Abstract
Segmentation is unquestionably a major factor in the evoluation of complex body plans, but how this trait itself evolved is unknown. Approaching this problem requires comparing the molecular mechanisms of segmentation in diverse segmented and unsegmented taxa. Notch/Hes signaling is involved in segmentation in sequentially segmenting vertebrates and arthropods, as judged by patterns of expression of one or more genes in this network and by the disruption of segmental patterning when Notch/Hes signaling is disrupted. We have previously shown that Notch and Hes homologs are expressed in the posterior progress zone (PPZ) from which segments arise, in the leech Helobdella robusta, a sequentially segmenting lophotrochozoan (phylum Annelida). Here we show that disrupting Notch/Hes signaling disrupts segmentation in this species as well. Thus, Notch/Hes functions in either the maintenance of the PPZ and/or the patterning processes of segmentation in representatives of all three super-phyla of bilaterally symmetric animals. These results are consistent with two evolutionary scenarios. In one, segmentation was already present in the ancestor of all three super-phyla. In the other, Notch/Hes signaling functioned in axial growth by terminal addition in an unsegmented bilaterian ancestor, and was subsequently exapted to function in segmentation as that process evolved independently in two or more taxa.
Keywords: Segmentation, Annelid, Leech, Notch, Hes, Lophotrochozoa
Introduction
Segmentation is seen in the annelid, arthropod and vertebrate phyla. Modifications of the basic segmented body plan, i.e. the emergence of segment-specific structures and functions, are associated with the success of all three groups. Thus, the evolutionary origin(s) of segmentation are of great interest.
Notch/Hes signaling is involved segmentation of representatives of the arthropods and vertebrates (Chipman and Akam 2008; Palmeirim et al. 1997; Stollewerk et al. 2003). This is consistent with three evolutionary scenarios (Davis and Patel 1999). One is that the similarity among distant phyla in Notch/Hes deployment, reflects homology of segmentation; i.e., that the last common ancestor of vertebrates and arthropods was segmented. A second scenario is that the similarity reflects convergence, i.e., that Notch/Hes signaling has been recruited independently to function in axial patterning as segmentation evolved separately in the arthropod and vertebrate ancestors. The third scenario is that Notch/Hes signaling functioned in axial growth of the ancestral bilaterian before segmentation evolved and then was exapted (in the parlance of Stephen J. Gould) to function in patterning as segmentation evolved independently in various bilaterian lineages.
In weighing these possibilities, knowledge of whether and how Notch/Hes signaling functions in segmented and unsegmented representatives of the third superphylum, Lophotrochozoa, will be informative. Here, we have performed the first knockdown of the presumptive Notch/Hes pathway in the leech Helobdella robusta (Annelida; Clitellata), and show that Notch/Hes signaling is also required for normal segmentation in this sequentially segmenting lophotrochozoan.
Methods and materials
Collection and handling of embryos, injections of lineage tracers and morpholino oligomers and semi-quantitative RT-PCR (sqRT-PCR)
These were all carried out as previously described (Gimlich and Braun 1985; Song et al. 2002). Antisense morpholino oligomers (AS Hro-hes MO1 and AS Hro-hes MO2) targeting the 5′ UTR and start site of Hro-hes (accession number AY144625) were designed with help from GeneTools, LLC : 5′-AACTCTGGAGGACATTTCTTTCCC-3′ and 5′-TTATTGTAACTATTCAAATCCGCG-3′, as was the control morpholino oligomer (MM Hro-hes MO) containing 4 mismatched bases relative to AS MO2: 5′-TTtTTcTAACTATTCAAAaCCcCG-3′.
Primer sequence information is as follows: forward and reverse primers for Hro-hes = 5′-ATCAAGAAGCCAATAATAGAG-3′ and 5′- CTGTAATCTCATAGCACGAAG-3′; forward and reverse primers for Hro-alpha-tubulin = 5′-GACATCGATAATACGACATCATTTTC-3′ and 5′-GCCAAAACTGTGGAAGATCAAGA-3′; forward and reverse primers for Hro-notch = 5′-GGATCTCCAGGACGAAACTCC-3′ and 5′-TGCTCTTCCTGTTCTCGCTTC-3′.
DAPT treatment
Experimental embryos were raised in media containing a final concentration of 62.5 microM DAPT and 0.25% DMSO, diluted 1/400 from a stock containing 25 mM DAPT in DMSO. Control embryos were raised in media containing 0.25% DMSO. Media was changed daily for all cultured embryos.
