Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) holds an important role in retaining lung function, but its association with lung cancer is unclear. A case-control study was conducted to determine the possible associations of the genetic variants in the CFTR gene with lung cancer risk. Genotypes of a most common deletion ΔF508, one functional SNP, and eight tag SNPs in the CFTR gene were determined in 574 lung cancer patients and 679 controls. A logistic regression model, adjusting for known risk factors, was used to evaluate the association of each variant with lung cancer risk, as confirmation haplotype and sub-haplotype analyses were performed. ΔF508 deletion and genotypes with minor alleles in one tag SNP, rs10487372, and one functional SNP, rs213950, were inversely associated with lung cancer risk. The results of haplotype and sub-haplotype analyses were consistent with single variant analysis, all pointing to deletion ΔF508 being the key variant for significant haplotypes and sub-haplotypes. Individuals with ‘deletion-T’ (ΔF508/rs10487372) haplotype had a 68% reduced risk for lung cancer compared to common haplotype ‘no-deletion-C’ (OR=0.32; 95% CI=0.15–0.68; p=0.01). Genetic variations in the CFTR gene might modulate the risk of lung cancer. This study, for the first time, provides evidence of a protective role of the CFTR deletion carrier in the etiology of lung cancer.
Keywords: Cystic fibrosis transmembrane conductance regulator, lung cancer, genetic variation
Introduction
Despite increasing knowledge of individual susceptibility, the genetic etiology of lung cancer remains ambiguous. Cystic fibrosis (CF) is a life-limiting autosomal recessive disorder in which progressive lung disease is common and early in life. The cystic fibrosis transmembrane conductance regulator (CFTR) functions as a chloride channel and controls the regulation of other transport pathways. CFTR gene mutations, resulting in severe dysfunction of the CFTR, are well-known to be responsible for CF [1, 2]. Although CF is rare, about 5% of the Caucasian populations are heterozygous mutation carriers of the CFTR gene. One theory for this high incidence of CFTR mutation carriers in the population is that these carriers may have a certain biological advantage [3].
The relationship between the CFTR gene and cancer risk has been investigated. A large cohort study in North American and European patients with CF found that while the overall risk of cancer was similar to that of the general population, there was an increased risk of digestive tract cancers in CF patients [4]. Individuals who were CFTR mutation carriers were found to be at an increased risk for young onset of pancreatic cancer [5]. An inverse association between CF gene mutations and incidence of several cancers, such as melanoma[3, 6], breast cancer[6–8], colon cancer[6] and prostate cancer[9], was also reported. However, no studies have been reported on the association between CFTR and lung cancer risk. Because of the important role of the CFTR in maintaining lung function, we hypothesize that the CFTR gene mutation may alter lung cancer susceptibility. In our case-control study, the genetic variations of the CFTR gene were systematically investigated by analyzing the ΔF508 deletion, one functional single nucleotide polymorphism (SNP), and eight tag SNPs. Our goal was to determine the possible association of CFTR gene alterations and lung cancer risk.
Materials and Methods
Study Subjects
Lung cancer patients were identified and enrolled at Mayo Clinic between 1997 and 2007. The detailed study design and the subject enrollment process were reported previously [10–12]. Briefly, new cases diagnosed with lung cancer are identified by a daily electronic pathology reporting system. Once identified, study consent was obtained from the patients for enrollment, their medical records abstracted, and interviews conducted. Controls were selected from community residents who were identified by having had a general medical examination and a leftover blood sample from routine clinical tests [10, 13]. Excluded were individuals who had been diagnosed with major organ failure (e.g., heart, brain, lung, kidney, or liver) on or prior to this visit. The controls were frequency matched to patients on age, sex, and race/ethnicity. A self-administered questionnaire with the same questions as obtained from patients with lung cancer was completed by the controls. The research protocol and consent form were approved by the Mayo Clinic Institutional Review Board.
Data Collection
Demographic and other risk information was obtained from all study subjects via a combination of a structured subject interview, self-administered questionnaire, and medical records [10–13]. Never smokers were defined as having smoked fewer than 100 cigarettes during their lifetimes. Detailed information on second hand smoking (SHS) history was collected on the source, amount, and duration of exposure. SHS was modeled as a dichotomized covariate (yes versus no). History of chronic obstructive pulmonary disease (COPD) was determined based on explicit diagnosis recorded in the medical history. Family history of lung cancer in first-degree relatives was also collected.
SNP Selection and genotyping
The most common deletion, ΔF508 (rs332), was the primary target alteration under evaluation. Tag SNPs for the CFTR were selected using Haploview software. Genotyping data of the CFTR gene for Caucasian (CEU) Hapmap samples was downloaded from HapMap (http://www.hapmap.org). Tag SNPs were identified using the following criteria: aggressive tagging using 2 and 3 marker haplotypes; a minor allele frequency ≥0.1; r2≥0.8; and LOD, 3.0. Nine tag-SNPs were selected to capture 103 of 103 (100%) alleles. One of tag-SNP, rs1800089, failed the test. Table 1 lists nine SNPs successfully genotyped and their genomic information; one of the SNPs was functional SNP (rs213950, 470 Met>Val), and the other eight SNPs were haplotype tag SNPs.
Table 1.
