in a collaborative framework), international data and metadata standards must be adopted to ensure interoperability between different software platforms, both within and between organizations. Standards play an important role in improving data quality and can liberate data structures from the specific aim for which they were collected. Adhering to such standards ensures that data can be reused for a wide range of purposes, maximizing the returns of research funding and facilitating multi-species, large-scale and long-term ecological studies.
Standards have been developed and applied to many forms of spatial data, but not yet for wildlife GPS data. For example, GPS devices from different vendors produce data outputs with no standardization, making different data sources hard to manage inside the same information system. At the moment, the recognized bodies developing standards related to GPS data and the spatial domain include: Dublin Core Metadata Initiative (http://dublincore.org); DarwinCore (http://www.tdwg.org/activities/darwincore); Access to Biological Collections Data (ABCD, http://rs.tdwg.org/abcd). Spatial functionality standards for database systems (De Smith et al. 2007) have been defined by the Open Geospatial Consortium (OGC 2006, http://www.opengeospatial.org). The International Organization for Standardization, Technical Committee for Geographic Information/Geomatics (ISO TC211, http://www.isotc211.org) is another organization that works on geospatial standardization, including spatial databases.
Standards for GPS tracking data urgently require further attention because the development and adoption of standards is a prerequisite for the integration of wildlife tracking data into national and international Spatial Data Infrastructures (SDI; Onsrud 2007).
5. At the core of the system: data modelling
As database systems grow in size and complexity, and user requirements get more sophisticated, (spatial) data modelling becomes more important. A data model in a database framework (typically, relational or object-relational) describes what type of data are stored and how they are organized. It can be defined as a conceptual representation of the real world in database structures that include data objects (i.e. tables), associations between data objects, and rules that govern operations on the objects, thus explicitly determining their spatial and non-spatial relationships (Shekhar & Chawla 2003; Arctur & Zeiler 2004; Yeung & Hall 2007).
The definition of a suitable data model for wildlife GPS data should take into account the research aims, the structure of GPS data, the technical environment, the policies governing the use of information and the expected performance of the application (Yeung & Hall 2007). Data modelling permits easy update, modification and adaptation of the database structure to accommodate changing goals, constraints and spatial scales of studies, and the evolution of wildlife tracking systems. Without a rigorous data modelling approach, an information system built for GPS tracking data might lose the flexibility to manage data efficiently in the long term, reducing its utility to a simple storage device for raw data, and thus failing to address many of the needs identified in the requirement analysis.
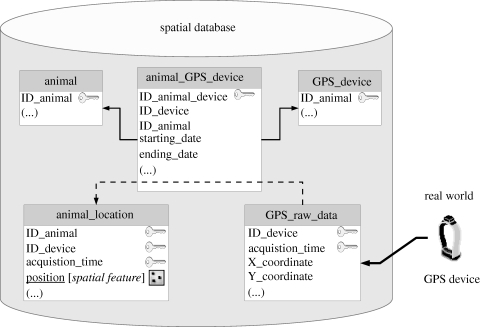
A reference conceptual data model in the context of GPS tracking should include at least two main objects, namely GPS devices and monitored individual animals. In figure 2, we propose a schema for the core elements of an example data model. Once GPS-based locations are stored in the database as spatial features (i.e. points), they can be intersected with other spatial layers using appropriate spatial SQL code, thus enriching the record with all the relevant environmental and socio-economic information describing the animal's habitat. Many other objects can be added and linked to these to extend the initial data model according to species, study objectives, information context, sensor devices, user environment and analysis. Thus, locations are managed not just as a pair of numbers (coordinates), but as complex multi-level objects. This perspective reduces the distance between physical reality and the way data are structured in the database, filling the semantic gap between the user's view of biological systems and its implementation in an information system (Shekhar & Chawla 2003; Nathan et al. 2008).
Figure 2.
General representation of a possible standard database data model for core wildlife GPS data. When the GPS device provides coordinates of a location at a specified time, they are uploaded in the database in a table (GPS_raw_data). A device is associated to an animal for a defined time range (table animal_GPS_device, connected to the tables animal and GPS_device with foreign keys, represented by solid arrows in the figure). According to the acquisition time, GPS-based locations are therefore assigned to a specific animal (i.e. the animal wearing the device in that particular moment) using the information of the table animal_GPS_device. Thus, the spatial table animal_location is filled (dashed arrow) with the identifier of the individual, its position (as a spatial object), the acquisition time and the identifier of the GPS device.