Results and discussion
As in vertebrates and many arthropods, the segments of clitellate annelids such as Helobdella arise in anterior to posterior progression. However, whereas segmentation in most other experimental systems proceeds via the formation of boundaries within fields of what are initially interchangeable cells, segmentation of the mesoderm and ectoderm in Helobdella and other clitellates is dependent on stereotyped lineages involving many asymmetric cell divisions. Thus, the segmentation process in Helobdella cannot be separated from the precisely regulated patterns of cell division and migration by which the segments arise.
The posterior progress zone (PPZ) from which segments arise in Helobdella comprises exactly five bilateral pairs of large, identifiable stem cells (teloblasts). Each teloblast undergoes many highly unequal divisions to generate a column of progeny called blast cells. There are actually seven distinct types of blast cells, because the N and Q lineages each generate two different blast cell types in exact alternation. Each blast cell type contributes a particular set of progeny to one or more segments by a stereotyped sub-lineage.
Thus, each left and right half-segment consists of five interdigitating groups of cells (M, N, O, P and Q kinship groups) derived from five corresponding segmentation stem cells (M, N, O, P and Q teloblasts) at the posterior of the embryo (Weisblat and Shankland 1985). The kinship groups are not clones because each kinship group arises by the partial interdigitation, across segment boundaries, of two or more blast cell clones (Weisblat and Shankland 1985). Previous studies have shown that this lineage-dependent segmentation process seen in Helobdella is largely, though not entirely, autonomous in each of the five lineages. Injected lineage tracers can be used to follow segmentation from the stem cell divisions of the teloblasts through the formation of segmentally iterated kinship groups of differentiated cells (Shankland and Savage 1997).
For the work presented here, we have made use of the following details of the segmentation process in Helobdella. First, morphological segmentation was assessed in the main neurogenic (N) lineage, where this process has been most carefully described (Shain et al. 1998). As alluded to above, blast cells in the N lineage assume two distinct fates (nf and ns) in exact alternation (Figure 1). A key step in dividing the continuous columns of blast cell progeny into discrete ganglionic primordia is the separation of cells in the nf.p clone (at the posterior margin of each ganglionic primordium) from cells in the ns.a clone (at the anterior margin of the next ganglionic primordium). This process, called fissure formation, appears to take place autonomously in the N lineage (Shain et al. 2000).
Figure 1.
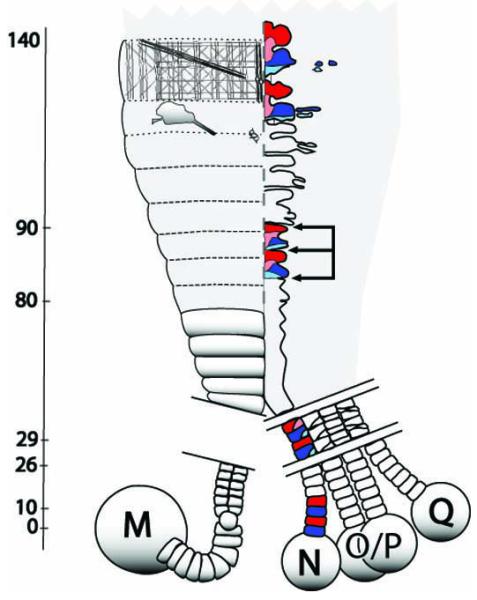
Schematic representation of the stem cell-mediated, lineage-dependent segmentation in the leech and other clitellate annelids. Anterior is to the top. Pairs of diagonal lines indicate discontinuities in the depicted structures. During cleavage (not illustrated), the D quadrant gives rise to one bilateral pair of mesodermal stem cells (M teloblasts) and four bilateral pairs of ectodermal stem cells (N, O/P, O/P and Q teloblasts). These constitute a cellularly defined posterior growth zone. Each teloblast undergoes several dozen highly unequal divisions producing columns (bandlets) of segmental founder cells (blast cells). Following the morphogenetic movements of gastrulation (not illustrated), the bandlets merge along the ventral midline (dotted line) in anteroposterior progression forming the germinal plate (gray shading), with older, more advanced blast cell clones at the anterior. Each of the five lineages contributes a stereotyped set of segmentally iterated progeny to the definitive segments. Thus, the timing of events in each lineage can be standardized by describing them in terms of the clonal age (cl.ag. indicated in hours on the axis), i.e., the time since the birth of the blast cell founding a given clone. For clarity, only the M lineage on the left side and the N lineage on the right side are shown here. The m blast cells undergo their first mitosis (indicated by a rounded cell in the bandlet) at 10 hours cl.ag.; the mitosis is oriented so that the daughter cells lie side by side within the bandlet. Prior to entering