Abstract
To monitor inaccuracy in gene expression in living cells, we designed an experimental system in the bacterium Bacillus subtilis whereby spontaneous errors can be visualized and quantified at a single-cell level. Our strategy was to introduce mutations into a chromosomally encoded gfp allele, such that errors in protein production are reported in real time by the formation of fluorescent GFP molecules. The data reveal that the amount of errors can greatly exceed previous estimates, and that the error rate increases dramatically at lower temperatures and during stationary phase. Furthermore, we demonstrate that when facing an antibiotic threat, an increase in error level is sufficient to allow survival of bacteria carrying a mutated antibiotic-resistance gene. We propose that bacterial gene expression is error prone, frequently yielding protein molecules that differ slightly from the sequence specified by their DNA, thus generating a cellular reservoir of nonidentical protein molecules. This variation may be a key factor in increasing bacterial fitness, expanding the capability of an isogenic population to face environmental challenges.
Keywords: Bacillus subtilis, translational errors, variations in living cells, translation fidelity
DNA is duplicated with remarkable fidelity to ensure that accurate genetic information is transmitted from one generation to the next. This information is passed from DNA to RNA and from RNA to protein during gene expression; however, the accuracy of these downstream events is relatively less understood. Although RNA and proteins are generally considered short-lived noninherited molecules, several diseases are now known to arise from errors occurring during transcription and translation (1–3), implying that faithful transfer of genetic information from DNA to proteins is crucial for maintaining proper cellular functions.
Interestingly, errors in gene expression can be beneficial, serving as a mechanism to promote noncanonical decoding. This strategy is mostly used by bacteria and viruses to increase diversity of particular proteins and to express alternative translational products (4). In Escherichia coli for example, programmed translational frame-shifting in the dnaX gene produces DNA polymerase subunits with different enzymatic activities. The canonical product of dnaX confers extreme processivity on DNA polymerase, whereas the truncated protein expressed because of translational frame-shifting tempers the polymerase activity (5). In more general terms, errors arising spontaneously during gene expression may increase protein variety and thereby enable genetically identical cells to display heterogeneous phenotypes. This phenomenon is likely to contribute to the robustness of unicellular organisms, allowing them to respond to fluctuating environments without changing their genotype (6–10).
Errors in gene expression emanate from inaccuracies during the processes of transcription or translation. Transcriptional fidelity is determined largely by the capability of the RNA polymerase to sense and discriminate between cognate and noncognate pairing to the DNA template before incorporation of the next nucleotide (11). Faithful translation relies mainly on correct aminoacylation of a given tRNA, selection of the proper tRNA, and maintenance of the correct ORF throughout synthesis (12). In vivo measurements of errors in gene expression in bacteria have been estimated to occur at a rate of 10−4 to 10−5 per nucleotide during transcription and ≈10−3 to 10−4 per codon during translation. These error rates are significantly higher than that attributed to the high-fidelity replication process, where the frequency of errors is as low as 10−8 to 10−9 per nucleotide (8, 9, 12–20).
For the most part, in vivo error rates in bacterial gene expression have been estimated by measuring the activity of a mutated reporter gene (such as lacZ) (14, 17, 18) in a large cell population. However, the resultant measurements represent the average error rates, and thus do not detect the actual error level per cell and the variability, if any, among individuals. Moreover, these assays do not discriminate between errors in gene expression and genetic mutations. Here, we designed an experimental system to visualize and quantify the rate of spontaneous errors in gene expression at a single-cell level in living Bacillus subtilis cells. Our strategy was to introduce mutations into the ORF of a chromosomally encoded gfp reporter allele, such that errors in protein production would yield functional GFP molecules, visible by fluorescence microscopy. Using this approach, we revealed that gene expression in bacteria is prone to high levels of errors reaching values that are much higher then previously estimated. Moreover, we demonstrate that bacteria have the capability to modify error levels in response to physiological and environmental conditions.
Results
Visualizing Errors in Gene Expression.
To visualize errors in gene expression at a single-cell level, we mutated a chromosomally encoded gfp reporter allele, such that errors in gene expression would generate functional GFP molecules. We reasoned that the use of highly sensitive fluorescence microscopy would enable us to detect small amounts of GFP molecules. Hence, we inserted frame-shift (gfpfs) or nonsense (gfpns) mutations at the beginning of the ORF of an inducible gfp gene. Ocher (TAA) was chosen as the nonsense mutation because it is the most abundant stop codon in the B. subtilis genome (SI Appendix).