In the software platform cited as an example in §3 (ISAMUD; Cagnacci & Urbano 2008), the system imports location data from GPS collars via GSM connection into the database. Then, triggers automatically raise functions that transform the coordinates in a spatial attribute, intersect new points with vector environmental layers stored in the database (e.g. land cover, protected areas) and update the relative attributes. Another SQL statement computes the linear distance, the time interval, and the relative and absolute angles between successive locations. A script connects GRASS to the database and intersect points with raster environmental layers (e.g. DEM, slope). PostGIS computes and stores trajectories and minimum convex polygons for selected points. Another script connects r to PostgreSQL to perform ecological spatial analysis (e.g. home range) and store the results back to the database. A set of additional tools combine functions from PostreSQL, PostGIS and GRASS to compute spatial statistics on locations, trajectories and home ranges based on their environmental attributes.
A further promising extension to spatial data models is the adoption of spatio-temporal data models. Animal locations are characterized by both a spatial and a temporal dimension, representing a unique double-faced attribute of animal movement. Spatio-temporal databases (Pelekis et al. 2004) offer the opportunity to extend the spatial data model by integrating data types and functions specifically related to the spatio-temporal nature of animal movements, thus fully satisfying requirements 5 and 6 (e.g. considering the movement as an attribute of the animal instead of relying on ‘location’ objects, and modelling the environment as an object changing over time). This approach would help to decipher the continuity of animal movement related to habitat use. Although commonly used DBMSs do not yet support an integrated spatio-temporal extension, we foresee that spatio-temporal databases, which are undergoing intense development (Pelekis et al. 2004), will be the natural evolution for wildlife tracking data management tools in the future.
6. Exploiting remote sensing: integration of wildlife location data with environmental datasets
Remote sensing is a rapidly advancing technology for gathering environmental data using a wide range of satellite and airborne platforms. As such, it plays a major role in spatio-temporal earth surface monitoring with a spatial resolution approaching GPS data precision. Different remote-sensing technologies are available, including optical (passive), thermal (passive), LiDAR (active) and microwave (active) systems (Lillesand et al. 2004). Table 1 lists a series of ecological indicators from selected remote-sensed data sources.
Table 1.
List of ecological indicators from selected data sources.
| ecological indicator | generated from | dataset source |
|---|---|---|
| daily/monthly/annual minimum, mean, maximum temperatures (°C), trends, growing degree days | MODIS land surface temperature (LST): land surface temperatures, 4 maps per day, spatial resolution 1 km, period 2000–today | NASA EOS, USA, https://wist.echo.nasa.gov (free of charge) |
| phenological status, duration of growing season | MODIS vegetation indices (NDVI/EVI): 1 map per 16 days, spatial resolution 250 m, period 2000–today; SPOT Vegetation VGT, 1 map per 10 days, spatial resolution 1 km, period 1998–today | NASA EOS, USA, https://wist.echo.nasa.gov; VITO Belgium, http://www.vgt.vito.be (free of charge) |
| snow extent | MODIS maximum snow extent: 1 map per 8 days, spatial resolution 500 m, period 2000–today | NASA EOS, USA, https://wist.echo.nasa.gov (free of charge) |
| daily/monthly/annual actual, mean, total precipitation (mm) | GPCP: worldwide accumulated daily precipitation in millimetres, spatial resolution 1 arc degree, period 1997–2008 | NOAA, USA, http://www1.ncdc.noaa.gov/pub/data/gpcp/1dd/data (free of charge) |
| fraction of absorbed photosynthetically active radiation and leaf area index | MODIS FPAR/LAI: 1 map per 8 days, spatial resolution 1 km, period 2000–today | NASA EOS, USA, https://wist.echo.nasa.gov (free of charge) |
| gross/net primary production (biomass) | MODIS GPP/NPP: 1 map per 8 days, spatial resolution 1 km, period 2000–today | NASA EOS, USA, https://wist.echo.nasa.gov (free of charge) |
| land cover/land use maps, derived habitat maps (high-level data processing) | LANDSAT satellite series: spatial resolution 15 m/30 m, period 1972 today; ASTER: spatial resolution 15 m/30 m, period 2000–today | USGS Earth Explorer, http://earthexplorer.usgs.gov (free of charge); NASA EOS, USA, https://wist.echo.nasa.gov (nominal fee) |