Information of genotyped variants
| SNP rs numbera | Contig position(bp) b |
Gene Position |
Minor allele frequency | p value for HWEd |
||
|---|---|---|---|---|---|---|
| In databasec | Cases | Controls | ||||
| ΔF508 (rs332) | 42383223 TCT> - | exon_11 | – | – | – | – |
| SNP1: rs17139943 | 42313067 A>G | Intron_1 | 0.33 | 0.21 | 0.23 | 0.05 |
| SNP2: rs17449197 | 42362330 A>G | Intron_7 | 0.18 | 0.13 | 0.14 | 0.43 |
| SNP3: rs213950 | 42383109 G>A (470 Met>Val) |
exon_11 | 0.49 | 0.39 | 0.43 | 0.49 |
| SNP4: rs10487372 | 42384475 C>T | Intron_11 | 0.13 | 0.10 | 0.14 | 0.82 |
| SNP5: rs1469486 | 42403411 C>T | Intron_11 | 0.11 | 0.10 | 0.10 | 0.21 |
| SNP6: rs17547853 | 42410480 G>A | Intron_11 | 0.23 | 0.15 | 0.17 | 0.11 |
| SNP7: rs213968 | 42413569 C>T | intron_12 | 0.48 | 0.39 | 0.43 | 0.43 |
| SNP8: rs11978434 | 42415428 T>C | intron_13 | 0.33 | 0.23 | 0.24 | 0.09 |
| SNP9: rs213988 | 42445880 C>T | intron_21 | 0.24 | 0.20 | 0.20 | 0.24 |
The rs number shown is the NCBI dbSNP cluster ID for each SNP and deletion.
The number indicates the location of the SNP relative to the start codon ATG according to the NCBI genomic contig NT_007933.
Minor allele frequency in the Hapmap database for Caucasian (CEU) population.
The Hardy-Weinberg equilibrium (HWE) in the control group was tested using a goodness-of-fit chi-square test.
–, Information is not available.
Each subject’s blood sample was assigned a blind identification number and tested at the Mayo Clinic Genomics Shared Resource laboratory. ΔF508 three-nucleotide deletion was assayed by fragment analysis using fluorescent primers tagged with 6-FAM and detected on the ABI 3730 Genetic Analyzer (Applied Biosystems, Foster City, CA). All SNP genotyping was conducted using a SNPstream Genotyping System (Beckman, Fullerton, CA). The detailed methods and quality control procedures are provided as an online supplement (Supplementary Methods).
Statistical Analysis
Only one subject had ΔF508 homozygous deletion (control group), and this individual was combined with the heterozygous deletion carriers. ΔF508 deletion was analyzed by comparing deletion carriers with non-carriers. The SNP genotyping data were analyzed by comparing the homozygotes and the heterozygotes of minor alleles with the homozygotes of common alleles. Individual SNP data were examined using an unconditional logistic regression model to estimate odds ratios (ORs) and corresponding 95% confidence intervals (CIs) with adjustment for the effect of covariates, i.e., age, gender, smoking status, SHS, history of COPD, and family history of lung cancer in first degree relatives. A logistic regression model was also applied to test for a linear trend by treating the genotypes as values of 0, 1, and 2 copies of the minor allele in one model. All above analyses were performed by SAS 8.0 (SAS Institute, Cary, NC, USA). Significant variants were further examined for their association with lung cancer risk stratified by gender, smoking status, and major histological subtypes of lung cancer. The heterogeneity between the stratas was analyzed by using a Q test. All above analyses were performed by SAS 8.0 (SAS Institute, Cary, NC, USA).
The linkage disequilibrium (LD) plot of the genotyped SNPs based on 679 Caucasian controls was constructed by the Haploview program [14] using the method described by Gabriel et al [15] with default settings. Haplotype analyses were performed by using the Haplo.stats package for R [16]. First, the haplo.score [16] was used to compute score statistics to test associations between haplotypes and lung cancer risk. Simulation p-values were computed with a minimum of 1000-time stimulations. Second, the haplo.glm [17] was used to compute the regression of a phenotype (lung cancer) on haplotypes adjusting for other potential risk factors. Third, a case-control haplotype analysis was performed to calculate the frequency of haplotypes between cases and controls and ORs by the haplo.cc [16]. Finally, to evaluate the association of sub-haplotypes (subsets of alleles from the full haplotype) with lung cancer risk, a ‘sliding window’ of 2-loci and 3-loci alleles was evaluated across the entire haplotype by the haplo.score.slide [16]. To confirm the result of sub-haplotype analysis, another approach, the sequential haplotype scan, was used to choose loci for haplotype associations by seqhap [18, 19]. This sequential haplotype scan method can search for combinations of adjacent markers that are jointly associated with disease status.
Results
Population Characteristics
Demographic and clinic characteristics of the subjects are described in Table 2. The percentage of ever smokers and second hand smokers was significantly higher in the cases than in the controls (P<0.001). Significantly more subjects with a history of COPD were present among the cases than among the controls (33.4% vs.2.9%; P<0.0001). The frequency of subjects with a family history of lung cancer was significantly higher in the cases than in the controls (12.4% vs. 3.4%; P < 0.0001). The lung cancer histological types for most of the patients (66.3%) were adenocarcinoma (46.3%) and squamous cell carcinoma (20.0%). Forty-four percent of the cases had stage I or II lung cancer, and 56% had stage III or IV.
Table 2.
General characteristics of cases and controls: A Mayo Clinic case-control study of lung cancer, 1998–2007
| Characteristics | Cases (574) N(%) |
Controls (679) N(%) |
p a |
|---|---|---|---|
| Race | |||
| Caucasian | 574 (100) | 679 (100) | |
| Gender | 0.327 | ||
| Female | 300(52.3) | 336(49.5) | |
| Male | 274(47.7) | 343(50.5) | |
| Age(years) | 0.771 | ||
| <=50 | 143(24.9) | 159(23.4) | |
| 50–79 | 360(62.7) | 439(64.7) | |
| >79 | 71(12.4) | 81(11.9) | |
| Cigarette smoking | <0.001 | ||
| Never | 202(35.2) | 304(44.8) | |
| Ever | 372(64.8) | 375(55.2) | |
| Second hand smoking | <0.001 | ||
| No | 76(13.2) | 134(19.7) | |
| Yes | 498(86.8) | 545(80.3) | |
| History of COPD | <0.0001 | ||
| No | 382(66.6) | 659(97.1) | |
| Yes | 192(33.4) | 20(2.9) | |
| Family history of lung cancer | <0.0001 | ||
| No | 503(87.6) | 656(96.6) | |
| Yes | 71(12.4) | 23(3.4) | |
| Histological types | |||
| Adenocarcinoma | 266(46.3) | – | |
| Squamous cell carcinoma | 115(20.0) | – | |
| Non-small cell carcinoma | 45(7.8) | – | |
| Small-cell carcinoma | 17(3.0) | – | |
| Large-cell carcinoma | 11(1.9) | – | |
| Bronchoalveolar carcinoma | 26(4.5) | – | |
| Carcinoid carcinoma | 40(7.0) | – | |
| Mixed histology/other | 54(9.5) | – | |
| Tumor stages | |||
| I +II | 255(44.4%) | – | |
| III+IV | 319(55.6%) | – |
Pearson’s chi-square test.