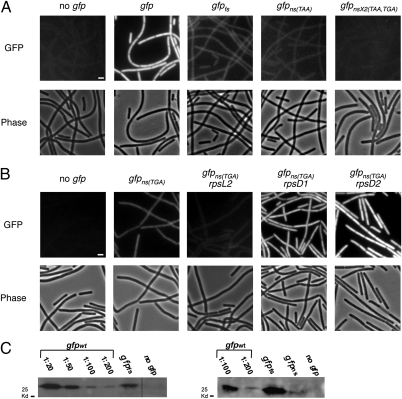
To monitor errors in gene expression arising during growth, cells bearing the different alleles were grown to mid-exponential phase and observed by fluorescence microscopy. As expected, a strong fluorescence signal was detected from cells expressing the wild-type gfp allele in the presence of the inducer (Fig. 1A). Remarkably however, a clear fluorescent signal was also observed from strains bearing either the gfpfs or the gfpns alleles upon induction. In both strains, the signal appeared as a uniformly distributed weak cytoplasmic haze present in all cells. This signal was above background level, as evidenced by the strain lacking the gfp gene or a strain containing two nonsense mutations within the gfp ORF (Fig. 1A). Moreover, fluorescence recovery after photobleaching experiments showed that 35 min after bleaching, the fluorescent signal displayed by the gfp mutant strains was almost restored to its original level, confirming that the signal derived from de novo inaccurate synthesis of protein molecules (SI Appendix, Fig. S1). Importantly, a similar GFP expression pattern was observed for mutations inserted into an endogenous B. subtilis gene (disA) fused to gfp (SI Appendix, Fig. S2) (21), and in strains carrying identical gfp alleles controlled by various promoters (SI Appendix, Fig. S3), implying that errors occur constantly regardless of the tested allele or the driving promoter.
Fig. 1.
Observing errors in gene expression at a single cell level. (A) B. subtilis strains were grown to mid-exponential phase at 32 °C in hydrolyzed casein (CH) medium supplemented with IPTG and observed by fluorescence microscopy. Fluorescence (Upper) and corresponding phase-contrast (Lower) images of the following strains: PY79 (no gfp), SB444 (gfpwt), SB448 (gfpfs), SB446 (gfpns(TAA)), and SB510 (gfpnsx2(TGA, TAA)). Fluorescence images have been normalized to a similar intensity range, apart from the high-intensity image of SB444 strain, which was scaled down for presentation purpose. (Scale bar, 1 μm.) (B) Fluorescence (Upper) and corresponding phase-contrast (Lower) images of the following strains: PY79 (no gfp), SB512 (gfpns(TGA)), SB632 (rpsL2, gfpns(TGA)), SB608 (rpsD1, gfpns(TGA)), and SB616 (rpsD2, gfpns(TGA)). Cells were grown to mid-exponential phase at 32 °C in CH medium. Fluorescence images have been normalized to a similar intensity range. (Scale bar, 1 μm.) (C) Immunoblot analysis of GFP protein extracts from strains: PY79 (no gfp), SB444 (gfpwt), SB448 (gfpfs), SB446 (gfpns(TAA)) grown at 32 °C to mid-exponential phase. Extracts were incubated with polyclonal anti-GFP antibodies. SB444 extract was diluted as indicated, to serve as a reference for SB446 and SB448 extracts. Right panel was exposed for a longer period than left panel to detect low GFP amounts (SI Appendix, SI Materials and Methods).
We validated the formation of erroneous GFP molecules using Western blot analysis (Fig. 1C). This examination revealed that full-length GFP molecules are produced by the gfpfs and the gfpns alleles, indicating that bypass of the mutations is carried out mainly at the beginning of the ORF in the vicinity of the mutations. Evidently, both fluorescent and immunoblot signals appeared to be stronger for the gfpfs strain than for gfpns (Fig. 1 A and C). This difference most likely emanates from the possibility that nonsense mutation can be read through specifically by erroneous introduction of an amino acid at the stop codon, whereas the frame-shift mutation can be bypassed by any compensatory frame-shift error at various sites in the vicinity of the mutation. Of note, both mutations can be bypassed by transcriptional or translational errors.
Next, we examined the strength and the consistency of our system by replacing the original nonsense mutation (TAA) with a TGA stop codon. TGA was shown to be decoded as tryptophan at low efficiency in B. subtilis and thus to increase errors in protein production (22). Accordingly, a strain carrying the gfpns(TGA) allele displayed greater fluorescence than the one containing the gfpns(TAA) allele (SI Appendix, Fig. S4). We then combined the gfpns(TGA) allele with mutations in the genes encoding for the RpsL and RpsD ribosomal proteins. Mutations perturbing RpsL or RpsD activity are known to influence translational fidelity, primarily by affecting the bypass of the TGA codon. Consistent with previous observations (14), a mutation in rpsL that represses translational errors decreased the fluorescence emanating from the gfpns(TGA) strain. Conversely, mutations in rpsD that elevate translational errors clearly increased the fluorescent signal exhibited by the gfpns(TGA) strain (Fig. 1B).