the germinal plate, m blast cell clones undergo several more divisions (not shown), giving rise to somites (solid and dotted contours). During subsequent development, each m blast cell clone interdigitates longitudinally with its neighbors, giving rise (by 140 hours cl.ag.) to a stereotyped complement of definitive progeny distributed over three segments; illustrated here are muscles (anteriormost meshwork), nephridia (irregular contour posterior to muscles) and a few ganglionic neurons (small contours near midline). In the N lineage, two types of blast cells, designated nf (blue) and ns (red) arise in exact alternation within the bandlet. These cells make their first divisions at different times (26 and 29 hours cl.ag., respectively) and with different degrees of asymmetry. Their definitive progeny comprise distinct sets of identifiable neurons and make up most of the segmental ganglia of the ventral nerve cord. A key event in the formation of discrete segmental ganglia is the partitioning of what are initially continuous columns of nf and ns progeny into separate ganglionic primorida. This occurs at roughly 80-90 hours cl.ag., as transverse fissures (3 arrows delimit 2 ganglionic primordia) separate the clones of cells derived from the posterior daughters of nf blast cells (nf.p, light blue) from those derived from the anterior daughters of ns blast cell (ns.a, pink).
Second, to examine the effects of disrupting Notch/Hes signaling at the level of individual cell divisions, we used the mesodermal (M) lineage, where the division pattern of the primary blast cells can be easily examined. As alluded to above, each m blast cell gives rise to one segment's worth of progeny, although individual m blast cell clones interdigitate longitudinally in a stereotyped manner (Weisblat and Shankland 1985). Divisions of the primary m blast cells can be assessed relatively conveniently for two reasons. Firstly, the mitotic axis is perpendicular to that of the blast cell column, so the column of m blast cells becomes two cells wide distal to the division and the last undivided cell is easily identified. Moreover, the m blast cells divide ~10 hours after they are born from the M teloblast, so variations in the positioning of this mitosis can be readily be determined by counting the number of undivided primary blast cells (Zackson 1984).
H. robusta embryos express both Notch and Hes homologs (Hro-notch and Hro-hes, respectively) in various cells of the early embryo, including the teloblasts and blast cells of the posterior growth zone (Song 2000; Song et al. 2004; Rivera et al. 2005). To test for a functional relationship between Notch signaling, Hro-hes expression and segmentation, we bathed stage 5 embryos for 36 hours in an inhibitor of Notch signaling, DAPT. DAPT (N-[N-(3,5-Difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester) is an inhibitor of a proteolytic processing step that is required for nuclear translocation of the intracellular domain of ligand-activated Notch and the ensuing up-regulation of Hes transcription. It has been shown to be a specific inhibitor of Notch signaling in protostomes, deuterostomes, and a cnidarian (Geling et al. 2002; Micchelli et al. 2002; Kasbauer et al. 2007). Alternatively, or in combination with DAPT treatment of embryos, individual teloblasts were injected with antisense morpholino oligomers targeting Hro-hes translation (AS Hro-hes MO). The efficacy of the AS MO knockdown approach in Helobdella has been previously demonstrated for even-skipped-, nanos-, and pax III-related genes (Song et al. 2002; Agee et al. 2006; Woodruff et al. 2007).
Neither treating embryos with DAPT alone, nor injecting individual teloblasts with AS Hro-hes MOs lead to detectable segmentation defects, as judged by the observed distribution of microinjected cell lineage tracer (data not shown and Figure 3). However, DAPT treatment alone did reduce global Hro-hes levels by about 30% as judged by semi-quantitative RT-PCR using template derived from whole embryos, while other control mRNA species, e.g. Hro-alpha-tubulin and Hro-notch, were not affected (Figure 2A). This specific decline in global Hro-hes levels in response to DAPT treatment was reproducible and statistically significant (p < 0.001), and is consistent with the hypothesis that Hro-hes is a target of Notch signaling. Localization of Hro-hes was not affected by DAPT treatment (Figure 2B, C).