Single variant associated analysis
The distributions of the CFTR genotypes among the cases and controls and the adjusted ORs associated with lung cancer are summarized in Table 3. The ΔF508 deletion and genotypes of rs213950 (GA and AA) and rs10487372 (TC) were found to be significantly associated with lung cancer risk. ΔF508 carriers had a lower risk for lung cancer compared with non-carriers (OR=0.37, P=0.01).
Table 3.
Frequency distribution of CFTR genotypes among cases and controls and their association with lung cancer risk: A Mayo Clinic case-control study of lung cancer, 1998–2007
| Mutations | Cases N(%) |
Controls N(%) |
Logistic regressiona |
||
|---|---|---|---|---|---|
| OR(95%CI) | p | Trend pb | |||
| ΔF508 deletion: rs332 | 0.010 | ||||
| Non-carriers | 535(98.3) | 603(94.5) | 1.00(Reference) | ||
| Carriers | 9(1.7) | 35(5.5) | 0.37(0.17–0.78) | 0.010 | |
| SNP1: rs17139943 | 0.800 | ||||
| AA | 347(60.6) | 413(60.8) | 1.00(Reference) | ||
| GA | 211(36.8) | 226(33.3) | 1.18(0.90–1.56) | 0.227 | |
| GG | 15(2.6) | 40(5.9) | 0.49(0.24–1.01) | 0.052 | |
| GA+GG | 226(39.4) | 266(39.2) | 1.08(0.83–1.41) | 0.557 | |
| SNP2: rs17449197 | 0.961 | ||||
| AA | 431(75.2) | 515(76.0) | 1.00(Reference) | ||
| GA | 136(23.7) | 149(22.0) | 1.02(0.75–1.40) | 0.892 | |
| GG | 6(1.1) | 14(2.0) | 0.57(0.17–1.88) | 0.356 | |
| GA+GG | 142(24.8) | 163(24.0) | 0.99(0.73–1.35) | 0.951 | |
| SNP3: rs213950 | 0.019 | ||||
| GG | 204(35.7) | 227(33.4) | 1.00(Reference) | ||
| GA | 292(51.1) | 323(47.6) | 0.54(0.36–0.80) | 0.001 | |
| AA | 76(13.3) | 129(19.0) | 0.58(0.39–0.88) | 0.004 | |
| GA+AA | 368(64.4) | 452(66.6) | 0.55(0.38–0.81) | 0.002 | |
| SNP4: rs10487372 | 0.003 | ||||
| CC | 459(80.4) | 500(74.5) | 1.00(Reference) | ||
| TC | 108(18.9) | 160(23.9) | 0.62(0.44–0.86) | 0.004 | |
| TT | 4(0.7) | 11(1.6) | 0.70(0.21–2.33) | 0.556 | |
| TC+TT | 112(19.6) | 171(25.5) | 0.63(0.45–0.87) | 0.005 | |
| SNP5: rs1469486 | 0.203 | ||||
| CC | 463(80.7) | 550(81.1) | 1.00(Reference) | ||
| TC | 108(18.8) | 125(18.4) | 0.89(0.63–1.25) | 0.948 | |
| TT | 3(0.5) | 3(0.4) | 0.56(0.07–4.79) | 0.853 | |
| TC+TT | 111(19.3) | 128(18.8) | 0.89(0.63–1.25) | 0.495 | |
| SNP6: rs17547853 | 0.669 | ||||
| GG | 404(70.6) | 476(70.1) | 1.00(Reference) | ||
| GA | 161(28.2) | 179(26.4) | 2.63(0.95–7.26) | 0.062 | |
| AA | 7(1.2) | 24(3.5) | 2.44(0.90–6.59) | 0.079 | |
| GA+AA | 168(29.4) | 203(29.9) | 2.61(0.99–6.89) | 0.053 | |
| SNP7: rs213968 | 0.056 | ||||
| CC | 207(36.1) | 228(33.6) | 1.00(Reference) | ||
| TC | 289(50.4) | 323(47.7) | 1.20(0.76–1.36) | 0.991 | |
| TT | 77(13.4) | 127(18.7) | 0.57(0.38–1.86) | 0.063 | |
| TC+TT | 366(63.8) | 450(66.4) | 0.89(0.68–1.17) | 0.404 | |
| SNP8: rs11978434 | 0.798 | ||||
| TT | 327(57.0) | 389(58.3) | 1.00(Reference) | ||
| TC | 228(39.7) | 233(34.9) | 1.95(0.99–3.83) | 0.059 | |
| CC | 19(3.3) | 45(6.8) | 1.60(0.89–3.33) | 0.068 | |
| TC+CC | 247(43.0 | 278(41.7) | 1.72(0.92–3.21) | 0.088 | |
| SNP9: rs213988 | 0.696 | ||||
| CC | 362(63.3) | 431(63.8) | 1.00(Reference) | ||
| TC | 196(34.3) | 214(31.6) | 1.17(0.89–1.55) | 0.163 | |
| TT | 14(2.4) | 31(4.6) | 0.59(0.27–1.32) | 0.198 | |
| TC+TT | 210(36.7) | 245(36.2) | 1.07(0.84–1.36) | 0.471 | |
Bold characters indicate corresponding p values that are less than 0.05.
ORs and p values were adjusted for gender, age, cigarette smoking, SHS, COPD, and family history of lung cancer.
P values were adjusted for gender, age, cigarette smoking, SHS, COPD, and family history of lung cancer with the genotypes as values of 0, 1, and 2 in one model.