| daily photoperiod, including cast shadows, geomorphological parameters (e.g. slope, aspect, curvature), flow accumulation | digital elevation model (DEM) | SRTM (spatial resolution 90 m), http://www2.jpl.nasa.gov/srtm governmental data at higher resolution (free of charge); ASTER GDEM, http://www.gdem.aster.ersdac.or.jp (free of charge) |
| habitat structure from 3D vegetation model (high-level data processing) | LiDAR data which permit to separate digital elevation model from digital surface model | governmental data (expensive, except part of US free of charge) |
Using classical methods of applied ecological remote sensing (e.g. statistical image classification or manual/semi-automated digitizing of patches), information on animal habitat use can be extracted from classified satellite or aerial images (Fuller et al. 1998). Also landscape metrics can be applied to the classified images in order to obtain information about connectivity, patchiness, diversity and more (e.g. Frohn 1998; Leyequien et al. 2007). Texture analysis and image segmentation are frequently used (e.g. Tuttle et al. 2006). Terrain modelling is another relevant source of information to enhance further the production of habitat maps (Hengl & Reuter 2009). The extracted ecological variables may then be used as input for habitat or population models (e.g. Handcock et al. 2009). However, it is important to use relevant environmental data at the appropriate temporal and spatial scales (Martin et al. 2009; Gaillard et al. 2010). For ecological applications, usually a choice must be made between very high-resolution data (spatial resolution of few metres, e.g. SPOT, or in the sub-metre range, e.g. QuickBird), which are often expensive and only generated on demand (acquisition requires prior data order, hence no continuous temporal and spatial coverage is granted), and high-resolution, low-cost (spatial resolution of 15/30 m, e.g. Aster and Landsat-ETM) or medium resolution (spatial resolution of 250 m or larger, e.g. MODIS) data, which are often available online for a nominal fee, or even for free. But while the latter have a high temporal and spatial coverage, they unfortunately do not usually line up well with the scale of animal home ranges. Ultimately, which kind of remote-sensing data and processing algorithm are selected for a wildlife tracking data management system strictly depends on study goals, scale and financial resources.
7. Concluding remarks
Data management is often not considered a core scientific issue in ecological studies, probably as a result of general reluctance to change work habits and undertake the initial costs for workflow and software updates, including hiring expert counselling and training of personnel. This paper advocates the view that good management of GPS-based locations is an essential step towards better science. Proving this statement empirically is difficult, although the best evidence is the enhanced efficiency and consistency in results. We suggest that dedicated management tools are needed and propose a client/server architecture based on a spatial database. This offers the opportunity to model location data as objects characterizing the presence of individuals in space and time within their habitat. We believe that the intrinsic consistency and integrity of spatial databases represents a necessary scientific infrastructure for rigorous science per se, preventing error propagation, optimizing performance of analysis and improving robustness of inferences. In particular, this favours the move from simple descriptive approaches towards mechanistic models with higher explanatory and predictive power (e.g. Millspaugh & Marzluff 2001; Morales et al. 2010; Smouse et al. 2010), focusing wildlife research on biological, rather than statistical, significance (Johnson 1999; Otis & White 1999; Ropert-Coudert & Wilson 2005).
To conclude, data management is increasingly becoming a necessary skill for ecologists, as has already happened with statistics and GIS. We expect further research towards innovative software solutions to assist the wildlife scientific community towards better data management techniques.
Acknowledgements
We thank Mathieu Basille and five anonymous reviewers for their important notes and suggestions, which considerably improved the paper. We also thank Kamran Safi, Bart Kranstauber and Hawthorne Beyer, who provided interesting input and comments on an early version of the manuscript. Most ideas presented in this paper were stimulated by discussions at the GPS-Telemetry Data: Challenges and Opportunities for Behavioural Ecology Studies workshop organized by the Edmund Mach Foundation (FEM) in September 2008, held in Viote del Monte Bondone, Trento, Italy. Funding of the workshop by the Autonomous Province of Trento is gratefully acknowledged.
Footnotes
One contribution of 15 to a Theme Issue ‘Challenges and opportunities of using GPS-based location data in animal ecology’.