Figure 3.
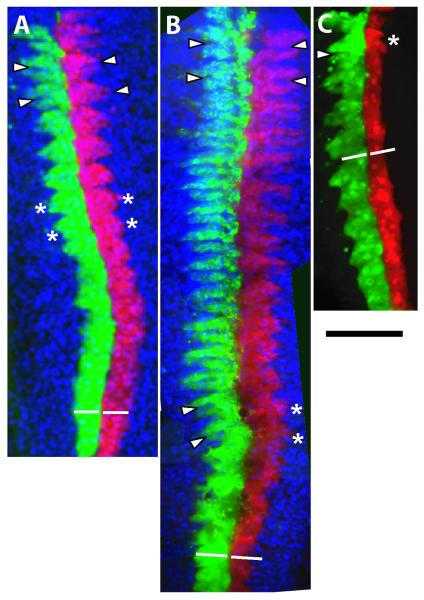
Fluorescence micrographs showing ventral views of germinal plates dissected from embryos 3 (C) to 4 (A, B) days after the right N teloblasts had been injected with a mixture of fluoresceinated aminodextran (FDA, green) and MM Hro-hes MO (MM MO; 4 mM in the injection needle), and the left N teloblasts had been injected with a mixture of rhodaminated aminodextran (RDA, red) and a cocktail of two AS Hro-hes MOs (AS MO; 4 mM in needle). Embryos shown in A and B were counterstained with Hoechst 33258 (blue) to reveal nuclei. A) In the absence of DAPT treatment, segmentation has proceeded normally in both the MM MO and AS MO-injected lineages, as judged by the formation of fissures (arrowheads) separating ganglionic primordia in the anterior segments, the formation of lateral bulges (asterisks) in middle segments and the uniform width of the labeled lineages in posterior segments (left and right bars of equal length). B and C) Experimental embryos raised in media containing DAPT. B) A moderate morphotype, in which the anteriormost ganglionic primoridia have separated normally (arrows). More posteriorly in the germinal plate, the lateral bulges on the AS MO-injected side have not yet formed fissures, and the AS MO injected lineage is narrower than the MM MO-injected lineage. C) A severe morphotype, in which the AS MO-injected lineage failed to form lateral bulges and is narrower along its entire length. Embryos with this phenotype did not survive to 4 days post injection. D) Graphical representation of a segment-by-segment comparison of the AS MO- and MM MO-injected N lineages in embryos such as those shown in panels A-C. Each bar corresponds to one embryo. Dark green indicates that segmentation had progressed to an equivalent extent on the two sides, light green indicates that segmentation was abnormal on the AS MO-injected side relative to the MM-MO-injected side, and white indicates the failure to form even a lateral bulge on the AS MO-injected side of a segment in which segmentation was visible on the MM-MO-injected side (see panels B and C for examples). Variability in the numbers of segments scored in various specimens depends primarily on the success of the dissection. Scale bar, 250 microns.
Figure 2.
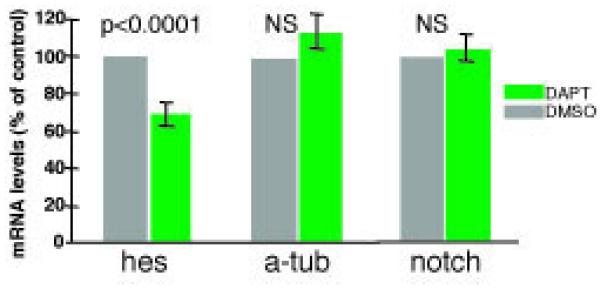
Hro-hes levels are reduced in response to DAPT treatment. A) Semi-quantitative RT-PCR was used to compare the levels of Hro-hes (hes), Hro-alpha-tubulin (a-tub), and Hro-notch (notch) transcripts in embryos treated with DAPT throughout stages 1-7 (~78 hours), relative to those in sibling control embryos treated with DMSO. Hro-hes levels were reduced by approximately 25% relative to controls (p < 0.0001), while those for Hro-alpha-tubulin and Hro-notch itself showed no statistically significant change (NS). B) Representative embryos grown in 62.5 microM DAPT or DMSO in parallel with the embryos used for RT-PCR, then fixed and processed to reveal Hro-hes by in situ hybridization at late stage 7. DAPT-treated embryos (left) and DMSO-treated sibling embryos processed in parallel show indistinguishable patterns of transcript localization. Staining is seen over the primary blast cell nuclei (black arrowheads) and in the germ bands (black arrows; slightly out of focus in these images). Staining is also evident over the mitotic apparatus of dividing primary m blast cells in both DAPT- and carrier -treated embryos (black arrows, insets); mitotic m blast cells were identified by their rounded up shape and by their position in the m-bandlet. Scale bar: 60 microns; 30 microns in insets.
