Abstract
Glioblastoma multiforme (GBM) is a fatal primary brain tumor harboring myriad genetic and epigenetic alterations. The recent multidimensional analysis of the GBM genome has provided a more complete view of the landscape of such alterations and their linked pathways. This effort has demonstrated that certain pathways are universally altered, but that the specific genetic events altered within each pathway can vary for each particular patient's tumor. With this atlas of genetic and epigenetic events, it now becomes feasible to assess how the patterns of mutations in a pathway influence response to drugs that are targeting such pathways. This issue is particularly important for GBM because, in contrast to other tumor types, molecularly targeted therapies have failed to alter overall survival substantially. Here, we combined functional genetic screens and comprehensive genomic analyses to identify CDK6 as a GBM oncogene that is required for proliferation and viability in a subset of GBM cell lines and tumors. Using an available small molecule targeting cyclin-dependent kinases (CDKs) 4 and 6, we sought to determine if the specific pattern of retinoblastoma pathway inactivation dictated the response to CDK4/6 inhibitor therapy. We showed that codeletion of CDKN2A and CDKN2C serves as a strong predictor of sensitivity to a selective inhibitor of CDK4/6. Thus, genome-informed drug sensitivity studies identify a subset of GBMs likely to respond to CDK4/6 inhibition. More generally, these observations demonstrate that the integration of genomic, functional and pharmacologic data can be exploited to inform the development of targeted therapy directed against specific cancer pathways.
Keywords: cyclin-dependent kinase inhibitor, INK4C, PD0332991, targeted therapy
Comprehensive genomic characterization of glioblastoma multiforme (GBM) has revealed major cancer-relevant pathways that are near-universally targeted for genetic or epigenetic alterations that regulate the activity of such pathways (1). Against the backdrop of an increasing appreciation for the highly heterogeneous and complex nature of the cancer genome, targeting these common pathways as a therapeutic strategy is appealing in comparison with sorting through the myriad of driver and passenger alterations for therapeutic targets that may only be relevant in a small subset of the tumors. Consistent with rapid and unrestrained proliferation that is characteristic of tumor cells, the retinoblastoma (RB) pathway is found to be nearly universally inactivated in human cancers, including glioblastoma (2). At the nexus of this pathway is the RB protein itself, the central regulator of cell cycle progression through its inhibitory effect on E2F transcription factors that in turn regulate key genes involved in proliferation. RB is tightly regulated by the opposing activating cyclin-dependent kinases 4 and 6 (CDK4/6) and their cyclin-D binding partners, as well as the inactivating CDK inhibitors (CKIs) such as p16INK4A(CDKN2A) and p18INK4C(CDKN2C) (3). Genetic alterations targeting each of these core components in addition to RB itself are observed in more than 78% of human GBM cases (2).
GBM is a fatal primary brain tumor that is resistant to conventional therapies. The current standard of care—surgical resection followed by radiation therapy and adjuvant temozolomide chemotherapy—yields a median survival of only 14.6 mo (4), underscoring the need for targeted therapies with improved efficacy. As a first step toward identification of novel targets for therapeutic development, we used an integrative functional genomics approach that leverages the power of RNAi screens and multidimensional cancer genomics by The Cancer Genome Atlas (TCGA) to identify “druggable” kinases that are essential and transforming in GBM. In this report, we showed that CDK6 is an essential oncogene in GBM, but that only a subset of GBM cells with a specific pattern of RB pathway lesions exhibited sensitivity to a pharmacological CDK4/CDK6 inhibitor, suggesting that different therapeutic strategies are likely required even for tumors with common pathway inactivation.
Results
Integrated Functional Genomics Identifies CDK6 as a GBM Oncogene.