For the three significant variants, we further examined their association with lung cancer risk stratified by gender, smoking status, and major histological subtypes of lung cancer. ΔF508 carriers had a decreased risk for lung cancer in females (OR=0.30, P=0.024) compared with non-carriers, and again this risk reduction was only found in squamous cell carcinoma (OR=0.34, P=0.037). For rs213950, lung cancer risk significantly decreased in females and never smokers with the GA and AA genotype compared with the GG genotype; this reduced risk was specific for squamous cell carcinoma. For rs10487372 genotype TC, the reduced risk for lung cancer was also found in females, never smokers, and squamous cell carcinoma. However, we tested the differences of ORs between the strata, and found no significant heterogeneity between the strata of gender, smoking, and histological types. Therefore, the subgroup analysis should be cautious to interpret.
Haplotype analysis
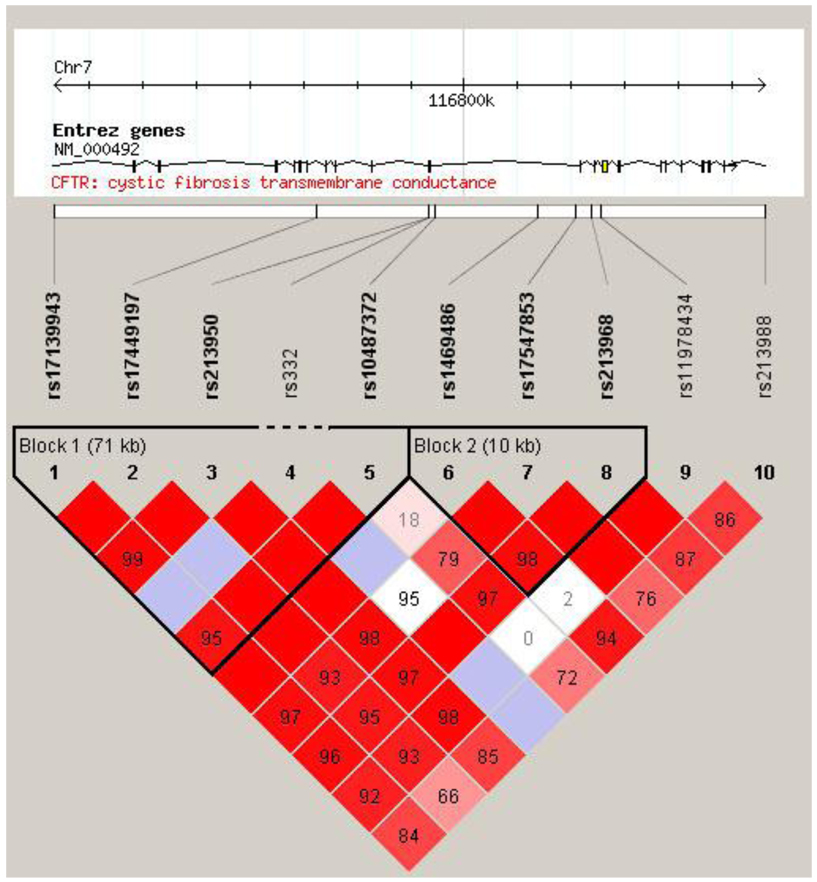
The LD structure of all variants was constructed. Two blocks were defined. ΔF508, rs213950, and rs10487372 were located in the LD block-1 (Figure 1). Table 4 summarizes the frequency of the haplotypes of each block and their association with lung cancer risk. The global score test showed significant differences in the block-1 haplotype distribution between the cases and controls (simulation P=0.008), but there was no significant difference in block-2 (simulation P=0.14). In block-1, the risk of lung cancer was significantly decreased among individuals carrying the haplotype ‘A-A-A-deletion-T’ (OR=0.33, 95% CI 0.16–0.69, adjust P=0.01) compared with individuals carrying the most common haplotype ‘A-A-G-no-deletion-C’.
Figure 1.
LD plot of one deletion and nine SNPs of the CFTR gene in 679 Caucasian controls. The dash line indicates the relative location of ΔF508 deletion (rs332). The numbers in the boxes indicate D′ values.
Table 4.
Association between CFTR haplotypes and lung cancer risk
| Haplotype | Case-control analysisb | Haplotype score test | Regression analysis adjusted pd |
||||
|---|---|---|---|---|---|---|---|
| Frequency | Simulation | ||||||
| Case | Control | OR(95% CI) | p | p | pc | ||
| Block-1 | |||||||
| (SNP1-SNP2-SNP3-ΔF508-SNP4)a | |||||||
| A-A-G-no-deletion-C | 0.610 | 0.569 | 1.00(Reference) | 0.027 | 0.036 | 0.042 | Reference |
| A-A-A-deletion-T | 0.008 | 0.023 | 0.33(0.16–0.69) | 0.002 | 0.013 | 0.012 | 0.010 |
| A-A-A- no-deletion-T | 0.091 | 0.110 | 0.74(0.59–1.10) | 0.063 | 0.061 | 0.062 | 0.083 |
| G-A-A- no-deletion-C | 0.079 | 0.089 | 0.83(0.62–1.12) | 0.365 | 0.417 | 0.409 | 0.408 |
| G-G-A- no-deletion-C | 0.130 | 0.136 | 0.89(0.71–1.13) | 0.607 | 0.854 | 0.855 | 0.860 |
| A-A-A- no-deletion-C | 0.079 | 0.068 | 1.07(0.79–1.46) | 0.281 | 0.466 | 0.448 | 0.834 |
| Block-2 | |||||||
| (SNP5-SNP6-SNP7) a | |||||||
| C-G-C | 0.614 | 0.574 | 1.00(Reference) | 0.033 | 0.044 | 0.051 | Reference |
| C-A-C | 0.155 | 0.182 | 0.80(0.65–0.98) | 0.066 | 0.061 | 0.053 | 0.039 |
| T-A-T | 0.154 | 0.168 | 0.86(0.69–1.06) | 0.874 | 0.415 | 0.428 | 0.923 |
| C-A-T | 0.078 | 0.076 | 0.95(0.71–1.28) | 0.316 | 0.931 | 0.921 | 0.318 |
| ΔF508-SNP4a | |||||||
| No-deletion-C | 0.900 | 0.861 | 1.00(Reference) | 0.004 | 0.004 | 0.005 | Reference |
| Deletion-T | 0.008 | 0.024 | 0.32(0.15–0.68) | 0.002 | 0.008 | 0.009 | 0.010 |
| No-deletion-T | 0.093 | 0.114 | 0.48(0.60–1.00) | 0.068 | 0.051 | 0.052 | 0.070 |
In Block-1, ten haplotypes were constructed, and four haplotypes with a frequency of less than 0.05 were excluded from the haplotype analysis. In Block-2, all four haplotypes with a frequency of greater than 0.05 were included in the analysis. Bold characters indicate corresponding p values that are less than 0.05.