In zebrafish, experimental and theoretical work has demonstrated the existence of homeostatic processes that counteract partial inhibition of the Notch/Hes network (Lewis 2003). Whether or not such processes operate in Helobdella, it seemed likely that combining DAPT treatment and AS Hro-hes MO would yield a more pronounced phenotype than either reagent by itself. To test this idea, we injected one N teloblast with AS Hro-hes MO (plus a rhodamine-conjugated dextran lineage tracer) and the contralateral teloblast with a control MO (plus a fluorescein-conjugated dextran lineage tracer) and treated the injected embryos with DAPT, or DMSO alone.
We found that segmentation was disrupted specifically in lineages subjected to the combined effects of DAPT and AS Hro-hes MO. Under these conditions, perturbing Notch/Hes signaling in Helobdella does not affect the overall production of blast cells by the teloblasts. The blast cells themselves also divide, but fail to generate the regular, segmentally iterated patterns of definitive progeny. Specifically, we monitored fissure formation in the N lineage, which normally leads to the formation of discrete ganglionic primordia (Figures 1 and 3). The severity of the observed defects in such embryos varies from mild to severe, presumably reflecting variability in the amount of AS Hro-hes MO injected. Moreover, the severity of the morphotype increases over time; in moderately affected lineages (34 of 42 embryos), blast cells produced soon after the injection generate discrete, seemingly normal ganglionic primordia at the anterior region of the germinal plate (compare Figure 3A and 3B). Posteriorly the AS Hro-hes MO-injected lineage fails to form the fissures that normally separate ganglionic primoridia and occupies a narrower space within the germinal plate, suggesting a disruption of normal cell proliferation and/or morphogenetic movements (Figure 3B). In more severely affected lineages (8 of 42 embryos), cell proliferation and ganglion morphogenesis are affected throughout the injected lineage (Figure 3C).
To gain a quantitative measure of the distribution of segmentation phenotypes, we evaluated morphogenesis on a segment by segment basis. To do this, we compared progeny of the AS MO-injected N teloblast to progeny of the contralateral MM MO-injected N teloblast, in embryos bathed in either DAPT or DMSO alone. We examined those portions of successfully dissected germinal plates in which segmentation was evident in the MM MO-injected side. Segmentation was judged in anterior portions of the germinal plate by the presence of fissures separating ganglionic primordia, and in the posterior portions by clearly defined lateral bulges, as expected morphology for the clonal age range of n blast cell clones at this stage. Twenty-six embryos were successfully dissected, 9 DMSO-treated embryos and 17 DAPT-treated embryos (14 of which were weakly affected and 3 of which were strongly affected). In the DAPT-treated embryos, a total of 186 segments were scored as present on the MM MO-injected sides, versus 133 on the AS MO-injected side, many of which were abnormal (Figure 3D). Statistically, this result was highly significant (P<0.0005; two-tailed t-test). In contrast, among the DMSO treated embryos all but one specimen showed equivalent degrees of segmentation on the MM MO-injected and AS MO-injected sides (Figure 3D). That one specimen may be attributed to misinjection of the teloblast, but even when it was included, the difference in the total numbers of segments produced on the MM MO-injected and MO MO-injected sides (101 and 95, respectively) was not statistically significant (P = 0.35).