All together, we established that under standard growth conditions each bacterial cell spontaneously introduces errors during gene expression to a significant level, which can be readily detected using a mutant fluorescent gene.
Quantifying the Rate of Spontaneous Errors in Gene Expression.
Next, we estimated the average amount of errors that occur during exponential growth. The GFP signal emanating from the gfpfs and gfpns mutant strains was quantified, averaged, and expressed as a percentage of the GFP signal detected in gfpwt cells (SI Appendix, SI Materials and Methods). Strikingly, the gfpfs cells contained 2.4 ± 0.4% GFP molecules, whereas gfpns contained 0.4 ± 0.1% fluorescent protein molecules relative to gfpwt cells. These values were reproducible, and showed little variation among cells of each strain. Importantly, estimates obtained from different promoters driving the transcription of the mutant gfp alleles were similar, and immunoblot signals from gfpfs and gfpns resembled that of gfpwt diluted 1:50 and 1:200, respectively (Fig. 1C). These measurements could even be underestimates, as mutations situated at the beginning of an ORF are predicted to minimize the error level as they predispose ribosomes to fall off the transcript (17, 23). To further examine if the error rates detected are exceptional or reflect a more standard level, we measured the fluorescence emanated from additional gfp mutated alleles. Replacing the nucleotide causing the frame-shift mutation from A to G did not affect the error level (SI Appendix, Fig. S4), suggesting that the location rather than the sequence is critical for determining the error level. Moreover, inserting frame-shift and nonsense mutations into additional positions within the gfp ORF revealed similar error frequencies (SI Appendix, Fig. S4).
A careful comparison of the values obtained using the gfpns alleles (SI Appendix, Fig. S4) in B. subtilis with those obtained using an ocher nonsense codon in E. coli (15, 17) revealed that our measurements are at least 10-fold higher than previous estimates, raising the possibility that an error occurs approximately once every 200 codons. Errors exceeding 0.1% have been previously reported for specific missense mutations and are generally considered exceptional (e.g. refs. 13 and 15). Our findings suggest that, at least for B. subtilis, these values may represent a standard error level, although the mechanisms for generating missense errors or bypassing nonsense codons may differ (24). In addition, we show that frame-shift errors are highly abundant, at least when located at the beginning of the ORF (SI Appendix, Fig. S4). Thus, we consider it likely that alternative translational products are produced frequently as a consequence of frame-shift errors. Taken together, the error rates exhibited by the gfp mutant alleles support the view that spontaneous errors have been underestimated.
Error Levels in Gene Expression Are Dynamic.
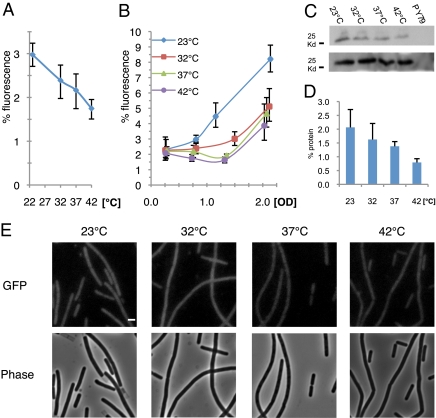
In view of our observations that errors are a prominent feature of bacterial gene expression, we reasoned that bacteria in natural habitats modulate error rates. This theory led us to survey the effect of temperature on the level of errors exhibited by B. subtilis cells, which naturally reside in the soil. The gfpfs strain was used for this analysis because the GFP signal therein is higher than that of the gfpns strain, allowing a greater range of variations to be detected. Samples of gfpfs cells at mid-exponential phase grown at temperatures ranging from 23 to 42 °C were collected and their fluorescent intensity was compared with that displayed by gfpwt cells incubated at the same temperatures (Fig. 2 A and E). We observed an inverse correlation between temperature and error level, such that bacteria grown at the lowest temperature exhibited 70% more errors than bacteria kept at the highest temperature, 3 ± 0.3 and 1.7 ± 0.2%, respectively. Consistently, Western blot analysis showed a similar correlation between the production of erroneous protein molecules and temperature (Fig. 2 C and D). Importantly, no significant difference was detected between the levels of wild-type GFP molecules produced at the different temperatures, suggesting that protein stability is not the cause for the observed tendency. Thus, growth temperature has a direct effect on the error level, implying that bacteria can modulate error rates in nature. The reduced error level measured at higher temperatures suggests that raising the temperature imposes the formation of more accurate protein molecules.
Fig. 2.