SNP number and polymorphic bases were listed in Table 1.
Global score test for Block-1: global-stat=17, d.f.=6, p =0.01, simulation p=0.008; global score test for Block-2: global-stat=5.5, d.f.=3, p=0.140, simulation p=0.140; global score test for ΔF508-SNP4: global-stat = 11.6, d.f. = 3, p = 0.009, simulation p=0.004.
P values were generated by permutation test with 1000 times simulation.
P values were adjusted by gender, age, cigarette smoking, SHS, history of COPD, and family history of lung cancer.
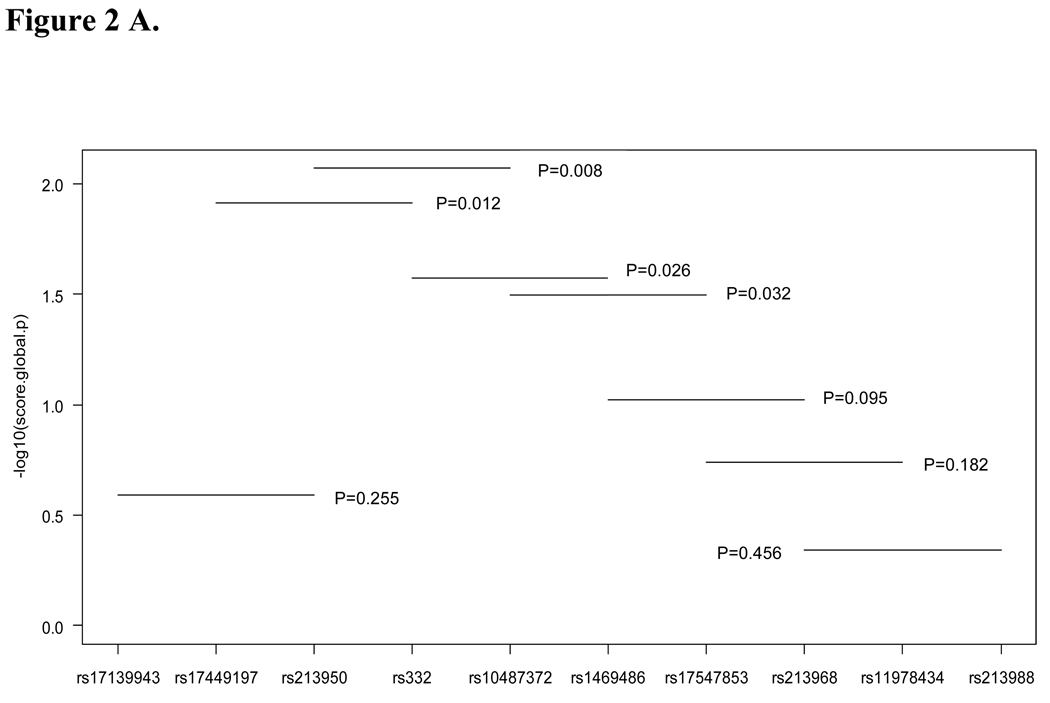
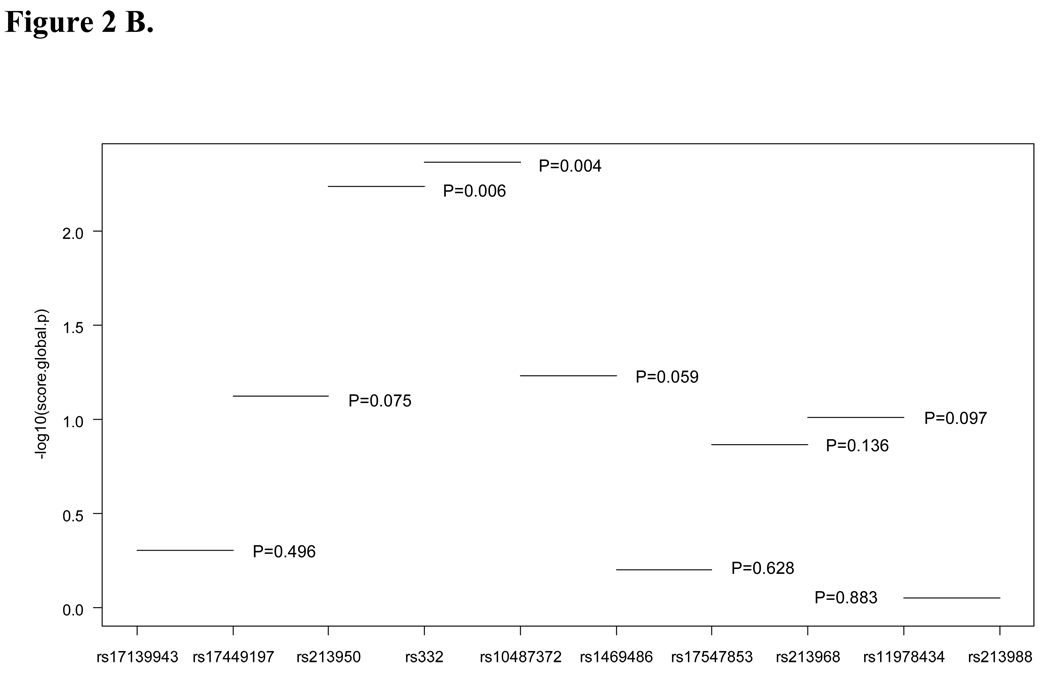
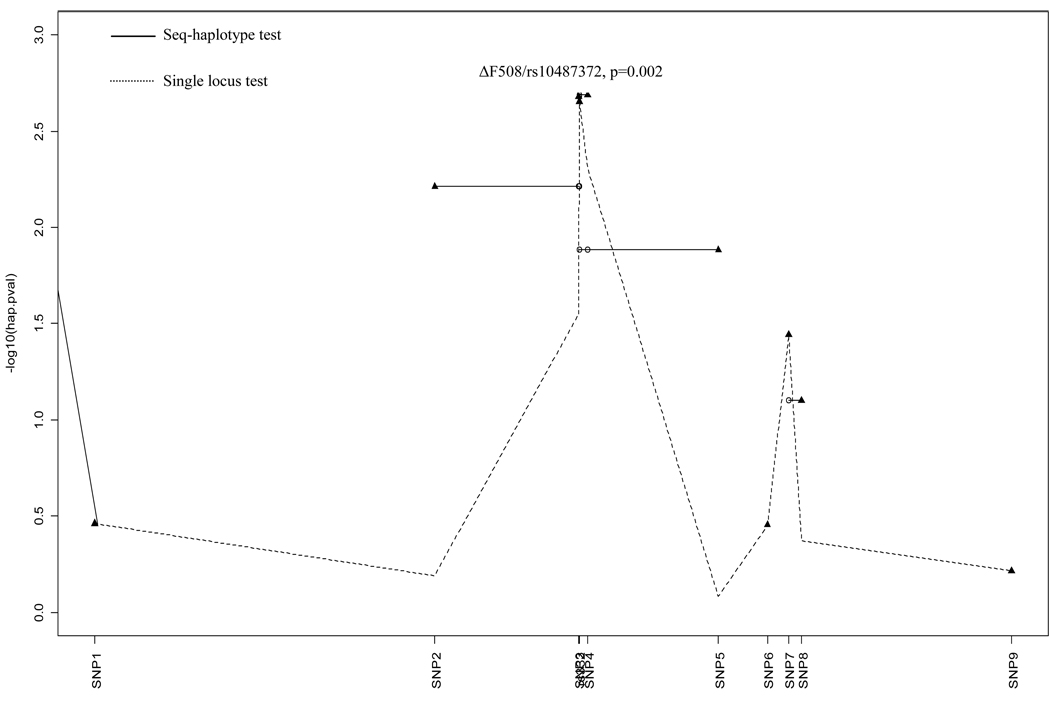
To evaluate the association of sub-haplotypes with lung cancer risk, we analyzed haplotypes from two and three loci. All significant sub-haplotypes included ΔF508 (rs332) and rs10487372 (Figure 2A and B). The sequential haplotype scan analysis also confirmed the above result: the haplotype with ΔF508 and rs10487372 showed the strongest statistical association (permutation P=0.002, Figure 3). Sub-haplotype analysis of ΔF508 and rs10487372 showed that individuals with the ‘deletion-T’ haplotype had a significantly lower risk for lung cancer compared with the common haplotype, ‘no-deletion-C’ (OR=0.32; 95% CI 0.15–0.68, adjust P=0.01) (Table 4).
Figure 2.
Plot of global p values for sub-haplotypes. A, three loci haplotypes; B, two loci haplotyps; Global p values were indicated.
Figure 3.
Plot of p values for the sequential haplotype scan. The start locus is indicated by a filled triangle, and the other loci combined with the start locus are indicated by a circle. SNP1-9 are listed in Table 1.
Discussion
In this study, for the first time, the association between the CFTR gene variations and lung cancer risk was investigated. ΔF508 deletion and genotypes with minor alleles in one tag SNP (rs10487372) and one functional SNP (rs213950) showed significantly inverse association with lung cancer risk. The results of the haplotype analysis were consistent with individual variant analysis. In particular, the sub-haplotype analysis indicated that all significant sub-haplotypes included ΔF508. Individuals with the ‘deletion-T’ haplotype (ΔF508/rs10487372) had a markedly lower risk for lung cancer by 68% compared with the common haplotype, ‘no-deletion-C’.