It has been shown previously that morphologically recognizable segments in Helobdella stem from the temporospatially stereotyped division patterns of the various blast cell sub-lineages, and Hro-notch and Hro-hes are expressed in these cells. We therefore, anticipated that the perturbation of morphological of segmentation by disrupting Notch/Hes signaling would be preceded by changes in primary blast cell division patterns. For ease of analysis, we chose to use the M lineage in testing this prediction. As described above, each m blast cell normally divides about 15 hours after its birth from the teloblast. At this time, the dividing cell is separated from the parent M teloblast by 9 or 10 younger blast cells, but has not yet entered the germinal band (Figure 1) and thus is easy to visualize. We injected M teloblasts with AS Hro-hes MOs, bathed the injected embryos in DAPT and examined the timing of the first division of the primary blast cells of the M lineage. DAPT-treated embryos in which the M teloblast was injected with AS Hro-hes MOs failed to even develop a normal germinal plate and so morphological segmentation was obviously disrupted (data not shown). At this time of its first division, the dividing M cell (Figure 1) lies near the surface where it is easy to visualize. After injecting M teloblasts with AS Hro-hes MOs and treating the injected embryos with DAPT, we observed a dramatic increase in the variability of the position in the blast cell column at which primary m blast cells underwent mitosis (Figure 4). In controls, the first m blast cell division occurred at a separation of 9 or 10 blast cells from the parent teloblasts in all cases [average 9.4 +/−0.5 (n=12)]. This range increased to between 7 and 13 cells in response to disruption of Notch signaling [average 9.2+/−2.0 (n=12)]. This increase in variability is statistically highly significant (P<10−5; F-test). Thus, in H. robusta, disrupting Notch/Hes signaling affects critical features of segmentation in both mesodermal and ectodermal tissues. Although we cannot yet exclude the possibility of an indirect effect, from mesoderm to ectoderm or vice versa, we have shown previously that Hro-notch and Hro-hes are expressed in both tissues (Song et al. 2004; Rivera et al. 2005).
Figure 4.
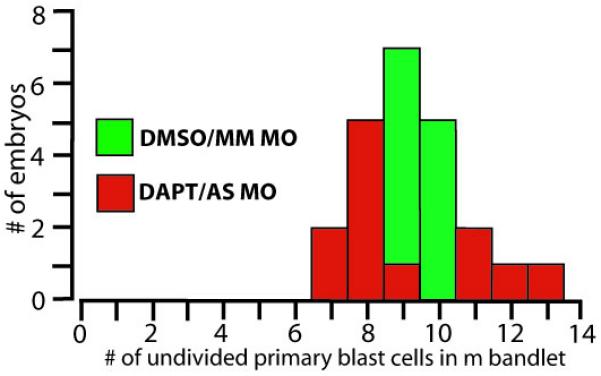
Disrupting Notch/Hes signaling increases the variability in the position of m blast cell mitoses. Graph illustrating the position in the bandlet (as judged by the number of cells away from the parent M teloblast) at which m blast cells underwent their first mitoses in 12 control embryos (green) and in 12 siblings (red) in which Notch/Hes signaling had been disrupted by a combination of DAPT treatment and AS MO injection of the M teloblast.
The functional experiments presented here, together with previous descriptions of the expression of Hro-hes and Hro-notch genes, indicate that Notch/Hes signaling is required for segmentation in the lophotrochozoan species Helobdella robusta, as has been shown previously for vertebrates and arthropods. Returning to the three evolutionary scenarios presented in the Introduction, finding that Notch/Hes signaling functions in segmentation in representatives of all three super-phyla makes it less likely that this similarity has arisen by convergence and favors the two other scenarios, i.e., that either the ur-bilaterian was already segmented, or that Notch/Hes signaling was exapted to function in segmentation from a role in axial growth and/or patterning in an unsegmented ancestor. Integrating the developmental analyses, with phylogenetic considerations (the paucity of segmented taxa and their scattered distribution among unsegmented modern phyla) and paleontological reasoning as to the ur-bilaterian body plan (Collins et al. 2000), we deem it more likely that Notch/Hes signaling was used in terminal elongation of an unsegmented ancestral bilaterian, from which it was exapted to function in segmentation as this trait evolved independently in the deuterostome, ecdysozoan and lophotrochozoan lineages. Resolving this fundamental evolutionary question requires further comparisons of cellular and molecular mechanisms of axial growth and patterning in both segmented and unsegmented taxa.
Acknowledgements
This work was supported by NIH RO1 GM 60240 to DAW. We thank members of our lab for many helpful discussions and comments on this manuscript.