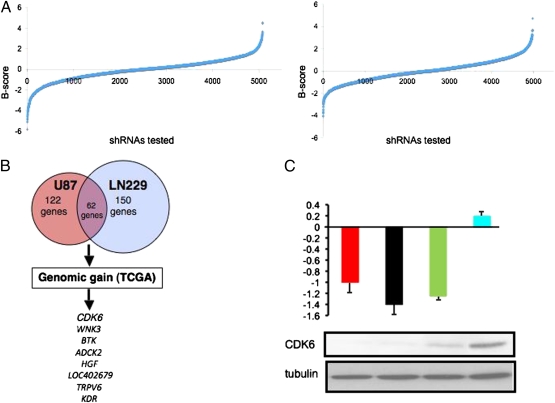
To identify genes required for the viability of GBM cells, we performed an RNAi-mediated, loss-of-function screen in two GBM cell lines, LN-229 and U87MG. Using a lentivirally delivered shRNA library composed of 4,849 shRNAs targeting 1,002 genes (Dataset S1A), including nearly all human kinases (5), we assessed which of these shRNAs induced alterations in cell proliferation and viability (hereafter designated viability) using a luminometric assay. B scores corresponding to relative viability conferred by individual shRNAs were plotted in a distribution curve for each cell line (Fig. 1A), where a B score of 0 represents average viability, positive B scores correlate with increased viability, and negative B scores indicate reduced viability. We considered genes targeted by at least one shRNA with a B score lower than −1.5 and one additional shRNA lower than −1.0 as potential candidates. From the U87MG cell line, 122 genes emerged as candidates, whereas 150 genes scored in LN-229 (Fig. 1B). To minimize cell line–specific artifacts, we considered only those hits that scored in both cell lines, producing a list of 62 candidate GBM viability genes (Dataset S1B).
Fig. 1.
Loss-of-function screens in two GBM cell lines. (A) Representative B score distribution curve representing cell proliferation for shRNAs tested in LN-229 cells (Left) and U87MG cells (Right). Negative B scores indicate shRNAs which reduced viability, whereas positive B scores correlate with increased viability. (B) Venn diagram illustrating the methodology of candidate gene selection. Genes found to be essential for GBM cell proliferation in both U87MG and LN-229 large-scale shRNA screens were analyzed by SGOL for evidence of copy number gain in the TCGA dataset (317 primary GBM tumors). CDK6, WNK3, BTK, ADCK2, HGF, LOC402679, TRPV6, and KDR ranked within the top 10%. (C) CDK6 is essential for LN-229 cell proliferation. (Upper) B scores from the LN-229 shRNA screen obtained after suppression of CDK6 expression with sh374 (red), sh696 (black), and sh720 (green) targeting CDK6 and with a control shRNA targeting GFP (shGFP, blue). (Lower) Reduced CDK6 protein expression in LN-229 cells expressing CDK6-targeting shRNAs.
Next, we triangulated these viability hits with genomic data from array-CGH profiles of 317 primary GBM tumors by TCGA (2). As copy number gain is a common mechanism that leads to activation of oncogenes, we determined which of the 62 candidates resided within regions of genomic gain with use of the segment gain or loss (SGOL) algorithm (Materials and Methods). Eight of the 62 candidates (CDK6, WNK3, BTK, ADCK2, HGF, LOC402679, TRPV6, and KDR) ranked among the top tenth percentile (Fig. 1B and Dataset S1C) and were thus considered candidates required for GBM cell viability and targeted for genomic amplification in human GBM.
Of these eight candidates, we focused on CDK6 because it was the top-ranking viability gene by SGOL analysis and is a known core component of the RB pathway (2). Moreover, as the primary binding partner, CDK6 showed impaired binding to loss-of-function variants of p18INK4C, a backup tumor suppressor codeleted with closely related CDK inhibitors p16INKA and p15INK4B in human GBM (6, 7). Here, we first validated that multiple shRNAs used in the viability screen were effective in extinguishing CDK6 expression on the protein level. Using LN-229 cells, we showed that three independent CDK6-targeting shRNAs—sh374, sh696, and sh720—that scored with significant B values in the initial viability screen were indeed effective in depleting CDK6 protein levels (Fig. 1C).