The CFTR functions in macromolecular complexes regulated by PKA and PKC phosphorylation. The CFTR gene encodes a chloride channel that transports negatively charged chloride ions into and out of cells. The transport of chloride ions helps control the movement of water in tissues, which is necessary for the production of thin, freely flowing mucus. Mucus is a slippery substance that lubricates and protects the lining of the airways and other organs and tissues. The CFTR protein also regulates the function of other channels, such as those that transport positively charged particles called sodium ions across cell membranes. These channels are necessary for the normal function of the lung [20, 21].
Over 1600 mutations in the CFTR have been reported. The most common mutation, ΔF508, is a deletion of phenylalanine residue at position 508 in the CFTR protein [22]. A worldwide mutation survey found that this mutation allele accounted for 66% of 43,849 tested CF chromosomes [23]. Many CFTR gene variants fail to be properly processed to a mature glycosylated form and transported to cell membranes. These CFTR variants are typically associated with severe CF phenotypes [24]. The CFTR with ΔF508, if correctly processed, possesses residual chloride ions channel activity, and may lead to a sustained normal or only mildly affected phenotype [25]. The efficiency of processing and trafficking of the ΔF508 deleted CFTR protein may vary considerably between different epithelial cells [24]. Therefore, the relative impact of the CFTR genotype on clinical phenotype is organ specific. In the current study, only one control subject was found to have the ΔF508 homozygote deletion. The heterozygote mutations may only result in subclinical (mildly affected) or normal phenotypes.