References
- Agee SJ, Lyons DC, et al. Maternal expression of a NANOS homolog is required for early development of the leech, Helobdella robusta. Developmental Biology. 2006;298:1–11. doi: 10.1016/j.ydbio.2006.04.473. [DOI] [PubMed] [Google Scholar]
- Chipman AD, Akam M. The segmentation cascade in the centipede Strigamia maritima: involvement of the Notch pathway and pair-rule gene homologues. Developmental Biology. 2008;319:160–169. doi: 10.1016/j.ydbio.2008.02.038. [DOI] [PubMed] [Google Scholar]
- Collins AG, Lipps JH, et al. Modern mucociliary creeping trails and the bodyplans of Neoproterozoic trace-makers. Paleobiology. 2000;26:47–55. [Google Scholar]
- Davis GK, Patel NH. The origin and evolution of segmentation. Trends Biochem Sci. 1999;24:M68–M72. [PubMed] [Google Scholar]
- Geling A, Steiner H, et al. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. Embo Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimlich RL, Braun J. Improved Fluorescent Compounds for Tracing Cell Lineage. Developmental Biology. 1985;109:509. doi: 10.1016/0012-1606(85)90476-2. [DOI] [PubMed] [Google Scholar]
- Kasbauer T, Towb P, et al. The Notch signaling pathway in the cnidarian Hydra. Developmental Biology. 2007;303:376–390. doi: 10.1016/j.ydbio.2006.11.022. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Current Biology. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, Esler WP, et al. gamma-secretase/presenilin inhibitors for Alzheimer's disease phenocopy Notch mutations in Drosophila. Faseb J. 2002;16:79–81. doi: 10.1096/fj.02-0394fje. [DOI] [PubMed] [Google Scholar]
- Palmerim I, Henrique D, et al. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Rivera AS, Gonsalves FC, et al. Characterization of Notch-class gene expression in segmentation stem cells and segment founder cells in Helobdella robusta (Lophotrochozoa; Annelida; Clitellata; Hirudinida; Glossiphoniidae) Evolution & Development. 2005;7:588–599. doi: 10.1111/j.1525-142X.2005.05062.x. [DOI] [PubMed] [Google Scholar]
- Shain DH, Ramirez-Weber FA, et al. Gangliogenesis in leech: morphogenetic processes leading to segmentation in the central nervous system. Dev Genes Evol. 1998;208:28–36. doi: 10.1007/s004270050150. [DOI] [PubMed] [Google Scholar]
- Shain DH, Stuart DK, et al. Segmentation of the central nervous system in leech. Development. 2000;127:735–744. doi: 10.1242/dev.127.4.735. [DOI] [PubMed] [Google Scholar]
- Shankland M, Savage RM. Annelids, the segmented worms. In: Gilbert SF, Raunio AM, editors. Embryology: Constructing the Organism. Sinauer; Sunderland, MA: 1997. pp. 219–235. [Google Scholar]
- Song MH. Biophysics. Berkeley, University of California; Berkeley: 2000. Characterization of hairy/E(spl)- and eve-class genes in the posterior growth zone of the leech Helobdella robusta, a segmented lophotrochozoan; p. 179. [Google Scholar]
- Song MH, Huang FZ, et al. Expression and function of an even-skipped homolog in the leech Helobdella robusta. Development. 2002;129:3681–3692. doi: 10.1242/dev.129.15.3681. [DOI] [PubMed] [Google Scholar]
- Song MH, Huang FZ, et al. Cell cycle-dependent expression of a hairy and Enhancer of split (hes) homolog during cleavage and segmentation in leech embryos. Developmental Biology. 2004;269:183–195. doi: 10.1016/j.ydbio.2004.01.025. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Schoppmeier M, et al. Involvement of Notch and Delta genes in spider segmentation. Nature. 2003;423:863–865. doi: 10.1038/nature01682. [DOI] [PubMed] [Google Scholar]
- Weisblat DA, Shankland M. Cell Lineage and Segmentation in the Leech. Philos T Roy Soc B. 1985;312:39. doi: 10.1098/rstb.1985.0176. [DOI] [PubMed] [Google Scholar]
- Woodruff J, Mitchell B, et al. Hau-Pax3/7A is an early marker of leech mesoderm involved in segmental morphogenesis, nephridial development, and body cavity formation. Dev Biol. 2007;306:824–837. doi: 10.1016/j.ydbio.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Zackson SL. Cell Lineage, Cell Cell-Interaction, and Segment Formation in the Ectoderm of a Glossiphoniid Leech Embryo. Developmental Biology. 1984;104:143–160. doi: 10.1016/0012-1606(84)90044-7. [DOI] [PubMed] [Google Scholar]