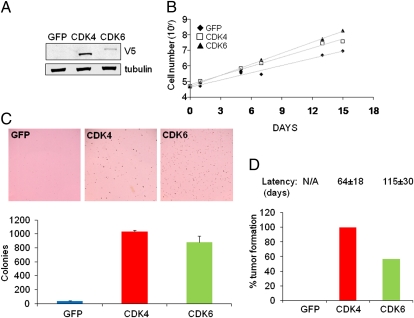
Next, we stably expressed GFP, V5-epitope–tagged CDK6 or CDK4 in primary nontransformed murine p16Ink4a/Arf-null astrocytes (8) at passage 13. Upon confirming expression of ectopic CDK4 or CDK6 by immunoblotting (Fig. 2A), we found that CDK6-expressing astrocytes, like the CDK4-expressing cells, exhibited enhanced proliferation relative to the control (Fig. 2B). In anchorage-independent growth assays, both CDK4 and CDK6 led to efficient transformation of p16Ink4a/Arf-null astrocytes (GFP, 37 ± 2 vs. CDK4, 1,031 ± 24 vs. CDK6, 877 ± 91; P < 0.00021; Fig. 2C). In vivo, GFP-expressing astrocytes were unable to form tumors s.c. In contrast, CDK6 expression conferred tumorigenicity in 57% of the injection sites with an average latency of 115 d, whereas CDK4 expression resulted in 100% penetrance with an average latency of 64 d (Fig. 2D). Taken together, we conclude that, like the classical GBM oncogene CDK4 (9), CDK6 also serves as a bona fide oncogene and represents a potential therapeutic target in GBM.
Fig. 2.
CDK6 enhances proliferation and tumorigenicity of p16Ink4a/Arf-null murine astrocytes. (A) p16Ink4a/Arf-null mouse astrocytes at passage 13 were lentivirally transduced with expression vectors for GFP, CDK4-V5, and CDK6-V5. Cells were selected with blasticidin for 1 wk and protein lysates were harvested from stable polyclonal populations. The Western Blot was probed with antibodies targeting V5 and tubulin. (B) Proliferation of p16Ink4a/Arf-null astrocytes expressing GFP, CDK4, or CDK6 was monitored over a period of 2 wk. Both CDK4 and CDK6 enhanced proliferation in vitro. (C) p16Ink4a/Arf-null astrocytes expressing GFP, CDK4, or CDK6 at passage 16 were seeded into soft agar. Colonies were stained and counted after 2 wk. Columns represent mean ± SD of triplicates. Both CDK4 and CDK6 increased colony formation. (D) p16Ink4a/Arf-null astrocytes expressing GFP, CDK4, or CDK6 were injected into the flanks of nude mice (n = 14 per cell line). Mice were monitored for tumor formation over a period of 6 mo. None of the sites injected with GFP-expressing cells developed tumors, whereas all sites injected with CDK4-expressing cells developed tumors with an average latency of 64 d. The tumor incidence of CDK6-expressing cells was 57%, accompanied by a longer average latency of 115 d.
Sensitivity to CDK4/6 Inhibition Is Influenced by RB Pathway Inactivation Pattern.
As a specific CDK4/CDK6 inhibitor PD0332991 is under development (10), we explored the therapeutic potential of CDK4/6 inhibition in GBM. Here, we subjected a panel of 25 glioma cell lines to treatment with PD0332991. Briefly, cells were exposed to varying concentrations of PD0332991 for 5 d and then assayed for cell number by ATP measurement, reflecting cell proliferation and/or viability (Fig. S1). IC50 values for individual cell lines were determined (Dataset S2).