Previous studies have suggested that one of the most frequent functional polymorphisms in the CFTR gene, rs213950 G>A (M470V) in exon 11, has a role in modulating the CFTR protein at both the transcriptional and translational levels [26]. It was reported that M470 (G allele) CFTR proteins have a 1.7-fold increased intrinsic chloride channel activity compared with V470 (A allele) CFTR proteins [26]. In this study, the genotypes (GA and AA) corresponding to the V variation of rs213950 were found to be significantly associated with reduced lung cancer by single variation analysis. However, the sub-haplotype and sequential haplotype scan analyses revealed that all significant sub-haplotypes included ΔF508. It is very likely that the association of rs213905 with lung cancer is due to a strong linkage disequilibrium with ΔF508.
Interestingly, deletion ΔF508 and all significant SNPs were found to have a protective effect on lung cancer risk. A protective effect of the heterozygous CTFR mutation against lung cancer concurs with a similar effect as previously suggested in breast cancer and prostate cancer. However, this is at variance with other data showing an increased risk of pancreatic and colorectal adenocarcinoma in CF patients. Moreover, cancer-specific hypermethylation of CFTR was recently found in hepatocellular carcinoma and in bladder cancer [27, 28]. These contradictory data indicated that the presumed differences in the biological effects in the CTFR mutations may depend on the tumor type.
The exact role of the CFTR in the airway and the mechanism for its direct participation in lung cancer pathology remain unclear. The results from very limited literature on the possible mechanisms are contested. Cohen et al [29] demonstrated that lungs showed decreased compliance and increased airway resistance in young CFTR−/− mice as compared to CFTR+/+ littermates. Surprisingly, the CFTR+/− animals exhibited a lung phenotype distinct from either the homozygous normal or knockout genotypes. The heterozygous mice showed increased lung compliance and decreased airway resistance when compared to the homozygous phenotype, suggesting a heterozygous advantage that might explain the high frequency of the mutation in certain populations. Previous observations have suggested many types of CFTR overlapping pathways, such as Na+, Ca2+, K+, and Cl− channels, contributed to the development of prostate cancer cell lines. Blockage of these channels can suppress the growth and proliferation of human prostate cancer cells [30, 31]. A dysfunctional CFTR may weaken the ability of cancer cells to survive by handling proliferative homeostasis and thereby strengthening their pro-apoptotic potential [9, 31]. On the other hand, it has been shown that a dysfunctional CFTR was responsible for elevated blood ATP concentrations in mice and in CF patients [8, 32]. Elevated extracellular ATP was found to inhibit tumor growth [33].
Several limitations of this study need to be noted. First, the patients were selected from a tertiary medical center; whereas, controls were selected from a community population. However, potential confounding bias was minimized by frequency matching of the controls to the cases by sex, age, and race/ethnicity. Second, for an assumed 0.1 frequency of a minor allele in the control population and a significance level of 0.05 with a power of 0.8, the sample size of this study was sufficient to detect a significant OR of 1.8, but the sample size was not sufficient for the subgroup analysis. Therefore, additional studies with larger sample sizes will be required to confirm our findings from the subgroup analysis. Third, because of a relatively low tag SNP density, some functional haplotypes might be missed in this study; the reconstructed LD blocks may not be very robust, and the blocks may continue to fall apart into sub-block if the SNP density was increased.
In conclusion, genetic variations in the CFTR gene may modulate the risk of lung cancer. ΔF508 deletion in the CFTR may be important protective variants for lung cancer risk. This finding, for the first time, provides evidence of a protective role of CFTR variations in the etiology of lung cancer. Further population-based and experimental studies are warranted to elucidate the mechanism on how an altered CFTR gene protects against lung cancer.
Supplementary Material
Acknowledgements
We thank Susan Ernst M. A., for her technical assistance with the manuscript. We thank W Edward Highsmith and Stephen N. Thibodeau for their helpful comments at various stages of this work. This study was supported by grants NIH-CA 77118, NIH-CA 80127, and NIH-CA 84354 and Mayo Foundation funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest The authors indicated no potential conflicts of interest.
References
- 1.Zielenski J, Tsui LC. Cystic fibrosis: genotypic and phenotypic variations. Annu Rev Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]
- 2.Durno C, Corey M, Zielenski J, Tullis E, Tsui LC, Durie P. Genotype and phenotype correlations in patients with cystic fibrosis and pancreatitis. Gastroenterology. 2002;123:1857–1864. doi: 10.1053/gast.2002.37042. [DOI] [PubMed] [Google Scholar]
- 3.Warren N, Holmes JA, al-Jader L, West RR, Lewis DC, Padua RA. Frequency of carriers of cystic fibrosis gene among patients with myeloid malignancy and melanoma. Bmj. 1991;302:760–761. doi: 10.1136/bmj.302.6779.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neglia JP, FitzSimmons SC, Maisonneuve P, Schoni MH, Schoni-Affolter F, Corey M, et al. The risk of cancer among patients with cystic fibrosis. Cystic Fibrosis and Cancer Study Group. N Engl J Med. 1995;332:494–499. doi: 10.1056/NEJM199502233320803. [DOI] [PubMed] [Google Scholar]