Error level in gene expression is temperature dependent. (A) Fluorescence levels (%) as a function of temperature in strain SB448 (gfpfs) grown to mid-exponential phase. Fluorescence levels (%) are the average fluorescence signal of strain SB448 (gfpfs) relative to the average fluorescence signal of strain SB444 (gfpwt); both strains were grown to the same OD and at the same temperature (SI Appendix, SI Materials and Methods). (B) Fluorescence levels (%) as a function of OD, when strain SB448 (gfpfs) was grown at different temperatures. Fluorescence levels (%) were calculated as in A. (C) Immunoblot analysis of GFP protein at different incubation temperatures. Extracts from strains SB448 (gfpfs) (Upper), SB444 (gfpwt) (Lower) grown at the indicated temperatures and PY79 grown at 23 °C were incubated with polyclonal anti-GFP antibodies. For SB444 only 1/20 of each extract was loaded (SI Appendix, SI Materials and Methods). (D) Protein levels (%) as a function of temperature in strain SB448 (gfpfs) as calculated from immunoblot analysis. Protein levels (%) are the calculated amount of GFP protein from strain SB448 (gfpfs) relative to that of strain SB444 (gfpwt); both strains were grown at the same temperature to the same OD600 (SI Appendix, SI Materials and Methods). (E) Fluorescence (Upper) and corresponding phase-contrast (Lower) images of strain SB448 (gfpfs) grown to mid-exponential phase at the indicated temperatures. All fluorescence images were normalized to a similar intensity range. (Scale bar, 1 μm.)
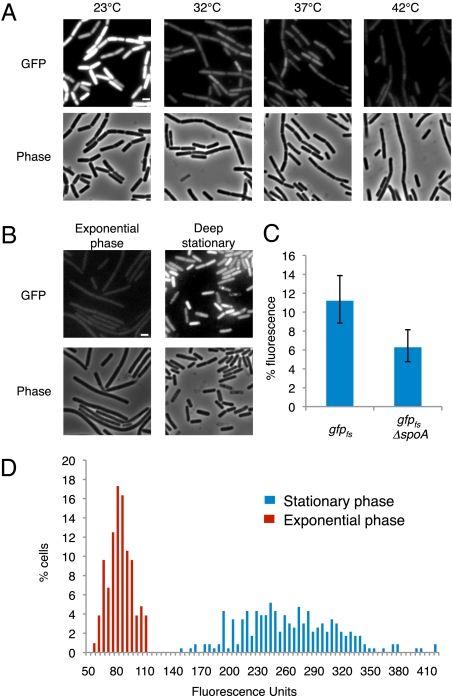
Next, we tested whether there is any correlation between growth phase and error level. A time-course experiment was performed whereby the error levels were determined at different growth phases at various temperatures. We found that the error level correlates with optical density, reaching the highest values at stationary phase during when the bacterium is challenged by unfavorable conditions (Fig. 2B). These results were corroborated by Western blot analysis showing a similar trend (SI Appendix, Fig. S5). The error rates observed during stationary phase were at least 2.1-fold higher than those observed during exponential phase for all temperatures, with cells grown at the lowest temperature reaching a mean level of more than 8% (Figs. 2B and 3A). To rule out the possibility that these high error levels were a consequence of protein accumulation during stationary phase, we induced gfpfs expression only during deep stationary phase (t = 20 h) for a short window of time. The resultant fluorescence was then compared with that of vegetative gfpfs cells induced for the same time period. As shown in Fig. 3B, cells residing in deep stationary phase evidently exhibited higher error levels than exponentially growing cells. Notably, cells unable to properly enter deep stationary phase, because of a mutation in the key transcription factor Spo0A (25), exhibited a lower level of errors than those observed in wild-type cells residing at this stage (Fig. 3C). This result indicates that proper entry into stationary phase is important for the observed increase in error levels. It is therefore possible that either an active induction of errors in gene expression takes place during stationary phase or, alternatively, a fidelity mechanism active during exponential growth shuts off.
Fig. 3.
Error level in gene expression increases during stationary phase. (A) Fluorescence (Upper) and corresponding phase-contrast (Lower) images of strain SB448 (gfpfs) during stationary phase at the indicated temperatures. All fluorescence images were normalized to a similar intensity range. (Scale bar, 1 μm.) (B) Fluorescence (Upper) and corresponding phase-contrast (Lower) images of strain SB448 (gfpfs) grown at 32 °C either to mid-exponential phase (Left) or during deep stationary phase (Right) 2 h after the addition of IPTG. Images were normalized to the same intensity range. (Scale bar, 1 μm.) (C) Fluorescence levels (%) of strains SB448 (gfpfs) and SB784 (gfpfs, Δspo0A) 11 h after entering into stationary phase at 32 °C. Fluorescence levels (%) are the average fluorescence signal of a gfp mutant strain relative to the average fluorescence signal of its corresponding wild-type gfp strain: SB444 (gfpwt) for SB448 and SB782 (gfpwt, Δspo0A) for SB784 (SI Appendix, SI Materials and Methods). (D) Fluorescence distribution of individual cells of strain SB448 (gfpfs) during mid-exponential phase (red bars) and during stationary phase (blue bars) grown at 32 °C. At least 100 cells were scored for each phase.