Contrary to our expectation that cell lines expressing high levels of CDK4 or CDK6 would be sensitive to PD0332991, the response to this inhibitor failed to correlate with the level of CDK6 or CDK4 expression (Fig. S2). Instead, of the seven sensitive cell lines [i.e., IC50 <1 μM, a dosage at which specific activity against CDK4/6 has been demonstrated (10)], five harbored codeletion of CDKN2A/B and CDKN2C (Dataset S2). Indeed, five of the nine cell lines with codeletion of CDKN2A/B and CDKN2C were sensitive, whereas only two of nine CDKN2A/B–deleted cell lines were susceptible (P = 0.073). In contrast, six of nine CDKN2A/B–deleted cell lines were highly resistant, exhibiting IC50 values greater than 4 μM, a level at which PD0332991 is known to affect other targets (10).
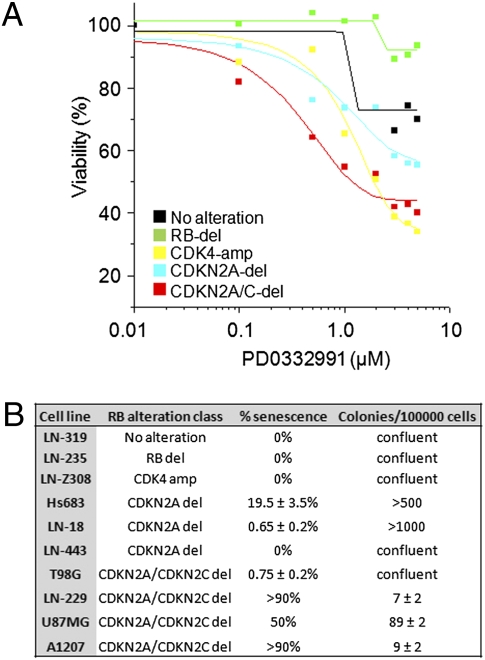
To show this differential sensitivity among cell lines with distinct patterns of RB pathway inactivation, we plotted the median IC50s for GBM cells with each pattern in Fig. 3A. As shown, only the CDKN2A/C–codeleted cells were enriched for responders (median IC50 value, 0.6 μM). Interesting, cell lines with mutation or deletion of RB1 itself, or cell lines without evidence of alterations in any of these RB pathway core components (Fig. S2), were highly resistant, all with IC50 values greater than 4 μM. This sensitivity pattern to pharmacological inhibition is recapitulated with genetic inhibition by RNAi, as CDKN2A/C–codeleted cell lines were sensitive to RNAi-mediated depletion of either CDK6 or CDK4 (Fig. S3 and Dataset S3), whereas RB-null or mutant cell lines and cell lines without apparent RB pathway alterations were unaffected by RNAi targeting CDK4 or CDK6.
Fig. 3.
Codeletion of CDKN2A and CDKN2C sensitizes glioma cells to pharmacological CDK4/6 inhibition. (A) Glioma cell lines representing different RB alteration classes were incubated in various concentrations of the CDK4/6 inhibitor PD0332991 for 5 d (IC50 values for individual cell lines are listed in Dataset S2). Relative cell number was determined by a luminometric assay, and the median IC50 value per RB alteration class was calculated (representative experiment shown). Cell lines without known RB pathway deletions and RB-del cell lines were least sensitive and CDKN2A/CDKN2C–codeleted cell lines were most sensitive to PD0332991. (B) Ten glioma cell lines representing different RB pathway alteration classes were seeded at a density of 100,000 cells per 10-cm dish and incubated in 1 μM PD0332991 for 14 d. Cells were assayed for colony formation and stained for senescence-associated β-gal activity. The proportion of senescent cells was at least 50% in three of four CDKN2A/CDKN2C–codeleted cell lines but not in six cell lines representing other RB pathway alteration classes. The last column states the number of resistant colonies per 100,000 cells after 14 d.