References
- Arctur D., Zeiler M.2004Designing geodatabases: case studies in GIS data modelling. Redlands, CA: ESRI Press [Google Scholar]
- Beyer H. L.2004Hawth's analysis tools for ArcGIS. Available at http://www.spatialecology.com/htools [Google Scholar]
- Beyer H. L., Haydon D. T., Morales J. M., Frair J. L., Hebblewhite M., Mitchell M., Matthiopoulos J.2010The interpretation of habitat preference metrics under use–availability designs. Phil. Trans. R. Soc. B 365, 2245–2254 (doi:10.1098/rstb.2010.0083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M. S., Pitt J., Northrup J. M., Morehouse A. T., Knopff K. H., Cristescu B., Stenhouse G. B.2010Temporal autocorrelation functions for movement rates from global positioning system radiotelemetry data. Phil. Trans. R. Soc. B 365, 2213–2219 (doi:10.1098/rstb.2010.0080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnacci F., Urbano F.2008Managing wildlife: a spatial information system for GPS collars data. Environ. Modell. Softw. 23, 957–959 (doi:10.1016/j.envsoft.2008.01.003) [Google Scholar]
- Cagnacci F., Boitani L., Powell R. A., Boyce M. S.2010Animal ecology meets GPS-based radiotelemetry: a perfect storm of opportunities and challenges. Phil. Trans. R. Soc. B 365, 2157–2162 (doi:10.1098/rstb.2010.0107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calenge C., Dray S., Royer M.2009The concept of animals' trajectories from a data analysis perspective. Ecol. Inform. 4, 34–41 (doi:10.1016/j.ecoinf.2008.10.002) [Google Scholar]
- Conway J.2008PL/R—R Procedural language for PostgreSQL. Version 8.3.0.6 [Google Scholar]
- Cooke S. J., Hinch S. G., Wikelski M., Andrews R. D., Kuchel L. J., Wolcott T. G., Butler P. J.2004Biotelemetry: a mechanistic approach to ecology. Trends Ecol. Evol. 19, 334–343 (doi:10.1016/j.tree.2004.04.003) [DOI] [PubMed] [Google Scholar]
- Coulombe M. L., Masse A., Cote S. D.2006Quantification and accuracy of activity data measured with VHF and GPS telemetry. Wildlife Soc. B. 34, 81–92 (doi:10.2193/0091-7648(2006)34[81:QAAOAD]2.0.CO;2) [Google Scholar]
- Cronin M. A., McConnell B. J.2008SMS seal: a new technique to measure haul-out behaviour in marine vertebrates. J. Exp. Mar. Biol. Ecol. 362, 43–48 (doi:10.1016/j.jembe.2008.05.010) [Google Scholar]
- De Smith M., Goodchild M., Longley P. Geospatial analysis: a comprehensive guide. Leicester, UK: Troubador Publishing Ltd; 2007. [Google Scholar]
- Dettki H., Ericsson G., Edenius L.2004Real-time moose tracking: an Internet based mapping application using GPS/GSM-collars. Alces 40, 13–21 [Google Scholar]
- Ecological Software Solutions LLC. Biotas, Version 1.03. Hegymagas, Hungary: Ecological Software Solutions: 2004. [Google Scholar]
- Elmasri R., Navathe S. B.2003Fundamentals of database systems, 4th edn.Reading, MA: Addison Wesley [Google Scholar]
- Fieberg J., Matthiopoulos J., Hebblewhite M., Boyce M. S., Frair J. L.2010Correlation and studies of habitat selection: problem, red herring, or opportunity? Phil. Trans. R. Soc. B 365, 2233–2244 (doi:10.1098/rstb.2010.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frair J. L., Nielsen S. E., Merrill E. H., Lee S. R., Boyce M. S., Munro R. H. M., Stenhouse G. B., Beyer H. L.2004Removing GPS collar bias in habitat selection studies. J. Appl. Ecol. 41, 201–212 (doi:10.1111/j.0021-8901.2004.00902.x) [Google Scholar]
- Frair J. L., Fieberg J., Hebblewhite M., Cagnacci F., DeCesare N. J., Pedrotti L.2010Resolving issues of imprecise and habitat-biased locations in ecological analyses using GPS telemetry data. Phil. Trans. R. Soc. B 365, 2187–2200 (doi:10.1098/rstb.2010.0084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohn R.1998Remote sensing for landscape ecology: new metric indicators for monitoring, modeling, and assessment of ecosystems (mapping sciences). Boca Raton, FL: CRC Press [Google Scholar]