A more detailed inspection of fluorescence levels emanating from individual cells revealed greater variation during stationary phase than during exponential phase for all tested temperatures (compare Fig. 2E with Fig. 3A). This increased heterogeneity of stationary cells was readily apparent when the distribution of GFP expression was plotted for cells grown at 32 °C (Fig. 3D). The distribution of fluorescence intensity originated from cells during stationary phase was considerably wider than that observed for exponentially growing cells. Thus, we conclude that individual cells in stationary phase exhibit varying degrees of accuracy in gene expression, expanding the capability of an isogenic population to display heterogeneous phenotypes, a feature that could serve to increase the ability of bacterial cells to face environmental challenges.
High Levels of Errors Can Be Selectively Advantageous.
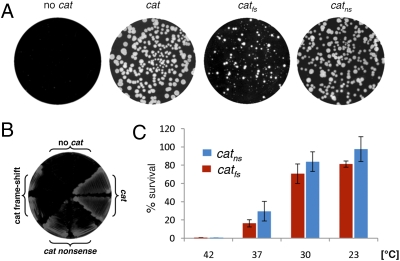
We reasoned that the high amount of errors monitored during bacterial gene expression may be beneficial under certain conditions. To simulate such a situation, we mutated an antibiotic resistance gene and assayed whether errors in gene expression could promote the production of protein molecules valuable for bacterial survival. To this end, mutations were introduced into the ORF of the cat gene encoding chloramphenicol resistance protein, and cells were plated on chloramphenicol-containing plates. Remarkably, at 30 °C catfs and catns mutant strains were able to grow slowly on the selective medium and to display numbers of resistant colonies similar to those of the catwt strain (Fig. 4 A and B). Importantly, we confirmed that resistant colonies derived from the mutant strains retained the mutated cat allele as well as the mutant phenotypes (Materials and Methods). Moreover, in line with the observed temperature-dependence of error levels, the survival rate on the selective medium increased as growth temperature was reduced. As shown in Fig. 4C, the number of resistant catfs and catns colonies appearing on the selective medium at 23 °C was similar to the number observed on the nonselective medium, whereas only a negligible number of colonies was obtained from selective plates incubated at 42 °C. In light of these data, we propose that a high error level could facilitate the expression of pseudogenes containing frame-shift and nonsense mutations, which are a common feature of bacterial genomes (e.g., ref. 26). Thus, pseudogenes may represent a valuable genetic reservoir that can be expressed and become beneficial under certain circumstances. The low amount of full-length proteins produced from pseudogenes may be a strategy used by some bacteria to maintain genetic information with a relatively low cost of protein synthesis.
Fig. 4.
Errors in gene expression enable survival of strains carrying a mutated antibiotic resistant gene. (A) Strains PY79 (no cat), SB463 (cat), SB477 (catfs), and SB475 (catns) were grown on 4-μg/mL chloramphenicol-containing plates. Plates were imaged after 40 h of incubation at 30 °C. (B) Strains PY79 (no cat), SB463 (cat), SB477 (catfs), and SB475 (catns) were streaked on 10-μg/mL chloramphenicol-containing plates and imaged after 40 h of incubation at 30 °C. (C) The viability (%) of SB477 (catfs) (red bars) and SB475 (catns) (blue bars) at different temperatures when plated on chloramphenicol-containing plates. Cells were grown in CH medium at the different temperatures in the absence of chloramphenicol, and plated at mid-exponential phase on either LB or LB supplemented with 4-μg/mL chloramphenicol plates. Percent-viability is the number of colonies growing on chloramphenicol-containing plates relative to the total number of colonies growing on LB plates incubated at the equivalent temperature (Materials and Methods). PY79 (no cat) strain did not grow on chloramphenicol-containing plates, whereas SB463 (cat) showed ≈100% viability at any growth temperature. The viability on LB plates of all tested strains was similar, with colony number ranging from 350 to 500.
Discussion
Our data reveal that protein production in the bacterium B. subtilis is prone to high levels of errors, a feature that may be widespread among bacteria. Therefore, we propose that in general each genetic locus generates an assortment of closely related protein molecules and that these inaccurate molecules become part of the cellular protein pool. Moreover, the high frequency of spontaneous frame-shifting monitored in our assays suggests that the cellular proteome could also contain large amounts of alternative translational products. Thus, the bacterial genetic code serves as a consensus sequence, on the basis of which protein production generates variations (Fig. 5). Accordingly, in vivo and in vitro experiments from different laboratories indicate that typically, proteins are robust to mutations and can tolerate a significant number of substitutions, even in highly conserved domains, with little change in structure, stability, or function (e.g., refs. 27–33). Furthermore, errors in protein production may be beneficial, as proteins can acquire new properties by only a few amino acid substitutions (e.g., refs. 27, 28, 30, 34). For example, certain mutations in DNA polymerase have been demonstrated to confer higher activity or an ability to incorporate nucleotide analogs (30).