On the mechanistic level, PD0332991 induced near-complete and lasting G1 arrest in sensitive cell lines (Fig. S4A and Dataset S4), in conjunction with a greatly reduced level of phosphorylated RB protein (Fig. S4B). G1 arrest was less complete in LN-18, a cell line with intermediate sensitivity to PD0332991, and nonexistent in two resistant cell lines (Dataset S4). Long-term exposure to PD0332991 (14 d at 1 μM) induced quantitative senescence in several CDKN2A/B/C–deleted cell lines with minimal residual colony formation (Fig. 3B) whereas resistant cell lines from other RB alteration patterns failed to undergo senescence and quickly became resistant to PD0332991. Taken together, these data suggest that a specific pattern of RB pathway alteration strongly correlates with sensitivity to pharmacological inhibition of CDK4/6 in GBM cell lines.
Pattern of RB Pathway Deregulation in Human GBM.
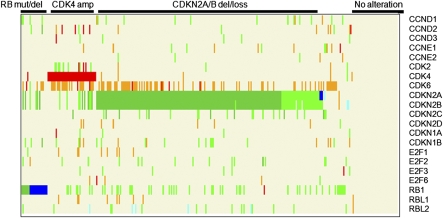
As a subset of GBM cells can undergo growth arrest and induction of senescence in response to pharmacological inhibition of CDK4/6, and such responders are enriched among cell lines with codeletion of the CDKN2A/B and CDKN2C loci, we next sought to determine whether similar patterns of RB pathway deregulation are observed in human GBM tumors. Here, we compiled a comprehensive overview of core and extended components of the RB pathway based on multidimensional genomic data from 317 primary GBM tumor samples characterized by TCGA. In particular, copy number loss, promoter methylation, or mutation of CDKN2A, CDKN2B, CDKN2C, CDKN2D, CDKN1A, CDKN1B, RB, RBL1, or RBL2 as well as copy number gain or mutation of CCND1, CCND2, CCND3, CCNE1, CCNE2, CDK2, CDK4, CDK6, E2F1, E2F2, E2F3, or E2F6 were considered RB pathway–compromising events (Fig. 4; see Materials and Methods for parameters).
Fig. 4.
Patterns of RB pathway inactivation in primary GBM. Genetic coalterations of RB pathway genes among 317 primary GBM specimens from the TCGA dataset. Each column represents an individual tumor sample. Copy number gains are depicted in orange (log2 ≥0.5), amplifications in red (log2 ≥1.0), copy number losses in light green (log2 <−0.5), deletions in dark green (log2 <−1.0), and mutations in dark blue. Promoter methylation (β ≥0.5) is depicted in light blue. RB-compromising events are defined in Materials and Methods.
Besides a subset of primary GBMs (43 of 317; 14%) with no detectable RB pathway-compromising alterations (hereafter classified as “no alteration”), the majority of the remaining 274 tumors harbor multiple RB pathway compromising lesions (average, 3.02 ± 1.42 events). Tumors with RB deletion (7 of 317; 2.2%) and RB mutation (15 of 317; 5%) are collectively grouped into the “RB-del/mut alteration class” (7.2%) regardless of presence or absence of other RB pathway events. As reported in the literature, deletion or loss of CDKN2A is found in 63% of all GBM (n = 201 of 317 tumors), making it the most frequent RB pathway alteration in primary GBM, similar to what we observed in glioma cell lines. In contrast, methylation of the CDKN2A promoter (in 1 of 150 specimens with promotion methylation data) or CDKN2A mutations (in 3 of 91 samples with mutation data; 3%) are relatively rare events. As observed among the cell lines, codeletion/loss of CDKN2A and CDKN2C occur in 6.3% of the cases (n = 20 of 317), whereas CDK6 gain in combination with CDKN2A loss is observed in 31% of the cases (n = 98 of 317). In contrast, amplification of the CDK4 gene is found in 41 of 317 tumors (13%) and appear to define a distinct subgroup having minimal overlap with the CDKN2A-deleted subgroup [4 of 160 tumors with log2(CDKN2A) less than 1 have concomitantly amplified CDK4]. In summary, analysis of the copy number profiles in TCGA GBM samples showed that 6% of all primary GBMs harbor codeletion/loss of CDKN2A and CDKN2C, representing a subset of GBM predicted to be especially sensitive to CDK4/6 inhibition based on our study in GBM cell lines.