- 5.McWilliams R, Highsmith WE, Rabe KG, de Andrade M, Tordsen LA, Holtegaard LM, et al. Cystic fibrosis transmembrane regulator gene carrier status is a risk factor for young onset pancreatic adenocarcinoma. Gut. 2005;54:1661–1662. doi: 10.1136/gut.2005.074534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Padua RA, Warren N, Grimshaw D, Smith M, Lewis C, Whittaker J, et al. The cystic fibrosis delta F508 gene mutation and cancer. Hum Mutat. 1997;10:45–48. doi: 10.1002/(SICI)1098-1004(1997)10:1<45::AID-HUMU6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Southey MC, Batten L, Andersen CR, McCredie MR, Giles GG, Dite G, et al. CFTR deltaF508 carrier status, risk of breast cancer before the age of 40 and histological grading in a population-based case-control study. Int J Cancer. 1998;79:487–489. doi: 10.1002/(sici)1097-0215(19981023)79:5<487::aid-ijc7>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 8.Abraham EH, Vos P, Kahn J, Grubman SA, Jefferson DM, Ding I, et al. Cystic fibrosis hetero- and homozygosity is associated with inhibition of breast cancer growth. Nat Med. 1996;2:593–596. doi: 10.1038/nm0596-593. [DOI] [PubMed] [Google Scholar]
- 9.Qiao D, Yi L, Hua L, Xu Z, Ding Y, Shi D, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) gene 5T allele may protect against prostate cancer: a case-control study in Chinese Han population. J Cyst Fibros. 2008;7:210–214. doi: 10.1016/j.jcf.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Yang P, Sun Z, Krowka MJ, Aubry MC, Bamlet WR, Wampfler JA, et al. Alpha1-antitrypsin deficiency carriers, tobacco smoke, chronic obstructive pulmonary disease, and lung cancer risk. Arch Intern Med. 2008;168:1097–1103. doi: 10.1001/archinte.168.10.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang P, Wentzlaff KA, Katzmann JA, Marks RS, Allen MS, Lesnick TG, et al. Alpha1-antitrypsin deficiency allele carriers among lung cancer patients. Cancer Epidemiol Biomarkers Prev. 1999;8:461–465. [PubMed] [Google Scholar]
- 12.Yang P, Allen MS, Aubry MC, Wampfler JA, Marks RS, Edell ES, et al. Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128:452–462. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 13.Taniguchi K, Yang P, Jett J, Bass E, Meyer R, Wang Y, et al. Polymorphisms in the promoter region of the neutrophil elastase gene are associated with lung cancer development. Clin Cancer Res. 2002;8:1115–1120. [PubMed] [Google Scholar]
- 14.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 16.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lake SL, Lyon H, Tantisira K, Silverman EK, Weiss ST, Laird NM, et al. Estimation and tests of haplotype-environment interaction when linkage phase is ambiguous. Hum Hered. 2003;55:56–65. doi: 10.1159/000071811. [DOI] [PubMed] [Google Scholar]
- 18.Yu Z, Schaid DJ. Application of sequential haplotype scan methods to case-control data. BMC Proc. 2007;1 Suppl 1:S21. doi: 10.1186/1753-6561-1-s1-s21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Z, Schaid DJ. Sequential haplotype scan methods for association analysis. Genet Epidemiol. 2007;31:553–564. doi: 10.1002/gepi.20228. [DOI] [PubMed] [Google Scholar]
- 20.Sheppard DN, Welsh MJ. Structure and function of the CFTR chloride channel. Physiol Rev. 1999;79:S23–S45. doi: 10.1152/physrev.1999.79.1.S23. [DOI] [PubMed] [Google Scholar]
- 21.Schwiebert EM, Benos DJ, Egan ME, Stutts MJ, Guggino WB. CFTR is a conductance regulator as well as a chloride channel. Physiol Rev. 1999;79:S145–S166. doi: 10.1152/physrev.1999.79.1.S145. [DOI] [PubMed] [Google Scholar]
- 22.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, et al. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–1080. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 23.Population variation of common cystic fibrosis mutations. The Cystic Fibrosis Genetic Analysis Consortium. Hum Mutat. 1994;4:167–177. doi: 10.1002/humu.1380040302. [DOI] [PubMed] [Google Scholar]
- 24.Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–133. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- 25.Tsui LC. The spectrum of cystic fibrosis mutations. Trends Genet. 1992;8:392–398. doi: 10.1016/0168-9525(92)90301-j. [DOI] [PubMed] [Google Scholar]
- 26.Cuppens H, Lin W, Jaspers M, Costes B, Teng H, Vankeerberghen A, et al. Polyvariant mutant cystic fibrosis transmembrane conductance regulator genes. The polymorphic (Tg)m locus explains the partial penetrance of the T5 polymorphism as a disease mutation. J Clin Invest. 1998;101:487–496. doi: 10.1172/JCI639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moribe T, Iizuka N, Miura T, Kimura N, Tamatsukuri S, Ishitsuka H, et al. Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. International Journal of Cancer. 2009;125:388–397. doi: 10.1002/ijc.24394. [DOI] [PubMed] [Google Scholar]
- 28.Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clinical Cancer Research. 2007;13:7296–7304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 29.Cohen JC, Lundblad LK, Bates JH, Levitzky M, Larson JE. The "Goldilocks effect" in cystic fibrosis: identification of a lung phenotype in the cftr knockout and heterozygous mouse. BMC Genetics. 2004;5:21. doi: 10.1186/1471-2156-5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foster CS, Cornford P, Forsyth L, Djamgoz MB, Ke Y. The cellular and molecular basis of prostate cancer. BJU Int. 1999;83:171–194. doi: 10.1046/j.1464-410x.1999.00954.x. [DOI] [PubMed] [Google Scholar]
- 31.Lemonnier L, Shuba Y, Crepin A, Roudbaraki M, Slomianny C, Mauroy B, et al. Bcl-2-dependent modulation of swelling-activated Cl- current and ClC-3 expression in human prostate cancer epithelial cells. Cancer Res. 2004;64:4841–4848. doi: 10.1158/0008-5472.CAN-03-3223. [DOI] [PubMed] [Google Scholar]
- 32.Lader AS, Prat AG, Jackson GR, Jr, Chervinsky KL, Lapey A, Kinane TB, et al. Increased circulating levels of plasma ATP in cystic fibrosis patients. Clin Physiol. 2000;20:348–353. doi: 10.1046/j.1365-2281.2000.00272.x. [DOI] [PubMed] [Google Scholar]
- 33.Humez S, Monet M, van Coppenolle F, Delcourt P, Prevarskaya N. The role of intracellular pH in cell growth arrest induced by ATP. Am J Physiol Cell Physiol. 2004;287:C1733–C1746. doi: 10.1152/ajpcell.00578.2003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.