- Frydman S., Gales N.2007HeardMap: Tracking marine vertebrate populations in near real time. Deep-Sea Res. Part II 54, 384–391 (doi:10.1016/j.dsr2.2006.11.015) [Google Scholar]
- Fuller R. M., Groom G. B., Mugisha S., Ipulet P., Pomeroy D., Katende A., Bailey R., Ogutu-Ohwayo R.1998The integration of field survey and remote sensing for biodiversity assessment: a case study in the tropical forests and wetlands of Sango Bay, Uganda. Biol. Conserv. 86, 379–391 (doi:10.1016/S0006-3207(98)00005-6) [Google Scholar]
- Gaillard J.-M., Hebblewhite M., Loison A., Fuller M., Powell R., Basille M., Van Moorter B.2010Habitat–performance relationships: finding the right metric at a given spatial scale. Phil. Trans. R. Soc. B 365, 2255–2265 (doi:10.1098/rstb.2010.0085) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall G. B., Leahy M. G.2008Open source approaches in spatial data handling Berlin, Germany: Springer [Google Scholar]
- Handcock R., Swain D., Bishop-Hurley G., Patison K., Wark T., Valencia P., Corke P., O'Neill C.2009Monitoring animal behaviour and environmental interactions using wireless sensor networks, GPS collars and satellite remote sensing. Sensors 9, 3586–3603 (doi:10.3390/s90503586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartog J. R., Patterson T. A., Hartmann K., Jumppanen P., Cooper S., Bradford R.2009Developing integrated database systems for the management of electronic tagging data. In Tagging and Tracking of Marine Animals with Electronic Devices (eds Nielsen J. L., Arrizabalaga H., Fragoso N., Hobday A., Lutcavage M., Sibert J.), pp. 367–380 Dordrecht, The Netherlands: Springer: (doi:10.1007/978-1-4020-9640-2_22) [Google Scholar]
- Hebblewhite M., Haydon D. T.2010Distinguishing technology from biology: a critical review of the use of GPS telemetry data in ecology. Phil. Trans. R. Soc. B 365, 2303–2312 (doi:10.1098/rstb.2010.0087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengl T., Reuter H. I.2009Geomorphometry: concepts, software, applications. Developments in soil science, vol. 33 Amsterdam, The Netherlands: Elsevier [Google Scholar]
- Hooge P. N., Eichenlaub B.2000Animal movement extension to Arcview. ver. 2.0. Anchorage, AK: Alaska Science Center, Biological Science Office, U.S. Geological Survey [Google Scholar]
- Hooker S. K., Biuw M., McConnell B. J., Miller P. J. O., Sparling C. E.2007Bio-logging science: logging and relaying physical and biological data using animal-attached tags. Deep-Sea Res. II 54, 177–182 (doi:10.1016/j.dsr2.2007.01.001) [Google Scholar]
- James J.2003Free software and the digital divide: opportunities and constraints for developing countries. J. Inf. Sci. 29, 25–33 (doi:10.1177/016555150302900103) [Google Scholar]
- Johnson D. H.1999The insignificance of statistical significance testing. J. Wildlife Manage. 63, 763–772 (doi:10.2307/3802789) [Google Scholar]
- Kenward R. E., Hodder K. H.1996Ranges V. An analysis system for biological location data Wareham, UK: Institute of Terrestrial Ecology [Google Scholar]
- Kie J. G., Matthiopoulos J., Fieberg J., Powell R. A., Cagnacci F., Mitchell M. S., Gaillard J.-M., Moorcroft P. R.2010The home-range concept: are traditional estimators still relevant with modern telemetry technology? Phil. Trans. R. Soc. B 365, 2221–2231 (doi:10.1098/rstb.2010.0093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyequien E., Verrelst J., Slot M., Schaepmanstrub G., Heitkonig I., Skidmore A.2007Capturing the fugitive: Applying remote sensing to terrestrial animal distribution and diversity. Int. J. Appl. Earth Obs. Geoinform. 9, 1–20 (doi:10.1016/j.jag.2006.08.002) [Google Scholar]
- Lillesand M., Kiefer T., Ralph W., Chipman W. J.2004Remote sensing and image interpretation. USA: The Lehigh Press [Google Scholar]