Discussion
Here, the integration of functional genetic screens with copy number information allowed for the identification of essential genes targeted for genomic amplification in human GBM, among them CDK6. Although CDK6 has been described as an occasional target of amplification in GBM (11), we provide functional in vitro and in vivo data validating CDK6, like CDK4, as a bona fide oncogene in GBM. The closely related kinases CDK4 and CDK6 are targets of pharmacological inhibitors, some of which have entered early clinical trials (reviewed in refs. 3, 12). In contrast to pan-CDK inhibitors, which exhibit general toxicity, CDK4/6-specific inhibitors such as PD0332991 are well tolerated by patients, possibly because of the observation that CDK4/6 are dispensable in normal, untransformed cells (13).
The effectiveness of targeted therapies is dictated by genetic context and by the activation status of cellular pathways (1). Although our observed response phenotypes in vitro (e.g., G1 arrest and senescence), along with the reported efficacy of PD0332991 in orthotopic models of human GBM (14), raise the exciting possibility of PD0332991 use in humans, successful clinical development of a targeted drug requires the delineation of a responder identification strategy. By defining four major patterns of RB pathway inactivation in GBM, and demonstrating in glioma cell line models that CDKN2A/CDKN2C codeletion predicts sensitivity to PD0332991 treatment, we thus provide a clinical path hypothesis for the deployment of CDK inhibitor-based therapies in GBM. Specifically, stratification of likely responders to CDK4/6 inhibition should be based on the genomic pattern of RB pathway inactivation, not merely the expression levels of its CDK targets. Mechanistically, PD0332991 caused G1 arrest in sensitive glioma cell lines, as seen in other tumor types (10, 15), and senescence in CDKN2A/CDKN2C–codeleted GBM cells, similar to reconstitution of p18INK4C to physiological levels (7). Although low p18INK4C expression had previously been reported to be an indicator of response to a pan-CDK inhibitor (16), we found the lowest median p18INK4C expression levels in GBM tumors without apparent RB pathway alterations (Fig. S5), and cell lines representing this subset were highly resistant to PD0332991.
Although the paradigm of exclusivity in pathway deregulation states that genetic alterations targeting core components of the RB pathway should occur in a mutually exclusive manner in human tumors (17), we have recently reported that CDKN2C is deleted in CDKN2A-null GBM, driven by its role as a back-up tumor suppressor; in particular, we found that CDKN2C can be transcriptionally activated by E2F1 in the setting of p16INK4A loss and inactivation of the RB checkpoint (6). Analysis of the pattern of RB pathway deregulation in the TCGA dataset indicates that such back-up events are the norm rather than an exception; on average there are three RB pathway–compromising events among GBM tumor samples with at least one RB pathway alteration. Although multiple RB pathway hits can explain reduced dependency on CDK4/6, hitherto unknown genetic and epigenetic alterations may further contribute to cell cycle deregulation and PD0332991 resistance.
In summary, this study supports the view that both the functional status of a cancer-relevant pathway and the specific alteration(s) within the molecular network of pathway members are critical modifiers of the response to targeted therapy, and that despite common pathway alterations, the specific mode of its deregulation in different tumors will drive different biology and dictate differential response to these pharmacological inhibitions.
Materials and Methods
RNAi Screens.
Large-scale RNAi screening was conducted in an arrayed format using the TRC subset of the Broad Institute RNAi Consortium shRNA library directed against kinases, phosphatases, and other cancer-related genes (5, 18, 19).