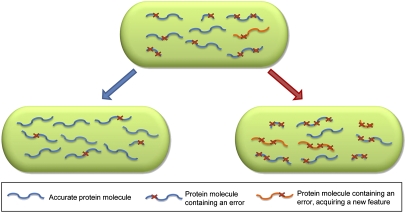
Fig. 5.
Heterogeneity of gene products in a bacterium. A schematic illustration of protein molecules produced by a single gene in a bacterium. Proteins are depicted as curly blue or orange lines, and each red X represents an error event in the production of the protein relative to the sequence dictated by the genetic code. Shorter protein molecules represent frame-shift events. (Upper) Under normal growth conditions, the bacterial cell contains a mixture of different protein species: accurate molecules (blue), inaccurate molecules (blue and red X), and inaccurate molecules acquiring a new feature (orange and red X). (Lower Left) During certain conditions, such as high temperatures, gene expression is more accurate resulting in a less heterogeneous protein pool. (Lower Right) During conditions such as low temperatures and stationary phase, an increase in the error level expands the protein pool, resulting in the formation of more protein molecules bearing unique features. These unique features can benefit the bacterium, allowing it to survive during stress conditions and consequently, increase its robustness.
In general, for any organism, the amount of errors represents a compromise between the “error cost,” namely the production of misfolded or dysfunctional proteins, and the “error payoff,” namely the production of beneficial protein variants and increased phenotypic heterogeneity. Several studies have reported large fluctuations in the level of errors among different codons encoding the same protein (14, 15, 17, 18), suggesting a different degree of susceptibility to errors along the coding sequence. In this view, evolutionary pressure that shapes codon usage could allow different genes to be prone to unequal error rates according to their cellular function (35).
We observed a significant decrease in the fidelity of protein production during stationary phase in response to other challenging conditions, such as minimal medium, and at lower temperatures. In a similar way, a decrease in translational fidelity was previously observed for E. coli cells grown under starvation conditions (36, 37). We hypothesize that in this way, errors in gene expression contribute to bacterial robustness, increasing survival in fluctuating environments or in response to sudden stress (Fig. 5). In addition, challenging conditions could promote expression of pseudogenes, thereby expanding the bacterial protein reservoir. Such plasticity enables cells to adapt to rapid changes that the slow evolutionary process cannot counter (38). Consistent with this idea, it has been proposed that gene-expression errors can contribute to the ability of an organism to develop genetically encoded traits that require multiple mutations. In such evolutionary process, intermediate steps could combine a genetic mutation in one gene with errors in gene expression in another, consequently bypassing the need to undergo two independent mutations simultaneously (10).
The temperature dependence of the error level observed in this study could be simply a result of reduced protein stability at high temperatures. Alternatively, this observation could be explained by temperature-induced expression of chaperones, proteins that were found to mask phenotypic diversity in both prokaryotes and eukaryotes (39–43). The best-studied example is the eukaryotic heat shock protein 90 (Hsp90) that was shown to buffer phenotypic variation in several organisms. Perturbing Hsp90 activity exposes diverse phenotypes and leads to increased population heterogeneity (39–42, 44).
It remains to be resolved whether the high error levels in gene expression observed in bacteria also exist in eukaryotes. It is tempting to speculate that multicellular organisms can modulate the level of errors in different cell types according to their function. In this regard, it has been shown that the proportion of proteins that are defective in certain eukaryotic cell types could exceed 30%; however, these faulty proteins are typically degraded by the proteasome (45, 46). In general, it seems that eukaryotic cells have developed mechanisms to reduce the production of truncated or inaccurate proteins, such as nonsense-mediated RNA decay (47). We suspect that the bacterial need to cope with rapid environmental changes imposes the existence of considerable diversity in their protein reservoir, an attribute that both ensures robustness and contributes to their ability to evolve.
Materials and Methods
Plasmids and Strains.
Plasmids, strains, and primers used in this study are described in SI Appendix, Tables S1–S4.
General Methods.
B. subtilis cells were grown in hydrolyzed casein (CH) growth medium at the indicated temperatures (23, 30, 32, 37, or 42 °C). Cultures were inoculated to an OD600 of 0.05 using an overnight culture grown in the same medium. Induction of Phyper-spank or Pspank promoters was carried out by adding IPTG to a final concentration of 0.5 mM for liquid medium and 1 mM for solid medium.