- Martin J., Tolon V., Van Moorter B., Basille M., Calenge C.2009Telemetry: research, technology and applications. Use of telemetry in habitat selection studies, pp. 1–19 Hauppage, NY: Nova Science Publishers [Google Scholar]
- Merrill E., Sand H., Zimmermann B., McPhee H., Webb N., Hebblewhite M., Wabakken P., Frair J. L.2010Building a mechanistic understanding of predation with GPS-based movement data. Phil. Trans. R. Soc. B 365, 2279–2288 (doi:10.1098/rstb.2010.0077) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller H. J., Han J.2001Geographic data mining and knowledge discovery London, UK: Taylor & Francis [Google Scholar]
- Millspaugh J. J., Marzluff J. M.2001Radio-tracking and animal populations: past trends and future needs. In Radio tracking and animal population (eds Millspaugh J. J., Marzluff J. M.), pp. 383–393 San Diego, CA: Academic Press [Google Scholar]
- Mitani Y., Watanabe Y., Sato K., Cameron M. F., Naito Y.20043D diving behavior of Weddell seals with respect to prey accessibility and abundance. Mar. Ecol. Progr. Ser. 281, 275–281 (doi:10.3354/meps281275) [Google Scholar]
- Moll R. J., Millspaugh J. J., Beringer J., Sartwell J., He Z.2007A new ‘view’ of ecology and conservation through animal-borne video systems. Trends Ecol. Evol. 22, 660–668 (doi:10.1016/j.tree.2007.09.007) [DOI] [PubMed] [Google Scholar]
- Moorcroft P. R., Lewis M. A., Crabtree R. L.2006Mechanistic home range models capture spatial patterns and dynamics of coyote territories in Yellowstone. Proc. R. Soc. B 273, 1651–1659 (doi:10.1098/rspb.2005.3439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales J. M., Haydon D. T., Frair J., Holsinger K. E., Fryxell J. M.2004Extracting more out of relocation data: building movement models as mixtures of random walks. Ecology 85, 2436–2445 (doi:10.1890/03-0269) [Google Scholar]
- Morales J. M., Moorcroft P. R., Matthiopoulos J., Frair J. L., Kie J. G., Powell R. A., Merrill E. H., Haydon D. T.2010Building the bridge between animal movement and population dynamics. Phil. Trans. R. Soc. B 365, 2289–2301 (doi:10.1098/rstb.2010.0082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan R., Getz W. M., Revilla E., Holyoak M., Kadmon R., Saltz D., Smouse P. E.2008A movement ecology paradigm for unifying organismal movement research. Proc. Natl Acad. Sci. USA 105, 19 052–19 059 (doi:10.1073/pnas.0800375105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- OGC (Open Geospatial Consortium) 2006OpenGIS implementation specification for geographic information—simple feature access—part 2: SQL option. See http://www.opengeospatial.org/standards/sfs [Google Scholar]
- Onsrud H. J.2007Research and theory in advancing spatial data infrastructure concepts Redlands, CA: ESRI Press [Google Scholar]
- Otis D. L., White G. C.1999Autocorrelation of location estimates and the analysis of radiotracking data. J. Wildlife Manage. 63, 1039–1044 (doi:10.2307/3802819) [Google Scholar]
- Owen-Smith N., Fryxell J. M., Merrill E. H.2010Foraging theory upscaled: the behavioural ecology of herbivore movement. Phil. Trans. R. Soc. B 365, 2267–2278 (doi:10.1098/rstb.2010.0095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pásztor B., Mottola L., Mascolo C., Picco G. P., Ellwood S. W., Macdonald D. A., McConnell B.2010Selective reprogramming of mobile sensor networks through social community detection. Proc. of the 7th European conference on wireless sensor networks (EWSN 2010), Coimbra, Portugal, 17–19 February 2010 [Google Scholar]
- Pelekis N., Theodoulidis B., Kopanakis I., Theodoridis Y.2004Literature review of Spatio-temporal database models. Knowl. Eng. Rev. J. 19, 235–274 (doi:10.1017/S026988890400013X) [Google Scholar]