Details of shRNA designs and protocols for high throughout lentiviral production and infection are available online (http://www.broad.mit.edu/rnai/trc/lib). In brief, cells were seeded in quadruplicate 384-well plates on day 0, followed by a media change with 8 μg/mL polybrene and lentiviral infection on day 1. Media was again changed on day 2, with and without puromycin selection for duplicate plates (concentration individualized per cell line; 2 μg/mL for most cell lines). An ATP-based luminescence assay (CellTiterGlo; Promega) was used to determine cell number 6 d after infection. Raw luminescence values from duplicate plates were averaged, and the ratio of puromycin-positive values to puromycin-negative values was used to assess infection efficiency. Data were normalized using the B score metric, a variant of the Z score that uses the median absolute deviation to account for plate-to-plate variability, as well as a two-way median polish to minimize row/column effects that accompany RNAi screens (20). After excluding shRNAs with low infection efficiency in each cell line, B score values from puromycin-positive and -negative replicates were averaged for each individual shRNA.
SGOL.
SGOL scores were calculated for genes using the SGOL function of the cghMCR package of the Bioconductor project. The algorithm applies the GISTIC (21) approach of summarizing copy number alterations across multiple samples to segmented data to speed up the computation for high density arrays and facilitate integration of data from different platforms (e.g., 244K and 415K custom arrays in this case; Agilent). We took the gene-centric approach by assigning the segment value to genes within the segment for all of the segments identified for each sample and then summarize the derived values per gene across all of the samples using thresholds of 0.4 and −0.4 for gains and losses, respectively, to generate the SGOL scores. High absolute SGOL scores indicate high frequency and/or magnitude of alterations across samples.
Proliferation and Tumorigenicity Assays.
Glioma cell lines (Dataset S2) and murine astrocytes were propagated in DMEM (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin/amphotericin B. Soft agar assays were performed in triplicate in six-well plates. Five thousand cells per well were seeded in regular medium containing 0.4% low-melting agarose on bottom agar containing 1% low-melting agarose in regular medium. After 14 or 21 d, colonies were stained with iodonitrotetrazolium chloride (Sigma) and counted. For in vivo tumorigenicity assays, 106 genetically engineered astrocytes comixed with Matrigel (Sigma) were transplanted s.c. into flanks of Ncr nude mice (Charles River or Taconic) and followed for tumor development. At termination of the experiment, tumors were harvested and processed for pathological and molecular analyses. All animal experiments were approved by the institutional animal care and use committee under protocol 04–136. To assay viability, 1,000 cells per well were seeded into 96-well plates. The next day, 0 h time points (i.e., T0) were assessed by a luminometric assay (CellTiterGlo; Promega) according to the manufacturer's protocol while experimental cells were exposed to PD0332991 or DMSO. Cells were scored after 5 d (i.e., T5). Relative viability was calculated as T5–T0, and the value for DMSO-treated cells was set to 100%. IC50 values were calculated using Origin 7.5 (OriginLab).
Genomic Analyses and Definition of RB-Compromising Events.