General Fluorescence Microscopy Methods.
Fluorescence microscopy was carried out as described previously (21). For all experiments, cells were visualized and photographed using an Axioplan2 microscope (Zeiss) equipped with a CoolSnap HQ camera (Photometrics; Roper Scientific) or an Axioobserver Z1 microscope (Zeiss) equipped with a CoolSnap HQII camera (Photometrics; Roper Scientific) and X-cite (120PC; EXFO) as a fluorescence illumination light source. All fluorescence-intensity measurements were performed using the latter system because the X-cite provides a more uniform and stable light source in comparison with Mercury lamps. System control and image processing were performed using MetaMorph software (version 7.5; Molecular Devices). Other fluorescence microscopy methods: fluorescence recovery after photobleaching and fluorescent images processing, are described in SI Appendix, SI Materials and Methods.
Monitoring Errors in Gene Expression in Strains Carrying a Mutated Chloramphenicol-Resistant Gene.
Strains PY79 (no cat), SB463 (cat), SB477 (catfs), and SB475 (catns) were grown in liquid nonselective CH medium at 32 °C to mid-exponential phase and plated on either LB or LB supplemented with 4-μg/mL chloramphenicol-containing plates. The plates were incubated at different temperatures and the percent-viability was determined by the number of colonies grown on chloramphenicol-containing plates relative to the total number of colonies grown on LB plates incubated at the equivalent temperature. To rule out the possibility that colonies from each mutant strain grown on the selective medium emanated from reversion, suppression, or other genetic modification, at least 100 colonies from each mutant strain were picked and retested for their ability to grow on chloramphenicol. All of the tested colonies (regardless of the colony size) maintained the mutant phenotypes as they were unable to grow on a high chloramphenicol concentration (20 μg/mL). In addition, cat alleles of at least 20 colonies from each mutant strain were sequenced and none of them contained any genetic alteration. Testing the frequency of colonies gaining the wild-type phenotype (probably by reversions, suppressor mutations, and so forth) was ≈10−7 for the frame-shift or the nonsense alleles, as estimated by plating the mutant cells on high chloramphenicol concentrations.
Supplementary Material
Acknowledgments
We thank R. Losick (Harvard University, Boston, MA), D. Kearns (Indiana University), A. Rouvinski (Hebrew University, Jerusalem), Y. Smith (Hebrew University, Jerusalem), and members of the Ben-Yehuda laboratory for valuable comments. We thank K. Ochi (Ibaraki, Japan) for providing Bacillus subtilis strains. This work was supported by the European Research Council Starting Grant 209130 and the Israel Science Foundation (696/07) (to S.B.-Y.)
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.0912989107/-/DCSupplemental.
References
- 1.Scheper GC, van der Knaap MS, Proud CG. Translation matters: Protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–723. doi: 10.1038/nrg2142. [DOI] [PubMed] [Google Scholar]
- 2.van Leeuwen FW, Burbach JP, Hol EM. Mutations in RNA: A first example of molecular misreading in Alzheimer's disease. Trends Neurosci. 1998;21:331–335. doi: 10.1016/s0166-2236(98)01280-6. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen FW, et al. Frameshift mutants of beta amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science. 1998;279:242–247. doi: 10.1126/science.279.5348.242. [DOI] [PubMed] [Google Scholar]
- 4.Farabaugh PJ. Programmed translational frameshifting. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]
- 5.Blinkowa AL, Walker JR. Programmed ribosomal frameshifting generates the Escherichia coli DNA polymerase III gamma subunit from within the tau subunit reading frame. Nucleic Acids Res. 1990;18:1725–1729. doi: 10.1093/nar/18.7.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–724. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldsmith M, Tawfik DS. Potential role of phenotypic mutations in the evolution of protein expression and stability. Proc Natl Acad Sci USA. 2009;106:6197–6202. doi: 10.1073/pnas.0809506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon AJ, et al. Transcriptional infidelity promotes heritable phenotypic change in a bistable gene network. PLoS Biol. 2009;7:e44. doi: 10.1371/journal.pbio.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ninio J. Connections between translation, transcription and replication error-rates. Biochimie. 1991;73:1517–1523. doi: 10.1016/0300-9084(91)90186-5. [DOI] [PubMed] [Google Scholar]