- R Development Core Team 2009R: a language and environment for statistical computing Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Rodgers A. R.2001Recent telemetry technology. In Radio tracking and animal population (eds Millspaugh J. J., Marzluff J. M.), pp. 79–121 San Diego, CA: Academic Press [Google Scholar]
- Rodgers A. R., Carr A. P.1998HRE: the home range extension for arcview. Ontario Ministry of natural resources Thunder Bay, Ontario: Centre for Northern Forest Ecosystem Research [Google Scholar]
- Rodgers A. R., Carr A. P., Smith L., Kie J. G.2005HRT: home range tools for ArcGIS. Ontario Ministry of natural resources Thunder Bay, Ontario: Centre for Northern Forest Ecosystem Research [Google Scholar]
- Ropert-Coudert Y., Wilson R. P.2005Trends and perspectives in animal-attached remote sensing. Front. Ecol. Environ. 3, 437–444 (doi:10.1890/1540-9295(2005)003[0437:TAPIAR]2.0.CO;2) [Google Scholar]
- Ropert-Coudert Y., Kato A., Wilson R. P.2006The Penguiness book. Version 1.0, June. See http://polaris.nipr.ac.jp/~penguin/penguiness. [Google Scholar]
- Rutz C., Hays G. C.2009New frontiers in biologging science. Biol. Lett. 5, 289–292 (doi:10.1098/rsbl.2009.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar S., Chawla S.2003Spatial databases: a tour. Upper Saddle River, NJ: Prentice Hall [Google Scholar]
- Smouse P. E., Focardi S., Moorcroft P. R., Kie J. G., Forester J. D., Morales J. M.2010Stochastic modelling of animal movement. Phil. Trans. R. Soc. B 365, 2201–2211 (doi:10.1098/rstb.2010.0078) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkiewicz S. M., Fuller M. R., Kie J. G., Bates K. K.2010Global positioning system and associated technologies in animal behaviour and ecological research. Phil. Trans. R. Soc. B 365, 2163–2176 (doi:10.1098/rstb.2010.0090) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufto J., Cavallini P.2005Should wildlife biologists use free software? Wildlife Biol. 11, 67–76 (doi:10.2981/0909-6396(2005)11[67:SWBUFS]2.0.CO;2) [Google Scholar]
- Tuttle E. M., Jensen R. R., Formica V. A., Gonser R. A.2006Using remote sensing image texture to study habitat use patterns: a case study using the polymorphic white-throated sparrow (Zonotrichia albicollis). Global Ecol. Biogeogr. 15, 349–357 (doi:10.1111/j.1466-822X.2006.00232.x) [Google Scholar]
- Watanabe S., Izawa M., Kato A., Ropert-Coudert Y., Naito Y.2005A new technique for monitoring the detailed behaviour of terrestrial animals: a case study with the domestic cat. Appl. Anim. Behav. Sci. 94, 117–131 (doi:10.1016/j.applanim.2005.01.010) [Google Scholar]
- Wilson R. P., Shepard E. L. C., Liebsch N.2008aPrying into the intimate details of animal lives: use of a daily diary on animals. Endangered Species Res. 4, 123–137 (doi:10.3354/esr00064) [Google Scholar]
- Wilson R. P., Vargas F. H., Steinfurth A., Riordan P., Ropert-Coudert Y., Macdonald D. W.2008bWhat grounds some birds for life? Movement and diving in the sexually dimorphic Galapagos cormorant. Ecol. Monog. 78, 633–652 (doi:10.1890/07-0677.1) [Google Scholar]
- Wong I. W., McNicol D. K., Fong P., Fillman D., Neysmith J., Russell R.2003The wildspace decision support system. Environ. Modell. Softw. 18, 521–530 (doi:10.1016/S1364-8152(03)00027-6) [Google Scholar]
- Wong I. W., Bloom R., McNicol D. K., Fong P., Russell R., Chen X.2007Species at risk: data and knowledge management within the WILDSPACE decision support system. Environ. Modell. Softw. 22, 423–430 (doi:10.1016/j.envsoft.2005.12.012) [Google Scholar]
- Worton B. J.1989Kernel methods for estimating the utilization distribution in home-range studies. Ecology 70, 164–168 (doi:10.2307/1938423) [Google Scholar]
- Yeung A. K. W., Hall G. B.2007Spatial database systems: design, implementation and project management. GeoJournal library, vol. 87. Dordrecht, The Netherlands: Springer [Google Scholar]