Data were downloaded from the TCGA website. For primary tumors, log2 copy number values determined by array-CGH were used to define gain (log2 ≥0.5), amplification (log2 ≥1.0), loss (log2 <−0.5), and deletion (log2 <−1.0). Mutation or promoter methylation (β ≥ 0.5, with β defined as Signalmethylated / (Signalmethylated + Signalunmethylated)) were considered inactivating events. Copy number loss, promoter methylation, or mutation of CDKN2A (p16INK4A), CDKN2B, CDKN2C (p18INK4C), CDKN2D, CDKN1A, CDKN1B, RB, RBL1, or RBL2 as well as copy number gain or mutation of CCND1, CCND2, CCND3, CCNE1, CCNE2, CDK2, CDK4, CDK6, E2F1, E2F2, E2F3, or E2F6 were considered RB pathway–compromising events in primary GBM tumors. Gain or mutation of CDK1, CCNA1, CCNA2, CCNB1, CCNB2, or CDC25A was very rare and therefore not included in Fig. 3. CCNB3 as an X-linked gene was excluded from this analysis. For glioma cell lines, log2 copy number values determined by array-CGH were used to define gain (log2 ≥0.5), amplification (log2 ≥1.4), loss (log2 <−0.7), and deletion (log2 <−1.4). We did not perform promoter methylation analyses or sequencing of RB pathway genes in the cell line cohort. However, none of the following common RB mutations were detected by OncoMap: E137*, L199*, R320*, R358*, R455*, R552*, R556*, R579*, L660fs*2, C706F, or E748*. Two cell lines that were classified p16INK4A-del (LN-382T) and “no alteration” (LN-319) had absent or very low levels of RB protein (Fig. S2) and may thus phenocopy RB-del cell lines.
Expression Analysis.
Western Blot and quantitative RT-PCR were performed using standard techniques as reported before (6).
Lentiviral Constructs.
ORFs of CDK6 and CDK4 were obtained from the Human ORFeome collection. The Gateway recombination system was used to transfer these cDNAs into the pLenti6/V5-DEST lentiviral expression vector (Invitrogen). Lentivirus was produced in 293T cells and target cells were infected at 48 h and 72 h after transfection in the presence of 5 μg/mL polybrene (Sigma). Infected cells were selected with 5 μg/mL blasticidin (Invitrogen) for 6 d before being assayed.
Supplementary Material
Acknowledgments
We thank Barbie Taylor-Harding for excellent technical assistance.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001613107/-/DCSupplemental.
References
- 1.Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 2.Atlas CG, Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: A changing paradigm. Nat Rev Cancer. 2009;9:153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 4.Stupp R, et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 5.Moffat J, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 6.Wiedemeyer R, et al. Feedback circuit among INK4 tumor suppressors constrains human glioblastoma development. Cancer Cell. 2008;13:355–364. doi: 10.1016/j.ccr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solomon DA, et al. Identification of p18 INK4c as a tumor suppressor gene in glioblastoma multiforme. Cancer Res. 2008;68:2564–2569. doi: 10.1158/0008-5472.CAN-07-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1:269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 9.He J, et al. CDK4 amplification is an alternative mechanism to p16 gene homozygous deletion in glioma cell lines. Cancer Res. 1994;54:5804–5807. [PubMed] [Google Scholar]
- 10.Fry DW, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 11.Costello JF, et al. Cyclin-dependent kinase 6 (CDK6) amplification in human gliomas identified using two-dimensional separation of genomic DNA. Cancer Res. 1997;57:1250–1254. [PubMed] [Google Scholar]
- 12.Malumbres M, Pevarello P, Barbacid M, Bischoff JR. CDK inhibitors in cancer therapy: What is next? Trends Pharmacol Sci. 2008;29:16–21. doi: 10.1016/j.tips.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 13.Santamaría D, et al. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–815. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- 14.Michaud K, et al. Pharmacologic inhibition of cyclin-dependent kinases 4 and 6 arrests the growth of glioblastoma multiforme intracranial xenografts. Cancer Res. 2010;70:3228–3238. doi: 10.1158/0008-5472.CAN-09-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn RS, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11:R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eguchi T, et al. Expression levels of p18INK4C modify the cellular efficacy of cyclin-dependent kinase inhibitors via regulation of Mcl-1 expression in tumor cell lines. Mol Cancer Ther. 2009;8:1460–1472. doi: 10.1158/1535-7163.MCT-08-1159. [DOI] [PubMed] [Google Scholar]
- 17.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 18.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129:1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 19.Scholl C, et al. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821–834. doi: 10.1016/j.cell.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 20.Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167–175. doi: 10.1038/nbt1186. [DOI] [PubMed] [Google Scholar]
- 21.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci USA. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.