- 10.Whitehead DJ, Wilke CO, Vernazobres D, Bornberg-Bauer E. The look-ahead effect of phenotypic mutations. Biol Direct. 2008;3:18. doi: 10.1186/1745-6150-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temiakov D, et al. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 12.Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 13.Bouadloun F, Donner D, Kurland CG. Codon-specific missense errors in vivo. EMBO J. 1983;2:1351–1356. doi: 10.1002/j.1460-2075.1983.tb01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inaoka T, Kasai K, Ochi K. Construction of an in vivo nonsense readthrough assay system and functional analysis of ribosomal proteins S12, S4, and S5 in Bacillus subtilis. J Bacteriol. 2001;183:4958–4963. doi: 10.1128/JB.183.17.4958-4963.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kunkel TA. DNA replication fidelity. J Biol Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberger RF, Foskett G. An estimate of the frequency of in vivo transcriptional errors at a nonsense codon in Escherichia coli. Mol Gen Genet. 1981;183:561–563. doi: 10.1007/BF00268784. [DOI] [PubMed] [Google Scholar]
- 18.Rosenberger RF, Hilton J. The frequency of transcriptional and translational errors at nonsense codons in the lacZ gene of Escherichia coli. Mol Gen Genet. 1983;191:207–212. doi: 10.1007/BF00334815. [DOI] [PubMed] [Google Scholar]
- 19.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 20.Springgate CF, Loeb LA. On the fidelity of transcription by Escherichia coli ribonucleic acid polymerase. J Mol Biol. 1975;97:577–591. doi: 10.1016/s0022-2836(75)80060-x. [DOI] [PubMed] [Google Scholar]
- 21.Bejerano-Sagie M, et al. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125:679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 22.Lovett PS, Ambulos NP, Jr, Mulbry W, Noguchi N, Rogers EJ. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis. J Bacteriol. 1991;173:1810–1812. doi: 10.1128/jb.173.5.1810-1812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennell D, Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977;114:1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- 24.Gesteland RF, Atkins JF. Recoding: Dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 25.Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Harrison PM, Kunin V, Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: Widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown BM, Sauer RT. Tolerance of Arc repressor to multiple-alanine substitutions. Proc Natl Acad Sci USA. 1999;96:1983–1988. doi: 10.1073/pnas.96.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo HH, Choe J, Loeb LA. Protein tolerance to random amino acid change. Proc Natl Acad Sci USA. 2004;101:9205–9210. doi: 10.1073/pnas.0403255101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesk AM, Chothia C. How different amino acid sequences determine similar protein structures: The structure and evolutionary dynamics of the globins. J Mol Biol. 1980;136:225–270. doi: 10.1016/0022-2836(80)90373-3. [DOI] [PubMed] [Google Scholar]
- 30.Patel PH, Loeb LA. DNA polymerase active site is highly mutable: Evolutionary consequences. Proc Natl Acad Sci USA. 2000;97:5095–5100. doi: 10.1073/pnas.97.10.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rennell D, Bouvier SE, Hardy LW, Poteete AR. Systematic mutation of bacteriophage T4 lysozyme. J Mol Biol. 1991;222:67–88. doi: 10.1016/0022-2836(91)90738-r. [DOI] [PubMed] [Google Scholar]
- 32.Suckow J, et al. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J Mol Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 33.Taverna DM, Goldstein RA. Why are proteins so robust to site mutations? J Mol Biol. 2002;315:479–484. doi: 10.1006/jmbi.2001.5226. [DOI] [PubMed] [Google Scholar]
- 34.Aharoni A, et al. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 35.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ballesteros M, Fredriksson A, Henriksson J, Nyström T. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001;20:5280–5289. doi: 10.1093/emboj/20.18.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Farrell PH. The suppression of defective translation by ppGpp and its role in the stringent response. Cell. 1978;14:545–557. doi: 10.1016/0092-8674(78)90241-6. [DOI] [PubMed] [Google Scholar]
- 38.Borenstein E, Meilijson I, Ruppin E. The effect of phenotypic plasticity on evolution in multipeaked fitness landscapes. J Evol Biol. 2006;19:1555–1570. doi: 10.1111/j.1420-9101.2006.01125.x. [DOI] [PubMed] [Google Scholar]
- 39.Fares MA, Ruiz-González MX, Moya A, Elena SF, Barrio E. Endosymbiotic bacteria: groEL buffers against deleterious mutations. Nature. 2002;417:398. doi: 10.1038/417398a. [DOI] [PubMed] [Google Scholar]
- 40.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 41.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 42.Sangster TA, et al. Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PLoS ONE. 2007;2:e648. doi: 10.1371/journal.pone.0000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang JD, Herman C, Tipton KA, Gross CA, Weissman JS. Directed evolution of substrate-optimized GroEL/S chaperonins. Cell. 2002;111:1027–1039. doi: 10.1016/s0092-8674(02)01198-4. [DOI] [PubMed] [Google Scholar]
- 44.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: Drug resistance in diverse fungi. Science. 2005;309:2185–2189. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 45.Reits EA, Vos JC, Grommé M, Neefjes J. The major substrates for TAP in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- 46.Schubert U, et al. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 47.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.